Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening of Saline–Alkaline-Tolerant Germplasm of Rice
2.1.1. Study Site
2.1.2. Rice Cultivation, Experimentation, and Data Collection
2.1.3. Germplasm Survey
2.2. Physiological Mechanism Analysis of Saline–Alkaline Tolerant Genotypes of Rice
2.2.1. Plant Material, Growth Conditions, and Stress Treatments
2.2.2. Measurement of Chlorophyll Content
2.2.3. Determination of Na+ and K+ Concentration
2.2.4. Determination of Proline and Soluble Sugars Content
2.2.5. Measurements of Total Antioxidant Capacity (T-AOC) and Malondialdehyde (MDA) Content
2.3. Statistical Analysis
3. Results
3.1. Screening of Saline–Alkaline Tolerant Germplasm of Rice
3.1.1. Initial Screening in 2011
3.1.2. Screening by Whole Growth Period in 2012
3.1.3. Further Verification in 2013
3.2. Evaluation of Salt and Alkali Tolerance and Physiological Mechanism Analysis of Jiudao-66
3.2.1. Effects of Saline–Alkaline Stress on Seedlings
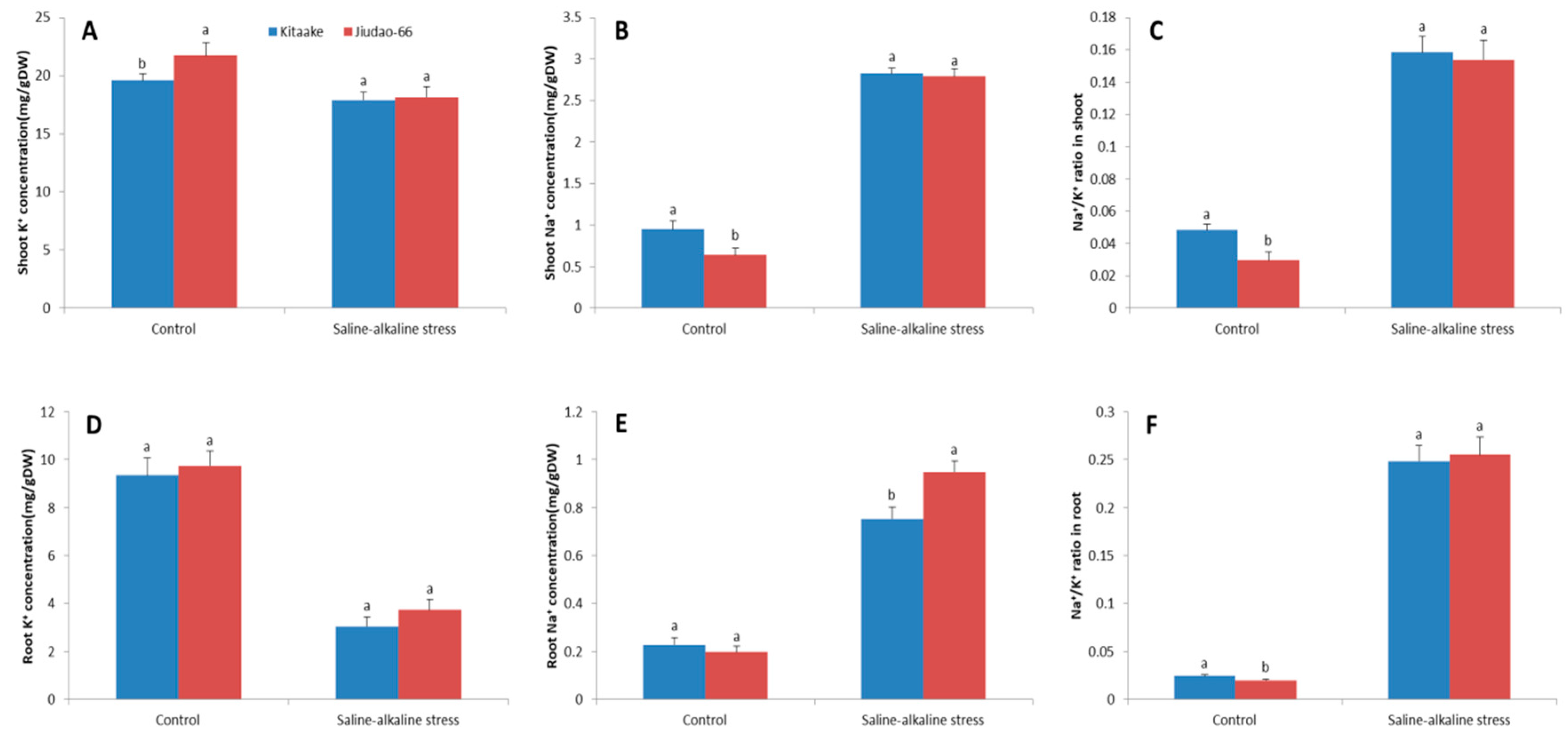
3.2.2. Effect of Saline–Alkaline Stress on Na+ and K+ Concentrations
3.2.3. Effect of Saline–Alkaline Stress on Osmoregulation
3.2.4. Effect of Saline–Alkaline Stress on Oxidative Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pascale, S.D.; Maggio, A.; Barbieri, G. Soil salinization affects growth, yield and mineral composition of cauliflower and broccoli. Eur. J. Agron. 2005, 23, 254–264. [Google Scholar] [CrossRef]
- Qadir, M.; Noble, A.D.; Schubert, S.; Thomas, R.J.; Arslan, A. Sodicity-induced land degradation and its sustainable management: Problems and prospects. Land Degrad. Dev. 2006, 17, 661–676. [Google Scholar] [CrossRef]
- Zhang, J.; Mu, C. Effects of saline and alkaline stresses on the germination, growth, photosynthesis, ionic balance and anti-oxidant system in an alkali-tolerant leguminous forage Lathyrus quinquenervius. Soil Sci. Plant Nutr. 2009, 55, 685–697. [Google Scholar] [CrossRef]
- Uddin, M.K.; Juraimi, A.S.; Ismail, M.R.; Hossain, M.A.; Othman, R.; Rahim, A.A. Effect of salinity stress on nutrient uptake and chlorophyll content of tropical turfgrass species. Aust. J. Crop Sci. 2011, 5, 620–629. [Google Scholar]
- Wei, L.X.; Lv, B.S.; Li, X.W.; Wang, M.M.; Ma, H.Y.; Yang, H.Y.; Yang, R.F.; Piao, Z.Z.; Wang, Z.H.; Lou, J.H.; et al. Priming of rice (Oryza sativa L.) seedlings with abscisic acid enhances seedling survival, plant growth, and grain yield in saline-alkaline paddy fields. Field Crops Res. 2017, 203, 86–93. [Google Scholar] [CrossRef]
- Yang, J. Development and prospect of the research on salt-affected soils in China. Acta Pedol. Sin. 2008, 45, 837–845. [Google Scholar]
- Feng, Y.J.; Zhang, W.; Chen, Q.; Ma, C.-H. Physico-chemical characteristics and microbial composition of saline-alkaline soils in Songnen Plain. Soils 2007, 39, 301–305. [Google Scholar]
- Chhabra, R. Classification of salt-affected soils. Arid Soil Res. Rehabil. 2004, 19, 61–79. [Google Scholar] [CrossRef]
- Bin, L.I.; Wang, Z.C.; Liang, Z.W.; Chi, C.M. Electrical conductivity and water quality evaluation of groundwater at sodic soil areas in the west of Jilin province. J. Agro-Environ. Sci. 2007, 26, 939–944. [Google Scholar]
- Huang, L.H.; Liang, Z.W. Ionic absorption characteristics of Leymus chinensis seeded in various pH soils. Chin. J. Grassl. 2008, 30, 35–39. [Google Scholar]
- Türkan, I.; Demiral, T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 2009, 67, 2–9. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shao, G.; Chang, R. The effect of salt stress on superoxide dismutase in various organelles from cotyledon of soybean seedling. Acta Agron. Sin. 1997, 23, 214–219. [Google Scholar]
- Zhu, D.F.; Cheng, S.H.; Zhang, Y.P.; Lin, X.Q.; Chen, H.Z. Analysis of status and constraints of rice production in the world. Sci. Agric. Sin. 2010, 73, 167–173. [Google Scholar]
- Koyama, M.L.; Yeo, A.R. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 2001, 125, 406–422. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Salam, M.A.; Hassan, L.; Collard, B.C.Y.; Singh, R.K.; Gregorio, G.B. QTL mapping for salinity tolerance at seedling stage in rice. J. Sci. Food Agric. 2011, 23, 137–146. [Google Scholar] [CrossRef]
- Javed, M.A.; Huyop, F.Z.; Wagiran, A.; Salleh, F.M. Identification of QTLs for morph-physiological traits related to salinity tolerance at seedling stage in indica rice. Procedia Environ. Sci. 2011, 8, 389–395. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Z.; Cheng, J.; Lai, Y.; Wang, J.; Bao, Y.; Huang, J.; Zhang, H. QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e51202. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhao, H.; Liu, H.; Wang, J.; Zou, D. Qtl analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice. Euphytica 2015, 201, 109–121. [Google Scholar] [CrossRef]
- Ammar, M.H.M.; Pandit, A.; Singh, R.K.; Sameena, S.; Chauhan, M.S.; Singh, A.K.; Sharma, P.C.; Gaikwad, K.; Sharma, T.R.; Mohapatra, T.; et al. Mapping of QTLs controlling Na+, K+ and Cl− ion concentrations in salt tolerant indica rice variety CSR27. J. Plant Biochem. Biotechnol. 2009, 18, 139–150. [Google Scholar] [CrossRef]
- Pandit, A.; Rai, V.; Bal, S.; Sinha, S.; Kumar, V.; Chauhan, M.; Gautam, R.K.; Singh, R.; Sharma, P.C.; Singh, A.K. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.). Mol. Genet. Genom. 2010, 284, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Sushma, T.; Krishnamurthy, S.L.; Vinod, K.; Balwant, S.; Rao, A.R.; Amitha, M.S.; Vandna, R.; Singh, A.K.; Singh, N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS ONE 2016, 11, e0153610. [Google Scholar]
- Qiao, K.; Takano, T.; Liu, S. Discovery of two novel highly tolerant NaHCO3 trebouxiophytes: Identification and characterization of microalgae from extreme saline-alkali soil. Algal Res. 2015, 9, 245–253. [Google Scholar] [CrossRef]
- Yu, Y.; Guan, L. Improving maize output through climate resources utilization. J. Anhui Agric. Sci. 2007, 35, 381–382. [Google Scholar]
- Li, Q.; Li, X.; Li, X.; Wang, Z.; Song, C.; Zhang, G. Sodium bicarbonate soil management and utilization in songnen plain. Resour. Sci. 2003, 25, 15–20. [Google Scholar]
- Wang, H.; Ahan, J.; Wu, Z.; Shi, D.; Liu, B.; Yang, C. Alteration of nitrogen metabolism in rice variety ‘nipponbare’ induced by alkali stress. Plant Soil 2012, 355, 131–147. [Google Scholar] [CrossRef]
- Lv, B.S.; Ma, H.Y.; Li, X.W.; Wei, L.X.; Lv, H.Y.; Yang, H.Y.; Jiang, C.J.; Liang, Z.W. Proline accumulation is not correlated with saline-alkaline stress tolerance in rice seedlings. Agron. J. 2015, 107, 51–60. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transprter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, A.; Zhang, W.H. Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 2016, 67, 6431–6444. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Sun, J.; Wang, J.; Liu, H.; Zheng, H.; Yang, L.; Liang, Y.; Li, X.; Zou, D. QTL analysis for alkaline tolerance of rice and verification of a major QTL. Plant Breed. 2017, 136, 881–891. [Google Scholar] [CrossRef]
- Li, Q.; Yang, A.; Zhang, W.H. Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Boil. 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jiang, C.J.; Liang, Z.W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Hossain, H.; Rahman, M.A.; Alam, M.S.; Singh, R.K. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Li, X.; Ma, L.; Bu, N.; Li, Y.; Zhang, L. Endophytic infection modifies organic acid and mineral element accumulation by rice under Na2CO3 stress. Plant Soil 2017, 420, 1–11. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Sako, K.; Matsui, A.; Suzuki, Y.; Mostofa, M.G.; Ha, C.V.; Tanaka, M.; Tran, L.P.; Habu, Y.; Seki, M. Ethanol enhances high-salinity stress tolerance by detoxifying reactive oxygen species in Arabidopsis thaliana and rice. Front. Plant Sci. 2017, 8, 1001. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.P.; Jian, S.; Suo, Y.N.; Liu, H.L.; Wang, J.G.; Zheng, H.L.; Sun, X.X.; Zou, D.T. QTL mapping and QTL × environment interaction analysis of salt and alkali tolerance-related traits in rice (Oryza sativa L.). Sci. Agric. Sin. 2017, 50, 1747–1762. [Google Scholar]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E. Mutmap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Mardani, Z.; Rabiei, B.; Sabouri, H.; Sabouri, A. Identification of molecular markers linked to salt-tolerant genes at germination stage of rice. Plant Breed. 2014, 133, 196–202. [Google Scholar] [CrossRef]
- Sakina, A.; Ahmed, I.; Shahzad, A.; Iqbal, M.; Asif, M. Genetic variation for salinity tolerance in Pakistani rice (Oryza sativa L.) germplasm. J. Agron. Crop Sci. 2016, 202, 25–36. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Boil. 2017, 17, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrão, S.; Courtois, B.; Ahmadi, N.; Abreu, I.; Saibo, N.; Oliveira, M.M. Recent updates on salinity stress in rice: From physiological to molecular responses. Crit. Rev. Plant Sci. 2011, 30, 329–377. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Boil. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Genc, Y.; Mcdonald, G.K.; Tester, M. Reassessment of tissue Na(+) concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 2007, 30, 1486–1498. [Google Scholar] [CrossRef] [PubMed]
- Babgohari, M.Z.; Niazi, A.; Moghadam, A.A.; Deihimi, T.; Ebrahimie, E. Genome-wide analysis of key salinity-tolerance transporter (HKT1; 5) in wheat and wild wheat relatives (A and D genomes). In Vitro Cell. Dev. Boil. Plant 2013, 49, 97–106. [Google Scholar] [CrossRef]
- Almeida, P.; Feron, R.; Boer, G.J.D.; Boer, A.H.D. Role of Na+, K+, Cl−, proline and sucrose concentrations in determining salinity tolerance and their correlation with the expression of multiple genes in tomato. AoB Plants 2014, 6, A354–A355. [Google Scholar] [CrossRef] [PubMed]
- Pires, I.S.; Negrão, S.; Oliveira, M.M.; Purugganan, M.D. Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiol. Plant 2015, 155, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, H.J.; Shen, B. Transformation and compatible solutes. Sci. Hortic. 1998, 78, 237–260. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.R. Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front. Plant Sci. 2017, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M. Crop plants and abiotic stresses. Biomol. Res. Ther. 2013, 3, e125. [Google Scholar] [CrossRef]
- Kumar, M.; Gho, Y.S.; Jung, K.H.; Kim, S.R. Genome-wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, J.; Zhang, Y.; Fan, F.; Li, W.; Wang, F.; Zhong, W.; Wang, C.; Yang, J. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 2018, 8, 2085. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.; Tripathi, A.; Sanan-Mishra, N. Comparative miRomics of salt-tolerant and salt-sensitive rice. J. Integr. Bioinform. 2017, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]




| Mixtures | EC 1:5 μS·cm−1 | TDS ppm | pH | Organic Matter % | Available N mg·kg−1 | Available P mg·kg−1 | Available K mg·kg−1 | CO32− mg·kg−1 | HCO3− mg·kg−1 | Na+ mg·kg−1 | Cl− mg·kg−1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal Soil (NS) | 220 | 96.0 | 8.0 | 2.2 | 45.5 | 35.4 | 164.0 | —— | 195.0 | 41.0 | 57.0 |
| 2/3 NS | 1000 | 445.0 | 8.8 | 1.5 | 66.5 | 32.2 | 218.0 | 192.0 | 512.0 | 181.0 | 114.0 |
| 1/2 NS | 1498 | 672.0 | 9.0 | 1.5 | 67.2 | 25.7 | 229.0 | 120.0 | 976.0 | 280.0 | 497.0 |
| Saline–Alkaline Soil (SAS) | 2540 | 1150.0 | 9.8 | 1.0 | 58.1 | 12.9 | 272.0 | 840.0 | 2562.0 | 700.0 | 398.0 |
| Variety No. | Survival Rate % | Variety No. | Survival Rate % | Variety No. | Survival Rate % | Variety No. | Survival Rate % |
|---|---|---|---|---|---|---|---|
| D10 | 97.78 ± 1.92 | D28 | 88.89 ± 6.94 | D25 | 77.78 ± 6.94 | D31 | 58.89 ± 1.92 |
| D70 | 96.67 ± 3.33 | D43 | 88.89 ± 3.85 | D20 | 76.67 ± 3.33 | D40 | 58.89 ± 1.92 |
| D36 | 96.67 ± 3.33 | D63 | 88.89 ± 5.09 | D45 | 76.67 ± 0.00 | D52 | 58.89 ± 3.85 |
| D30 | 95.56 ± 5.09 | D3 | 87.78 ± 5.09 | D60 | 76.67 ± 6.67 | D24 | 56.67 ± 3.33 |
| D57 | 95.56 ± 3.85 | D19 | 87.78 ± 1.92 | D61 | 75.56 ± 1.92 | D11 | 56.67 ± 3.33 |
| D68 | 95.56 ± 3.85 | D46 | 87.78 ± 5.09 | D62 | 75.56 ± 1.92 | D14 | 56.67 ± 3.33 |
| D1 | 94.44 ± 1.92 | D5 | 86.67 ± 3.33 | D15 | 73.33 ± 6.67 | D54 | 54.44 ± 5.09 |
| D47 | 94.44 ± 5.09 | D17 | 86.67 ± 3.33 | D33 | 73.33 ± 3.33 | D22 | 53.33 ± 3.33 |
| D71 | 93.33 ± 3.33 | D21 | 86.67 ± 6.67 | D41 | 73.33 ± 3.33 | D53 | 53.33 ± 0.00 |
| D8 | 93.33 ± 5.77 | D34 | 86.67 ± 0.00 | D26 | 72.22 ± 5.09 | D39 | 52.22 ± 5.09 |
| D49 | 93.33 ± 6.67 | D42 | 86.67 ± 5.77 | D44 | 72.22 ± 1.92 | D37 | 51.11 ± 1.92 |
| D64 | 93.33 ± 5.77 | D66 | 85.56 ± 1.92 | D18 | 65.56 ± 5.09 | D4 | 50.00 ± 3.33 |
| D23 | 92.22 ± 5.09 | D51 | 85.56 ± 5.09 | D48 | 65.56 ± 5.09 | D6 | 50.00 ± 3.33 |
| D35 | 92.22 ± 3.85 | D69 | 84.44 ± 1.92 | D32 | 65.56 ± 1.92 | D13 | 50.00 ± 0.00 |
| D2 | 91.11±5.09 | D67 | 84.44 ± 6.94 | D29 | 64.44 ± 1.92 | D38 | 48.89 ± 3.85 |
| D27 | 90.00±3.33 | D7 | 82.22 ± 5.09 | D56 | 64.44 ± 1.92 | D65 | 48.89 ± 1.92 |
| D50 | 90.00±0.00 | D12 | 81.11 ± 5.09 | D59 | 62.22 ± 5.09 | D55 | 46.67 ± 3.33 |
| D72 | 90.00±3.33 | D58 | 81.11 ± 5.09 | D9 | 61.11 ± 3.85 | D16 | 45.56 ± 3.85 |
| Variety No. | MD | Variety No. | MD | Variety No. | MD | Variety No. | MD |
|---|---|---|---|---|---|---|---|
| D36 | 0.90 | D57 | 0.73 | D3 | 0.68 | D66 | 0.56 |
| D68 | 0.88 | D1 | 0.73 | D63 | 0.68 | D5 | 0.53 |
| D34 | 0.83 | D47 | 0.71 | D17 | 0.67 | D42 | 0.51 |
| D30 | 0.81 | D43 | 0.71 | D23 | 0.67 | D51 | 0.50 |
| D50 | 0.81 | D71 | 0.70 | D35 | 0.65 | D12 | 0.47 |
| D27 | 0.79 | D49 | 0.70 | D2 | 0.64 | D69 | 0.47 |
| D8 | 0.78 | D64 | 0.69 | D46 | 0.64 | D58 | 0.45 |
| D10 | 0.76 | D72 | 0.68 | D21 | 0.62 | D67 | 0.45 |
| D70 | 0.74 | D28 | 0.68 | D19 | 0.60 | D7 | 0.32 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Takano, T.; Liu, S. Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil. Agronomy 2018, 8, 205. https://doi.org/10.3390/agronomy8100205
Wang H, Takano T, Liu S. Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil. Agronomy. 2018; 8(10):205. https://doi.org/10.3390/agronomy8100205
Chicago/Turabian StyleWang, Hao, Tetsuo Takano, and Shenkui Liu. 2018. "Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil" Agronomy 8, no. 10: 205. https://doi.org/10.3390/agronomy8100205
APA StyleWang, H., Takano, T., & Liu, S. (2018). Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil. Agronomy, 8(10), 205. https://doi.org/10.3390/agronomy8100205




