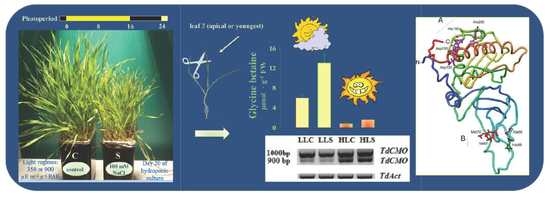
Unveiling the Enigmatic Structure of TdCMO Transcripts in Durum Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Isolation of TdCMOTranscripts
2.3. cDNA Sequence Analysis
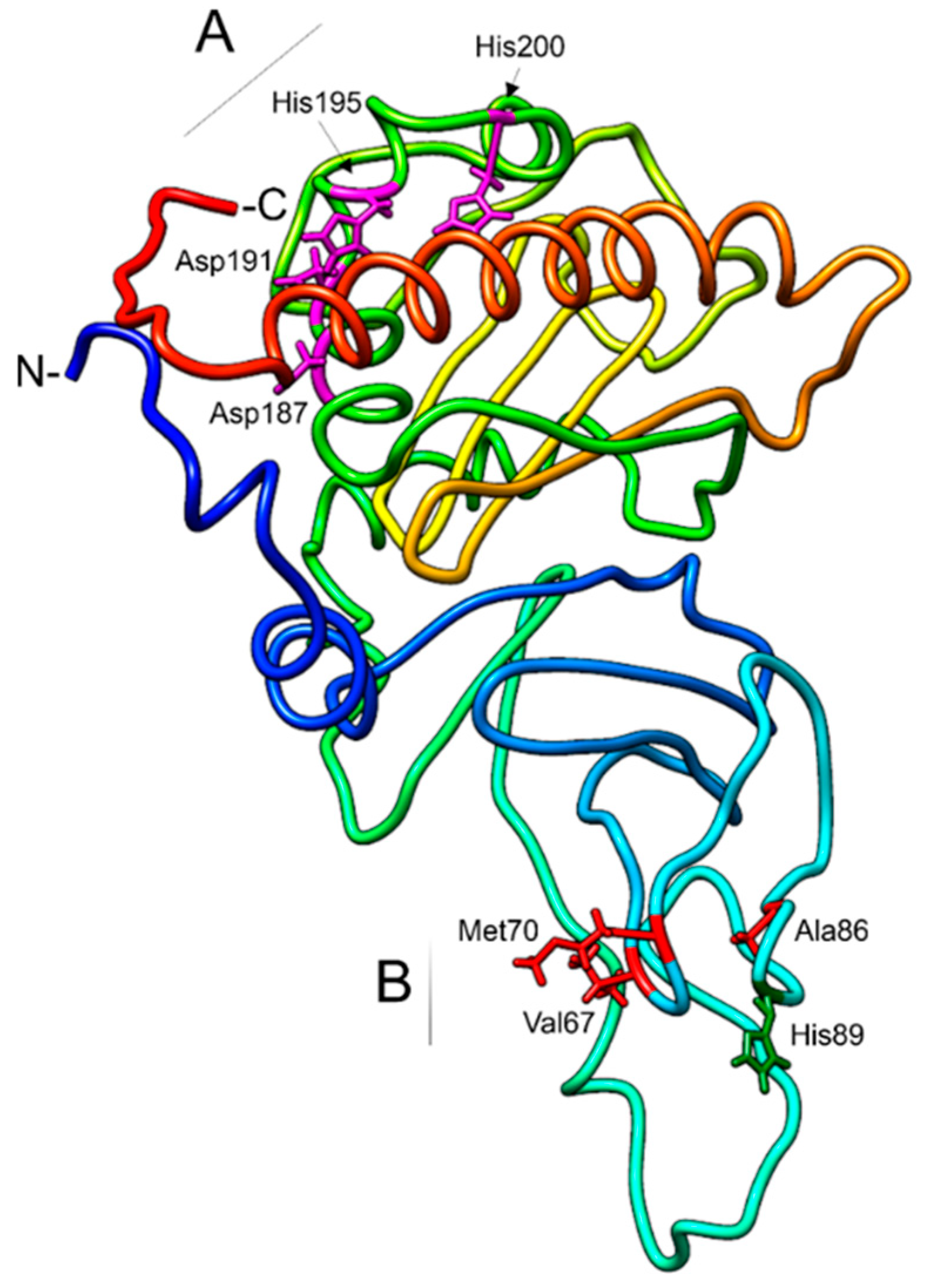
2.4. Sequence Analysis and Homology Modeling
3. Results
3.1. Choline Monooxygenase Transcripts Isolation
3.2. Influence of Salt and Light Stresses of the TdCMO1a6 Expression
3.3. ChromosomalLocalization and Functional Domain Characterization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gao, J.P.; Chao, D.Y.; Lin, H.X. Understanding abiotic stress tolerance mechanisms: Recent studies on stress response in rice. J. Integr. Plant Biol. 2007, 49, 742–750. [Google Scholar] [CrossRef]
- Rodziewicz, P.; Swarcewicz, B.; Chmielewska, K.; Wojakowska, A.; Stobiecki, M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiol. Plant 2014, 36, 1–19. [Google Scholar] [CrossRef]
- FAOSTAT (Food and Agriculture Organization Corporate Statistical Database). FAO Online Statistical Database on Annual Population Estimation and Year of Projection. Available online: http://faostat3.fao.org/download/O/OA/E (accessed on 7 May 2018).
- Ciarmiello, L.F.; Woodrow, P.; Piccirillo, P.; De Luca, A.; Carillo, P. Transcription Factors and Environmental Stresses in Plants. In Emerging Technologies and Management of Crop Stress Tolerance, 1st ed.; Paraviz, A., Saiema, R., Eds.; Elsevier: Eastbourne, UK, 2014; Volume 1, pp. 19–40. [Google Scholar]
- Kafantaris, I.; Woodrow, P.; Carillo, P. R gene expression changes related to Cercosporahydrangeae L. Mol. Biol. Rep. 2013, 40, 4173–4180. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Pontecorvo, G.; Ciarmiello, L.F.; Annunziata, M.G.; Fuggi, A.; Carillo, P. Transcription factors and genes in abiotic stress. In Crop Stress and Its Management: Perspectives and Strategies; Springer Science: London, UK, 2012; pp. 317–357. [Google Scholar]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Parisi, D.; Woodrow, P.; Pontecorvo, G.; Massaro, G.; Annunziata, M.G.; Fuggi, A.; Sulpice, R. Salt-induced accumulation of glycine betaine is inhibited by high light in durum wheat. Funct. Plant Biol. 2011, 38, 139–150. [Google Scholar] [CrossRef]
- Guerriero, G.; Behr, M.; Hausman, J.F.; Legay, S. Textile Hemp vs. Salinity: Insights from a targeted gene expression analysis. Genes 2017, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Maximova, E.; Fuggi, A.; Carillo, P. Durum wheat roots adapt to salinity remodeling the cellular content of nitrogen metabolites and sucrose. Front. Plant Sci. 2016, 7, 2035. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Mastrolonardo, G.; Nacca, F.; Fuggi, A. Nitrate reductase in durum wheat seedling as affected by nitrate nutrition and salinity. Funct. Plant Biol. 2005, 32, 209–219. [Google Scholar] [CrossRef]
- Ferchichi, S.; Hessini, K.; Dell’Aversana, E.; D’Amelia, L.; Woodrow, P.; Ciarmiello, L.F.; Fuggi, A.; Carillo, P. Hordeum vulgare and Hordeummaritimumrisponde to extended salinità stress displaying different temporal accumulation pattern of metabolites. Funct. Plant Biol. 2018, 45, 1096. [Google Scholar] [CrossRef]
- Sakamoto, A.; Murata, N. The role of glycine betaine in the protection of plants from stress: Clues from transgenic plants. Plant Cell Environ. 2002, 25, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, C.; Delfine, S.; Pizzuto, R.; Loreto, F.; Fuggi, A. Free amino acids and glycine betaine in leaf osmoregulation of spinach responding to increasing salt stress. New Phytol. 2003, 158, 455–463. [Google Scholar] [CrossRef] [Green Version]
- Carillo, P.; Mastrolonardo, G.; Nacca, F.; Parisi, D.; Verlotta, A.; Fuggi, A. Nitrogen metabolism in durum wheat under salinity: Accumulation of proline and glycine betaine. Funct. Plant Biol. 2008, 35, 412–426. [Google Scholar] [CrossRef]
- Chen, W.P.; Li, P.H.; Chen, T.H.H. Glycinebetaine increases chilling tolerance and reduces chilling-induced lipid peroxidation in Zea mays L. Plant Cell Environ. 2000, 23, 609–618. [Google Scholar] [CrossRef]
- Chen, T.H.H.; Murata, N. Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 2011, 34, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, S.; Kuwahara, J.; Ozaki, K.; Saeki, E.; Fujiwara, T.; Takabe, T. Isolation and characterization of a novel peroxisomal choline monooxygenase in barley. Planta 2011, 234, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Rathinasabapathi, B.; Burnet, M.; Russell, B.L.; Gage, D.A.; Liao, P.C.; Nye, G.J.; Scott, P.; Golbeck, J.H.; Hanson, A.D. Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: Prosthetic group characterization and cDNA cloning. Proc. Natl. Acad. Sci. USA 1997, 94, 3454–3458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, B.L.; Rathinasabapathi, B.; Hanson, A.D. Osmotic Stress induces expression of choline monooxygenase in sugar beet and Amaranth. Plant Physiol. 1998, 116, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.L.; Wang, Y.M.; Zhang, D.B.; Naosuke, N.I.I. Isolation of a choline monooxygenase cDNA clone from Amaranthus tricolor and its expressions under stress conditions. Cell Res. 2001, 11, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.W.; Showalter, A.M. Cloning and salt-induced, ABA-independent expression of choline mono-oxygenase in Atriplexprostrata. Physiol. Plant. 2004, 120, 405–412. [Google Scholar] [CrossRef] [PubMed]
- ISTAT (Istituto Nazionale di Statistica). Available online: https://www.istat.it/it/agricoltura (accessed on 9 January 2018).
- Cuin, T.A.; Zhou, M.; Parsons, D.; Shabala, S. Genetic behavior of physiological traits conferring cytosolic K+/Na+ homeostasis in wheat. Plant Biol. 2012, 14, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; d’ Amelio, L.; Dell’ Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum wheat seedling responses to simultaneous high light and salinity involve a fine reconfiguration of amino acids and carbohydrate metabolism. Phys. Plant. 2016, 159, 290–312. [Google Scholar] [CrossRef]
- Carillo, P. GABA shuont in durum wheat. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Pontecorvo, G.; Fantaccione, S.; Fuggi, A.; Kafantaris, I.; Parisi, D.; Carillo, P. Polymorphism of a new Ty1-copia retrotransposon in durum wheat under salt and light stresses. Theor. Appl. Genet. 2010, 121, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Pontecorvo, G.; Ciarmiello, L.F.; Fuggi, A.; Carillo, P. Ttd1a promoter is involved in DNA-protein binding by salt and light stresses. Mol. Biol. Rep. 2011, 38, 3787–3794. [Google Scholar] [CrossRef] [PubMed]
- Blast. Available online: http://www.ncbi.nlm.nih.gov/BLAST/.
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.J.; Gakhar, L.; Ramaswamy, S. Rieskebussiness:Structure-function of rieskenon hemeoxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Iwata, S.; Saynovits, M.; Link, T.A.; Michel, H. Structure of a water soluble fragmentof the ‘Rieske’ iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution. Structure 1996, 4, 567–579. [Google Scholar] [CrossRef]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Borrelli, G.; Ficco, D.; Giuzio, L.; Pompa, M.; Cattivelli, L.; Flagella, Z. Durum wheat salt tolerance in relation to physiological, yield and quality characters. Cereal Res. Commun. 2011, 39, 525–534. [Google Scholar] [CrossRef]
- Landergot, U.; Naciri, Y.; Schneller, J.J.; Holderegger, R. Allelic configuration and polysomic inheritance of higly variable microsatellites in tetraploid gynodioecious Thymus praecox agg. Theor. Appl. Genet. 2006, 113, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kalyna, M.; Simpson, CG.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J.; et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012, 40, 2454–2469. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W.S. Alternative splicing in plants-coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Hayashi, M.; Nishimura, M. A leaf-peroxisomal protein, hydroxypyruvate reductase, is produced by light-regulated alternative splicing. Cell Biochem. Biophys. 2000, 32, 147–154. [Google Scholar] [CrossRef]
- Fisher, A.J.; Franklin, K.A. Chromatin remodelling in plant light signalling. Physiol. Plant. 2011, 142, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Yamaguchi, K.; Hayashi, M.; Nishimura, M. Stromal and thylakoidbound ascorbate peroxidases are produced by alternative splicing in pumpkin. FEBS Lett. 1997, 413, 21–26. [Google Scholar] [CrossRef]
- Wu, H.P.; Su, Y.S.; Chen, H.C.; Chen, Y.R.; Wu, C.C.; Lin, W.D.; Tu, S.L. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 2014, 15, R10. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 6194. [Google Scholar] [CrossRef]
- Koszul, R.; Cabouret, S.; Dujon, B.; Fischer, G. Eukaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004, 23, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 2000, 97, 7051–7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cronn, R.C.; Small, R.L.; Wendel, J.F. Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 1999, 96, 14406–14411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hibino, T.; Waditee, R.; Araki, E.; Ishikawa, H.; Aoki, K.; Tanaka, Y.; Takabe, T. Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J. Biol. Chem. 2002, 277, 41352–41360. [Google Scholar] [CrossRef] [PubMed]


| Treatments | Clones | Fragments Size | Rieske 2Fe-2S | Non-Heme Fe Binding Motif |
|---|---|---|---|---|
| LLC | 1a2, 1a3 | 1000 bp | HNVCRHHASLLACGSGQKTCFQCPYHGWT | VFCDNYLDGGYHVPYAHGALA |
| LLS | 2a1,2a2,2a4, 2a5 | |||
| HLS | 4a1 | |||
| LLC | 1a2, 1a3, 1a4, 1a5, 1a6 | NOT FOUND | ||
| LLS | 2a1, 2a2, 2a3,2a5 | |||
| HLC | 3a6 | |||
| HLS | 4a6 | |||
| LLC | 1b1, 1b5 | 900 bp | ||
| HLC | 3b4 | |||
| LLC | 1b6 | VFCDNYLDRGYHFSYSHGANA | ||
| HLC | 3b5 | VFCDNYLDRGYHVPYAHGALA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciarmiello, L.F.; Di Maro, A.; Woodrow, P.; Annunziata, M.G.; Kafantaris, I.; Mirto, A.; Iannuzzi, F.; Fuggi, A.; Carillo, P. Unveiling the Enigmatic Structure of TdCMO Transcripts in Durum Wheat. Agronomy 2018, 8, 270. https://doi.org/10.3390/agronomy8110270
Ciarmiello LF, Di Maro A, Woodrow P, Annunziata MG, Kafantaris I, Mirto A, Iannuzzi F, Fuggi A, Carillo P. Unveiling the Enigmatic Structure of TdCMO Transcripts in Durum Wheat. Agronomy. 2018; 8(11):270. https://doi.org/10.3390/agronomy8110270
Chicago/Turabian StyleCiarmiello, Loredana F., Antimo Di Maro, Pasqualina Woodrow, Maria Grazia Annunziata, Ioannis Kafantaris, Antonio Mirto, Federica Iannuzzi, Amodio Fuggi, and Petronia Carillo. 2018. "Unveiling the Enigmatic Structure of TdCMO Transcripts in Durum Wheat" Agronomy 8, no. 11: 270. https://doi.org/10.3390/agronomy8110270
APA StyleCiarmiello, L. F., Di Maro, A., Woodrow, P., Annunziata, M. G., Kafantaris, I., Mirto, A., Iannuzzi, F., Fuggi, A., & Carillo, P. (2018). Unveiling the Enigmatic Structure of TdCMO Transcripts in Durum Wheat. Agronomy, 8(11), 270. https://doi.org/10.3390/agronomy8110270