1. Introduction
A growing global population drives the need to increase agronomic output using less land and agricultural inputs [
1,
2]. At the same time, increases in climate variability require crops of the future be more resilient. These are challenging problems, requiring the development of sustainable, systems-level approaches to agricultural improvement that must evolve over time. Integrated research and development efforts that seek to address these needs involve expertise in engineering to develop novel sensing devices that can measure environmental parameters and plant traits; genetics to identify causal genes; and data sciences to predict genetic combinations that will result in crop improvement.
With this framework in mind, we devised a new way to train graduate students (
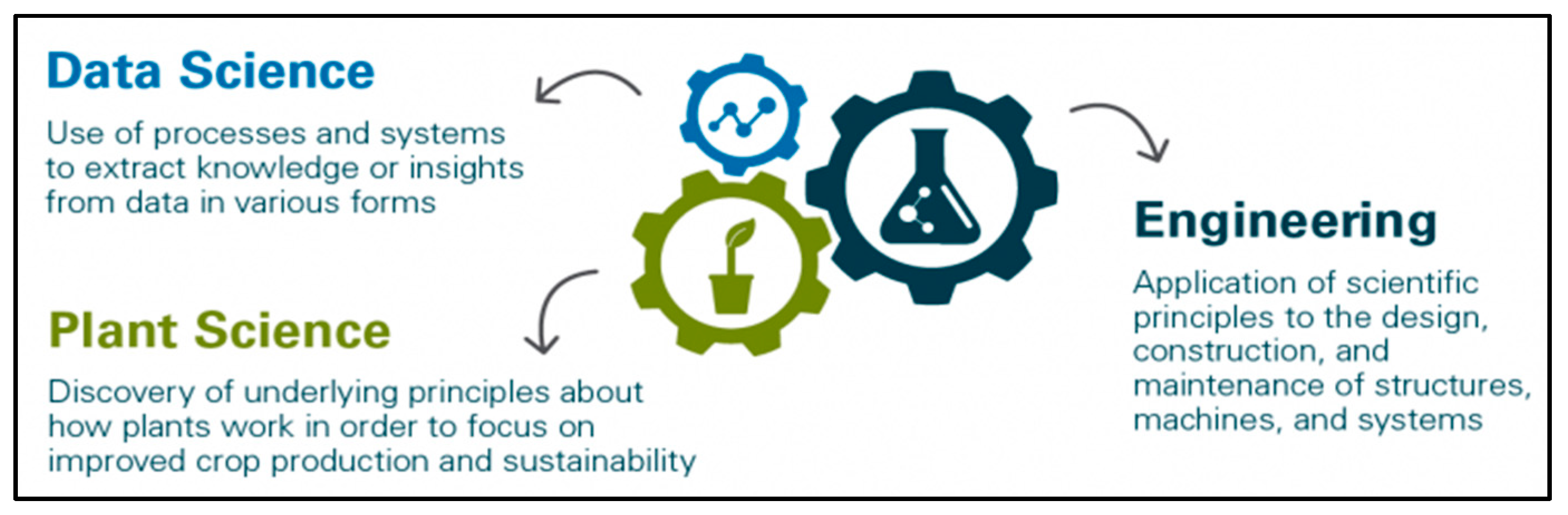
Table 1) and developed a graduate specialization in Predictive Plant Phenomics (P3). P3 is designed to train students broadly across the disciplines of plant sciences, data sciences, and engineering (
Figure 1). The core concepts for P3 training are based on the T-training model (
Figure 2), which was proposed by the American Society of Plant Biology (ASPB) and described in “Unleashing a Decade of Innovation in Plant Science: A Vision for 2015–2025” [
3]. This approach to graduate education requires exposure to multiple STEM (science/technology/engineering/mathematics) disciplines, development of effective communication and collaboration skills, and engagement with diverse institutions (industry, federal, academic, etc.). This must all occur without affecting the time needed to graduate the course.
2. Program Organization
In overview, the P3 degree specialization draws students from various departments and majors across the Iowa State campus and involves faculty members working in the colleges of Agriculture and Life Sciences, Liberal Arts and Sciences, and Engineering. Participating majors include Bioinformatics and Computational Biology, Electrical and Computer Engineering, Genetics and Genomics, Plant Biology, Plant Breeding, and Mechanical Engineering.
Students participate in a two-week boot camp before their first semester of graduate school begins. They are required to take the P3 core course entitled “Fundamentals of Predictive Phenomics” during their first semester and to rotate through three different research groups (i.e., one in plant science, one in data science, and one in engineering) over the course of the first and second semesters. For many students, these research rotations not only expose them to different areas of expertise, but they also serve as the foundation for matching students with a major professor and home laboratory for their graduate training.
As described in a previous report [
4], the P3 core course entitled “Fundamentals of Predictive Plant Phenomics” is a four-credit course that aims to ensure that all P3 students have a common knowledge base in engineering, plant sciences, and data sciences, irrespective of their undergraduate disciplinary focus. Through course activities, students learn to communicate using appropriate and discipline-specific terminology. Course planning took nearly a year and was based on guidance and help from faculty across diverse disciplinary backgrounds. The course is taught through a series of guest lecturers covering plant science, data science, and engineering topics over a 15-week period, and is supplemented with a laboratory session that meets once a week where students apply the knowledge they learn in the lectures to problem-solving situations.
Beyond the P3 core course, additional P3-specific course requirements are designed to work with existing requirements of the student’s major to ensure that increases in course loads are minimized. Students follow the curricular requirements of their primary graduate program or department, and supplement coursework with P3-required courses as technical electives. The program prescribes a 3-2-1 model where students take three courses in their primary discipline in keeping with the student’s major, two courses in a second area, and one course in the third area. For example, a student in the Interdepartmental Plant Biology program would take three courses in the broad area of plant sciences and might select two data science courses and a single engineering course.
Given the interdisciplinary nature of the P3 specialization, each participating major was consulted while devising P3 curricular requirements. Specific courses in the list acceptable for the P3 specialization were selected from participating majors. Based on our experiences, we recommend formal coursework to gain proficiency in subject areas shown in
Table 2, where topics are organized with the most critical toward the top of each section.
Each P3 student’s graduate committee includes at least three faculty members participating in the P3 program, with expertise drawn from all three major areas of expertise (i.e., plant sciences, data sciences, and engineering). Additional components of the program include matching students with internships in industry to better equip them to understand the similarities and differences between academic and industrial career options and training in entrepreneurship, which requires students to enroll in an entrepreneurship course or propose an alternate entrepreneurship experience for approval by the program leadership, including the P3 curriculum committee. More detailed information describing how the P3 program is organized can be found in Dickerson et al. [
5].
3. Assessment
Because P3 is a new concept in graduate training, we considered it important to assess the attitudes and perspectives of students to document the benefits of the training and to ensure that any program difficulties could be dealt with as they emerged. The details of this assessment appeared in a previous report [
5]. In brief, the P3 program features both internal and external evaluation. The P3 evaluation framework is recursive by design so that the external evaluators continuously inform the P3 leadership team about program performance and implementation. Quantitative assessments to measure the performance on program goals include online interviews with project leadership, P3 students, affiliated program faculty and university administrators. For the student group, a comparison group design was incorporated to allow stronger evaluation of the P3 student outcomes. A cohort of graduate students who are in the P3-participating majors but are unaffiliated with P3 has been recruited and participated in the evaluation activities so that the P3 cohort has a matched comparison group for analyses.
Qualitative data collection activities for the external evaluation include in-depth interviews and focus groups of students, which serve to enrich the findings of the quantitative assessments and to gain insights from students and faculty on the perceptions of and experiences with the P3 program. Feedback obtained during qualitative data collection helps the P3 leadership team to learn more about how students, faculty, and administrators feel about the program, obtain insights on how well it is working, and determine where it could be improved or revised during the planning for future activities. The external evaluation includes formative (i.e., related to implementation and progress) and summative dimensions to assess the quality and success of the P3 program.
With two student cohorts now participating in the program, preliminary assessment outcomes indicate that the project itself is progressing as anticipated. The feedback from the participating faculty indicates a good understanding of program goals and activities. P3 students report positive views of the program itself and their experiences in it. The interpersonal relationships of students in the P3 program with affiliated faculty and other students in the program were primarily positive. Most P3 students rated themselves favorably on various communication and employability skills. Both P3 and non-P3 cohorts reported positive views on their experiences thus far with peers, faculty, and staff. However, there was a notable difference. Although P3 students described rich connections beyond their departments and programs, such broad interactions were virtually non-existent for the non-P3 cohort.
Critical assessments by students are evaluated and program changes are implemented based on student feedback. For example, based on student feedback, we have increased coordination with the orientation activities offered by participating majors, involved more faculty members in activities and training sessions, expanded data carpentry components of the boot camp, expanded the statistical course requirement options, and developed specific exercises that are more project-based and hands-on for both the boot camp and the P3 core course.
4. Lessons Learned: Challenges and Opportunities for Training in Plant Phenomics
Agricultural and societal demands coupled with technological advancements are driving scientists and engineers to use sensors, robots, and drones to collect agricultural and environmental data for crop improvement. Individual research labs are leading not only research, but also educational opportunities by offering novel coursework and hands-on experiential learning through research. The next step is to infuse university systems with transdisciplinary thinking and training programs so that these technologies and methods become the norm for agricultural research. For that, a few obstacles must be overcome.
Departments, programs, and majors must be willing to consider flexibility in their curricular requirements and offerings. For example, some disciplinary departments at Iowa State have decided not to participate in the P3 specialization because they felt that they would lose some control over their curricular requirements.
New ways to recruit the best and the brightest minds to agricultural research must be developed. It is uncommon to find an individual who has the capacity and interest to learn across plant sciences, data sciences, and engineering. Better methods to identify and encourage candidates to join these programs are essential.
Industry partnerships are needed for internships, making it potentially difficult for institutions without agricultural industry nearby to offer such opportunities.
Despite these difficulties, it is clear that combining skills across these disciplines promises to advance agricultural research and to develop data-driven solutions to challenges in agriculture. Novel thinking on how to overcome these obstacles is needed, and proof-of-concept training programs like P3 are a good environment for formulating and sharing lessons learned and ‘best practices’ to advance this field.
5. Current Status and Future Plans for P3
We are in the third year of the P3 program. At this point, we can report that the students from the first two P3 cohorts have taken an active interest in driving many aspects of the program directly. P3 students are currently organizing a DuPont Pioneer-funded symposium (see
https://www.pioneer.com/home/site/about/research/PlantSciSymposiaSeries/). They have also taken over the leadership of many of the program’s boot camp activities, including the data carpentry workshop, and are volunteering their time to teach sections of the P3 core course. It is anticipated that by the end of support from the National Science Foundation for the P3 program, many activities developed to support the predictive plant phenomics at Iowa State University will be self-sustaining, demonstrating that the program itself will have been a source of institutional change. For that to happen, each of the challenges outlined in
Section 4 must be solved.
Author Contributions
J.A.D. is the Principal Investigator for the National Science Foundation grant supporting the P3 NRT. C.J.L-D., T.J.H., and P.S.S. are coPIs. M.E.L. leads the assessment team, which also includes S.J.S. and J.W. T.J.H. organizes and teaches the P3 core course with assistance from C.J.L-D. All authors contributed to writing this manuscript.
Acknowledgments
We are grateful to MaryAnn Grapp who serves as the P3 Program Coordinator. We could not manage this program without her constant efforts to keep things running smoothly. We also thank members of the P3 External Advisory Committee including and Tabare Abadie, Stephen Graham, Greg Heck, Edgar Spalding (chair), Abe Stroock, and Sriram Sundararajan who offer program guidance and opportunities to P3 students for professional development and internship opportunities. The work reported here is supported by the National Science Foundation under Grant Number DGE-1545463. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- The Royal Society of London; The US National Academy of Sciences; The Brazilian Academy of Sciences; The Chinese Academy of Sciences; The Indian National Science Academy; The Mexican Academy of Sciences; The Third World Academy of Sciences. Transgenic Plants and World Agriculture; The National Academies Press: Washington, DC, USA, 2000; p. 40. [Google Scholar]
- National Science and Technology Council. National Plant Genome Initiative Five Year Plan: 2014–2018; Office of Science and Technology Policy: Washington, DC, USA, 2014. [Google Scholar]
- American Society of Plant Biologists. Unleashing a decade of innovation in plant science–a vision for 2015–2025. In Plant Science Decadal Vision; American Society of Plant Biologists: Rockville, MD, USA, 2013; p. 36. [Google Scholar]
- Heindel, T.J.; Dickerson, J.A.; Dill-Lawrence, C.J.; Schnable, P. An interdisciplinary graduate course for engineers, plant scientists, and data scientists in the area of predictive plant phenomics. In Proceedings of the ASEE Annual Conference and Exposition, Columbus, OH, USA, 25–28 June 2017. Paper Number: 17970. [Google Scholar]
- Dickerson, J.A.; Heindel, T.J.; Lawrence-Dill, C.J.; Schnable, P.S. Training Students with T-Shaped Interdisciplinary Studies in Predictive Plant Phenomics. In Proceedings of the 2017 ASEE Annual Conference and Exposition, Columbus, OH, USA, 25–28 June 2017. Paper Number: 20006. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).