Abstract
Laurel is a medicinally important plant and is known to the world for its essential oil. Turkey is the main market in the laurel leaf trade by sharing about 90% of the world trade. Here we made an effort to elucidate genetic diversity and population structure of 94 Turkish laurel genotypes collected from 26 provinces and four geographical regions using inter-primer binding site (iPBS) retrotransposon markers. A total of 13 most polymorphic primers were selected which yielded 195 total bands, of which 84.10% were found polymorphic. Mean polymorphism information content (PIC) was (0.361) and diversity indices including mean effective number of alleles (1.36), mean Shannon’s information index (0.35) and overall gene diversity (0.22) revealed the existence of sufficient amount of genetic diversity in the studied plant material. Most diversity was found in genotypes collected from the Mediterranean region. Analysis of molecular variance (AMOVA) revealed that most of the variation (85%) in Turkish laurel germplasm is due to differences within populations. Model-based structure, principal coordinate analysis (PCoA) and neighbor-joining algorithms were found in agreement and clustered the studied germplasm according to their collection provinces and regions. This is a very first study exploring the genetic diversity and population structure of laurel germplasm using iPBS-retrotransposon marker system. We believe that information provided in this work will be helpful for the scientific community to take more interest in this forgotten but the medicinally important plant.
1. Introduction
There is an estimation that the current loss of plant species is between 100 and 1000 times more than the expected natural extinction rate [1] and it is believed that an increase in genetic erosion will be observed during the upcoming years. The rapid expansion of plant breeding activities during the 20th century for the development of improved varieties replaced or minimized the utilization of conventional landraces and resulted in a lower level of genetic diversity in in crop species [2]. To minimize the genetic erosion and maintain sustainability in agriculture, the Global Crop Diversity Trust initiated a consultation process for the development of 30 global crops and made strategies for the ex situ conservation and utilization of germplasm [3]. Germplasm corresponds to the living tissues, which can be utilized for the generation of new plants and is therefore the most important component in the maintenance of plant genetic resources [4]. Gene banks comprise of plant germplasm in the form of seed collection, nursery, pollen and in vitro [5]. These gene banks are important by reflecting the genetic diversity of cultivated and wild relatives of various crops having unique phenotypic and genotypic characteristics [6]. Characterization of these genetic resources is one of the important alternatives for the scientific community to deal with the several challenges like food scarcity, various biotic and abiotic stresses through the investigation of genetic diversity [7,8].
Genetic diversity is the essence of the biological world and serves as a source of natural variations, which can be helpful to deal with various challenges to the world [7]. Medicinal plants kept the continuous attention from the human being through their multiple uses [9]. In 2008, WHO (World Health Organization) issued a report confirmıng the role of these plants in routine life and stated that 80% of the world population directly or indirectly depended on these plants as traditional medicine [10]. More than 1300 medicinal plants are in use by Europe and 90% of it is harvested from the wild and natural resources. Among the 150 topmost used drugs, 118 are based on the natural resources [11]. International Union for Conservation of Nature and the World Wildlife Fund stated in a report that between 50,000–80,000 flowering plant species are in use by the world for medicinal purposes. Nearly 15,000 of these flowering plants are threatened with extinction due to habitat destruction and over-harvesting [1]. There is a need to collect, characterize and conserve the plants having medicinal value to maintain sustainability in the ecosystem [12].
Mediterranean region contains a good diversity of plants and laurel (Laurus nobilis L.) is one of the important plants of this region [9]. Laurel belongs to the Asia and Balkan region and later it spread to various Mediterranean countries like Italy, Spain, Israel, France Corsica Island and North Africa [13]. Laurel belongs to the Lauraceae family and this family comprises of 2500–3000 species with a total of 50 genera mainly distributed in the tropic and subtropics of the world [9]. Laurus nobilis and L. azorica (Seub) Franco are two most economically important species. Laurel has been traditionally used for the treatment of epileptic events, neuralgia and Parkinsonism [14]. Laurel is very popular and well known due to its essential oil, which has been proven very effective for the treatment of epileptic, convulsion and flatulent colic problems [15,16]. Beside medicinal applications, laurel leaves are used as a flavoring agent and to improve the shelf life of food due to their high antimicrobial and antioxidant activities [15,16].
Turkey is considered the cradle of agriculture because it is the origin and distribution center for various crops, due to its geographic [7,17]. This plant arrived in Turkey in 1655 and was known by the name of “Daphne”. The Mediterranean, Aegean, West and Central Black Sea and Marmara region are the most important areas of Turkey where this plant naturally grows [18]. Turkey is ruling the laurel market and shares the 90% of world production [15,16,17,18,19]. According to Nadeem et al. [9], European countries imported 77% of laurel (HS code, 09109950) and 72% of these were imported from Turkey. As compared to the others plant, the laurel market is different because consumer herein is very concerned with the quality rather than the price [9].
Advancement in the molecular markers changed the fate of breeding and boost up the breeding activities. Several types of molecular markers have been developed by the scientific community according to their feasibility [20]. Retrotransposon, sometimes called the jumping element comprises 50%–90% of plant genome [21]. Long terminal repeat (LTR) and non-LTR retrotransposons are two types of retrotransposon, and the former are the most represented in plant genomes. However, Kalendar et al. [22] developed a new marker system named the “inter primer binding site (iPBS)” and suggested it a universal method for DNA fingerprinting, which can be employed both for plant and animals. This method overcomes all application limits of LTR and non-LTR retrotransposons. Inter primer binding sites primers are designed on the primer binding site (PBS) sequences having conserved parts of the tRNAs for reverse transcription during the replication cycle of retrotransposons [22,23]. These tRNAs complement primer binding sites (PBS) in LTR retrotransposons and are used as genomic regions during the PCR (polymerase chain reaction) amplification [22]. The universality of the iPBS-retrotransposon marker has been proven because it can be successfully utilized for any plant for the diversity assessment as well for the phylogenetic and evolutionary study [8,24,25]. Inter primer binding site (iPBS) markers are dominant markers. Previous studies have confirmed that the dominant marker system likely precludes obtaining unbiased estimates of many genetic parameters [26,27]. Besides of its dominant nature, it becomes the marker of choice for the genetic diversity assessment due to its universal nature [8,24,25]. Moreover, its lower cost gained the attentions of scientific laboratories having funding problems.
Very few efforts have been done to conserve the genetic resources of laurel and small germplasm collections are created and maintained by individual breeders. Most of the research is aimed to investigate the composition and effects of laurel essential oil. Yalçın et al. [28] collected the laurel leaves from Northern Cyprus and Caputo et al. [29] used the Italian laurel leaves for the investigation of composition and activities of essential oil. Arroyo-Garcia et al. [30] used the amplified fragment length polymorphism (AFLP) markers to investigate the genetic similarity in this important plant. Arroyo et al. [31] used the simple sequence repeats (SSR) markers for the characterization of genetic diversity among various species of the Lauraceae family. Rodríguez-Sánchez et al. [32] applied the chloroplast DNA (cpDNA) sequences to understand the phylogenetic history of laurel trees. Besides the germplasm characterization, almost nothing has been done regarding the breeding perspective of Laurel. There was a need to explore the genetic variation and investigate the population structure using a good number of germplasm. Therefore, we aimed to investigate the genetic diversity and population structure of Turkish laurel germplasm through the iPBS-retrotransposon marker system.
2. Materials and Methods
2.1. Plant Material and DNA Isolation
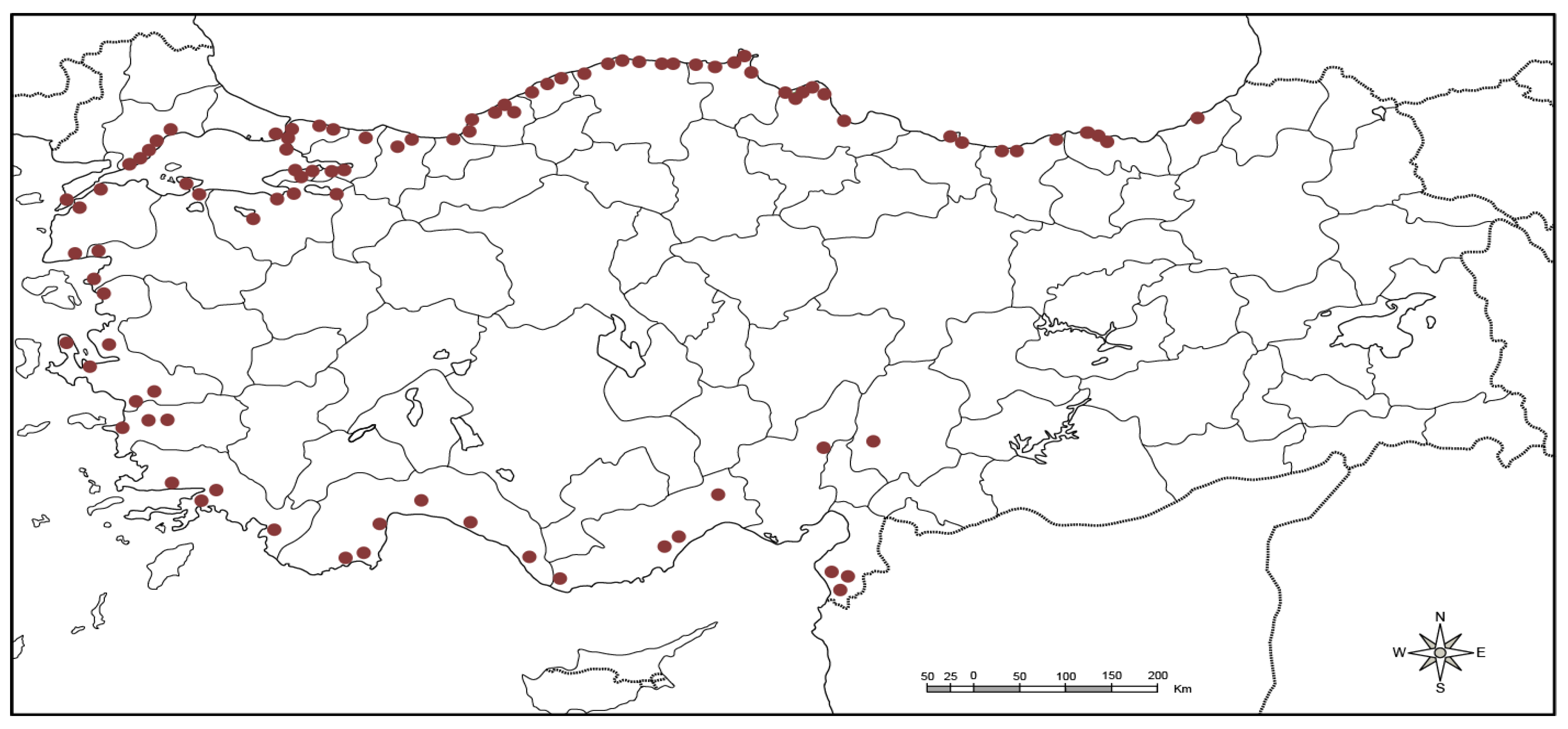
A total of 94 laurel genotypes collected from the 26 provinces from four geographical regions of Turkey were used as plant material for this study (Table S1; Figure 1). A nursery of collected germplasm was established at the Aegean Agricultural Research Institute of İzmir. Plant DNA was isolated by taking young and fresh leaves from each genotype. Genomic DNA was extracted by following the CTAB protocol [33] with some modifications [34]. Agarose gel (0.8%) was used for the assessment of DNA concentration and further quantification was done with NanoDrop (DeNovix DS-11 FX, USA). Five ng/μL was maintained as final DNA concentration for the further usage in a polymerase chain reaction (PCR).
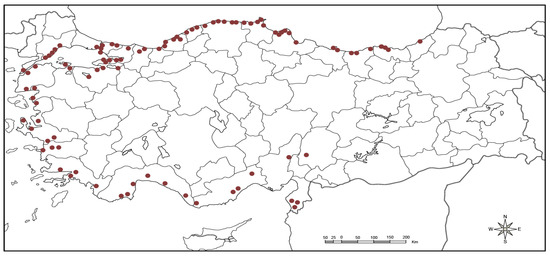
Figure 1.
Collection points of 94 Turkish laurel genotypes.
2.2. iPBS-Retrotransposon Analysis
Eight laurel genotypes were selected randomly for the screening purpose and a total of 83 iPBS-retrotransposons primers were used in the screening. All these primers were derived from a study by Kalendar et al. [22]. From this screening, 13 of the most polymorphic primers producing perfect banding profiles were selected for the fingerprinting of 94 laurel genotypes. Table 1 explains the name, sequence and annealing temperature of iPBS primers used in this study. For the confirmation of reproducibility of iPBS-retrotransposon primers, PCR amplification was repeated two times for two primers (Figure S1). All PCR amplifications were performed in 20 μL reaction containing 4 uL template DNA, 2 µL dNTPs (Thermo Scientific, Waltham, MA, USA), 0.2 µL U Taq DNA polymerase (Thermo Scientific), 4 µL primer, 2 µL 1 × PCR buffer (Thermo Scientific), 1.8 µL MgCl2 and 6 µL distilled water. By following the protocol suggested by the Kalendar et al. [22], PCR conditions were arranged which contains denaturation at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 15 s, annealing temperature 50–65 °C depending on primers used for 1 min; and a final extension at 72 °C 5 min. Amplified products were electrophoresed on a 2% (w/v) agarose gel, using 0.5 × Tris-borate-EDTA (TBE) buffer for 230 min; ethidium bromide was used for gel staining after electrophoresis and Imager Gel Doc XR+ system (Bio-Rad, Hercules, CA, USA) was used for the visualization and photographing. A 100 bp+ ladder was used as a molecular weight marker.

Table 1.
List of 13 inter-primer binding site (iPBS)-retrotransposon primers with their sequence and annealing temperature used to elucidate genetic diversity among 94 Turkish laurel genotypes.
2.3. Data Analysis
PCR products were scored as binary fashion; 0 or 1 for the absence and presence of specific bands with respect to 100 bp+ DNA ladder (Figure S2) because iPBS-retrotransposon is a dominant marker system. Reproducibility of DNA profiles for iPBS-retrotransposon marker systems was investigated by repeating two times the PCR amplification with some of the selected primers. Polymorphism information contents (PICs) were calculated by following the criteria set by Baloch et al. [24]. To investigate the level of genetic variations in the 94 laurel genotypes, various diversity parameters like the number of effective alleles (Ne), gene diversity (h) and Shannon’s information index (I) and Nei’s genetic distance were measured by PopGene ver. 1.32 [35]. The germplasm evaluated in this work was collected from four Turkish regions i.e., the Mediterranean, Black Sea, Aegean and Marmara region. Therefore, various diversity metrics were calculated on a per-region basis using PopGene ver. 1.32 [35]. An analysis of molecular variance (AMOVA) and principal coordinate analysis (PCoA) were investigated through GenAlEx 6.5 software [36]. To understand the level of the relationship among the 94 laurel genotypes, neighbor-joining analysis was performed using the R statistical software (version 3.4.1, Vienna, Austria). To explore the population structure of Turkish laurel germplasm, the Bayesian clustering model was applied in STRUCTURE software (version 2.3.4, Stanford, CA, USA). A continuous series of K were tested from 1 to 10 in five independent runs. For each run, the initial burn-in period was set to 50,000 followed by 50,000 Markov Chain Monte Carlo (MCMC) iterations as stated by the Baloch et al. [17]. To determine the proper numbers of the cluster (number of K; the number of subpopulations), criteria set by Evanno et al. [37] was followed and plotted the number of clusters (K) against logarithm probability relative to the standard deviation (ΔK).
3. Results
To characterize the 94 laurel genotypes, 13 selected primers resulted in a total of 195 scoreable bands with an average of 15 bands for each primer (Table 2). Among the 195 bands, 84.10% (164) were found polymorphic with an average of 12.61 fragments per primer. iPBS2398 produced maximum (23) and iPBS2232 and iPBS2295 resulted minimum (11) numbers of total bands. iPBS2402 was found much informative because all 14 bands were found polymorphic and iPBS2295 was found least informative by producing minimum (8) number of polymorphic bands. These 13 primers resulted in an average of 83.98% polymorphism, which ranged from 72.72% to 100% for iPBS2295, and iPBS2256 and iPBS2402, respectively. PIC values ranged between 0.163–0.585 for the iPBS2277 and iPBS2230 and 0.361 was the average PIC value. The maximum (1.57) and the minimum (1.15) number of effective alleles were produced by iPBS2230 and iPBS2277, respectively, and 1.36 was the mean effective number of alleles. Maximum and minimum gene diversity was 0.33 and 0.12 reflected by iPBS2230 and iPBS2277 respectively, while 0.22 was the mean gene diversity. Shannon’s information index ranged between 0.4803 (iPBS2230) and 0.22 (iPBS2277) and 0.35 was the found average Shannon’s information index. Mean pairwise genetic distance for 94 laurel genotypes was 0.228. Maximum Nei’s genetic distance was 0.607 found between Canakkale1 and Mersin1 genotypes followed by Izmir6 and Mersin1 having 0.598 genetic distance. Bartin3 and Zonguldak3 genotypes reflected the minimum (0.089) Nei’s genetic distance and Kocaeli1 and Kocaeli2 followed them having 0.096 genetic distance. To confirm the reproducibility of DNA profiles for the iPBS-retrotransposon marker system, PCR was repeated for two primers and there were the same total and polymorphic bands (Figure S2).

Table 2.
Various diversity indices computed to explore genetic diversity in Turkish laurel germplasm.
Diversity indices were also calculated according to the collection regions of 94 laurel genotypes (Table 3). Genotypes from the Mediterranean region showed a higher number of effective alleles (1.41), gene diversity (0.24) and expected heterozygosity (0.19). Genotypes from the Marmara region showed a minimum number of effective alleles (1.426), gene diversity (0.16) and expected heterozygosity (0.11). Genotypes from the Aegean and Mediterranean region reflected the maximum (0.31) and minimum (0.24) Shannon’s information index respectively. Genotypes from the Mediterranean and Marmara regions reflected the maximum (0.28) and minimum (0.18) mean Nei’s genetic distance.

Table 3.
Region based diversity indices evaluation for Turkish laurel germplasm.
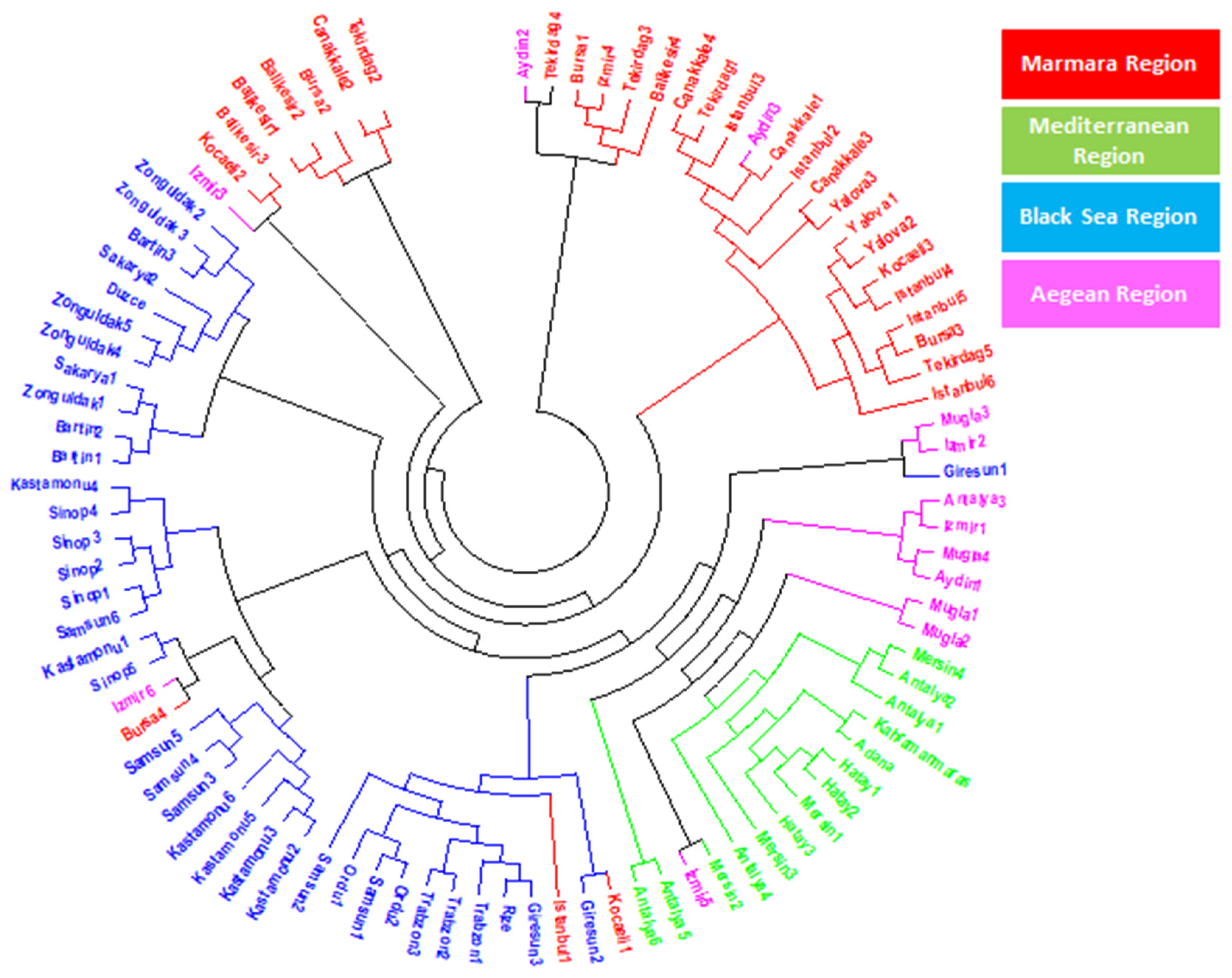
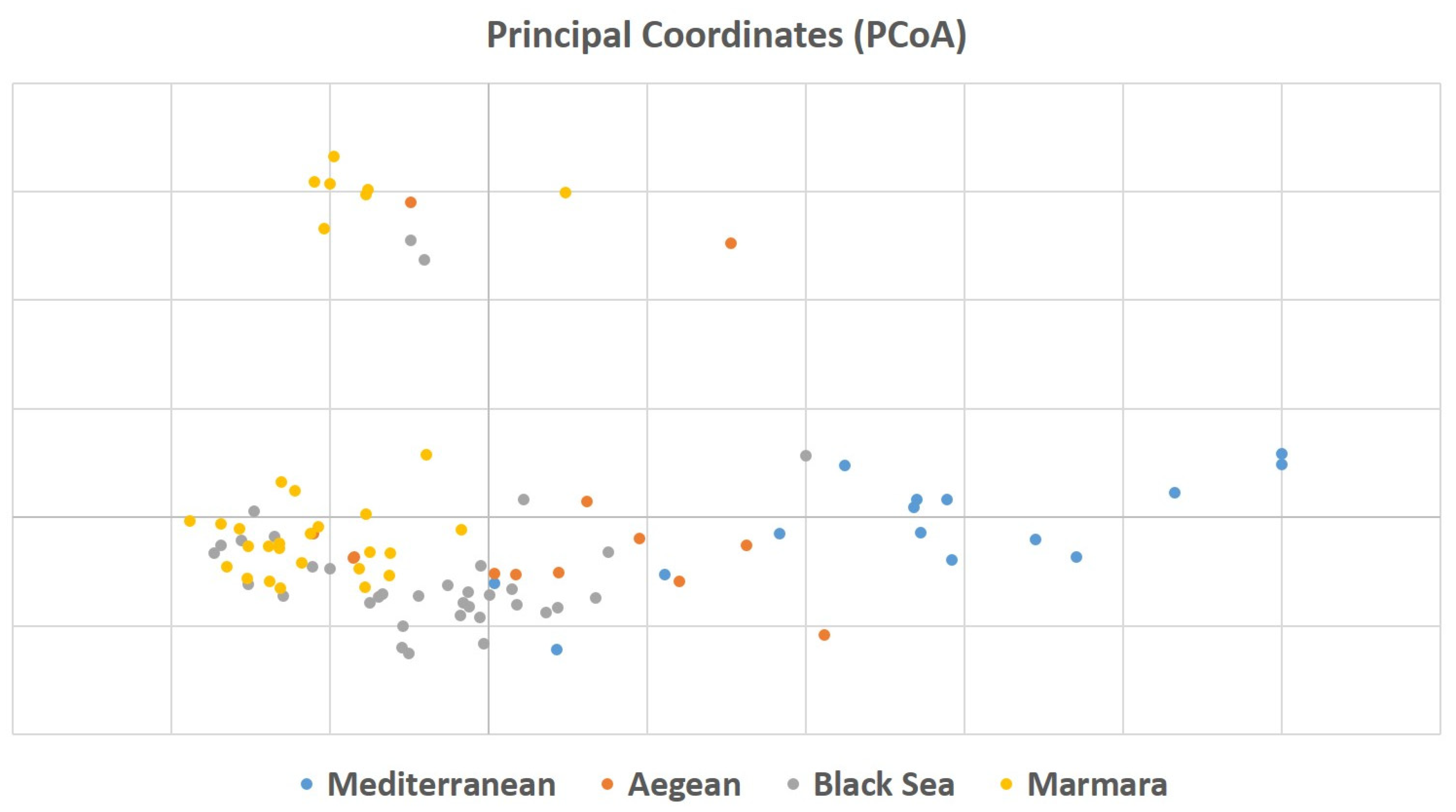
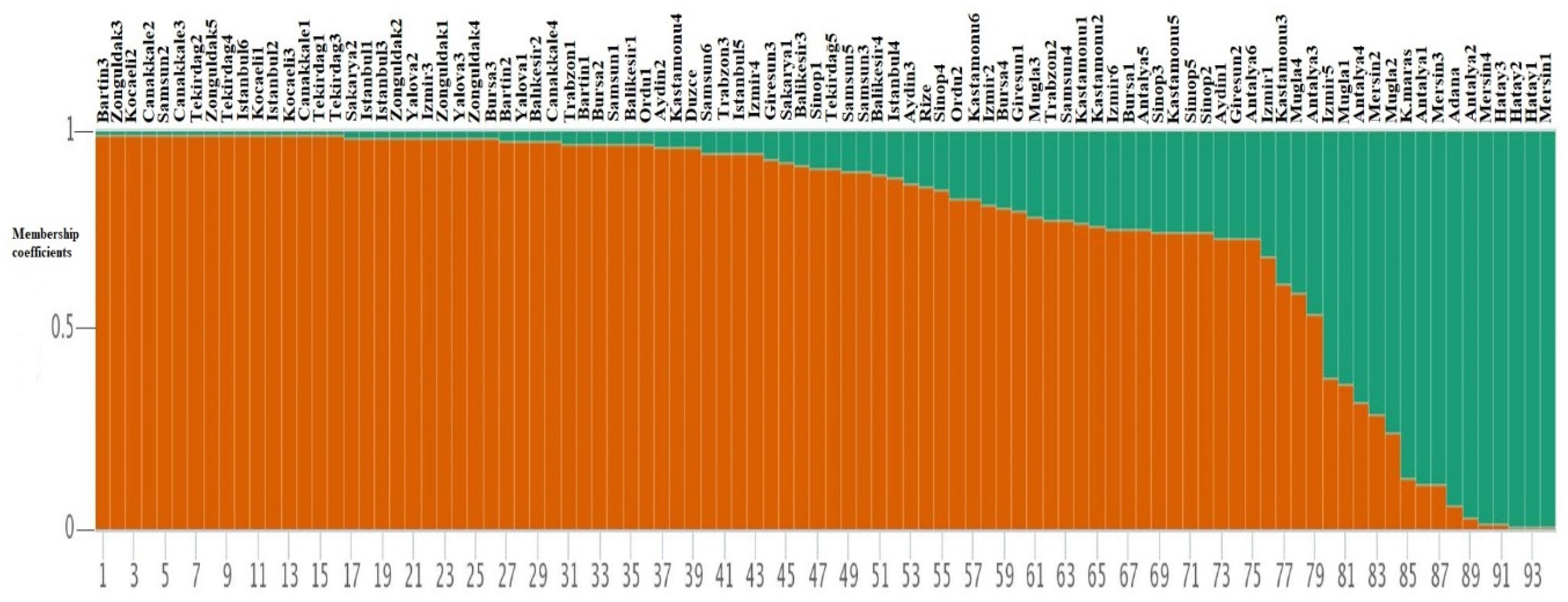
To understand the relationship among 94 laurel genotypes, the neighbor-joining clustering was performed, which separated the germplasm based on their geographical regions and provinces (Figure 2). Principal coordinate analysis (PCoA) also separated the 94 laurel genotypes based on their geographical regions (Figure 3). For the understanding of population structure of the Turkish laurel germplasm, Bayesian-based clustering algorithm was performed, which divided the genotypes into two main populations A and B (Figure 4). Population A contained mainly genotypes from the Marmara, Black Sea and Aegean region. Most of the genotypes from the Mediterranean region were present in population B. Analysis of molecular variance (AMOVA) was performed, which revealed higher variations (85%) within populations as compared to among the population (15%; Table 4).
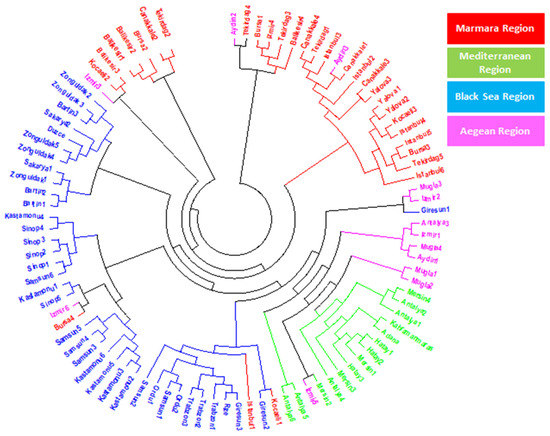
Figure 2.
Neighbor-joining clustering of Turkish laurel germplasm revealed by 13 iPBS-retrotransposon primers.
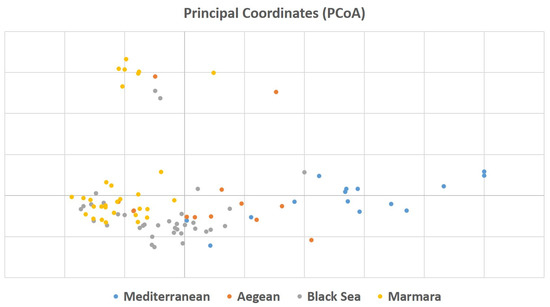
Figure 3.
Principal coordinate analysis of Turkish laurel germplasm revealed by 13 iPBS-retrotransposon primers.
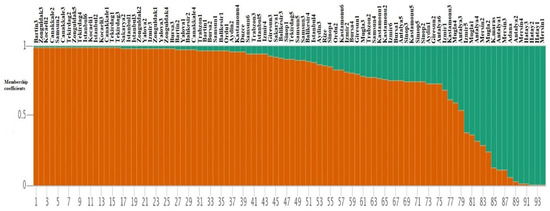
Figure 4.
Structure-based clustering among 94 Turkish laurel genotypes using 13 iPBS-retrotransposon markers.

Table 4.
Analysis of molecular variance (AMOVA) revealing genetic diversity in the Turkish laurel germplasm.
4. Discussion
4.1. Polymorphism in Turkish Laurel Germplasm Revealed by iPB-Retrotransposon Primers
A total of 13 most polymorphic iPBS-retrotransposon primers were used to elucidate genetic diversity and population structure of Turkish laurel germplasm (Table 2). Total and polymorphic bands obtained in this study were found much higher than reported by Arroyo-Garcia et al. [30] using AFLP markers and Bulut et al. [38] using an SSR marker for laurel. The average number of polymorphic bands obtained in this study were found much greater than the reported by Baránek et al. [39] (7.1 bands), Guo et al. [23] (6.6 bands) and Nemli et al. [40] (3.8 bands). This could be due to laurel having a larger genome size as compared to plant genomes earlier studied. These results may also indicate that iPBS-retrotransposon are more conserved for laurel compared to guava [41], Cicer species [42] and saffron [43]. Additionally, we came to know that iPBS-retrotransposon markers are more informative as compared to other molecular marker systems, like inter-retrotransposon-amplified polymorphism (IRAP) [44], random amplification of polymorphic DNA (RAPD) and inter simple sequence repeat (ISSR) [45] and sequence-related amplified polymorphism (SRAP) [46] and can be suggested for any crop to elucidate genetic diversity.
Mean polymorphism obtained in this study was 83.98%, which varied from 72.72% to 100%. Mean polymorphism obtained in this study was found greater than reported by Sevindik [47] for laurel, Yaldiz et al. [25] for tobacco and Aydin and Baloch [48] using iPBS-retrotransposons and thus confirming greater variations in Turkish laurel germplasm. The PIC value is used to understand the efficiency of polymorphic loci for the identification of genetic diversity [43] and explore the discriminating power marker among genotypes [40]. In this study, the mean PIC value was 0.361 which varied 0.163–0.585. Mean PIC value obtained in this study was found much greater than reported by earlier studies using iPBS-retrotransposons markers [25,49].
Previous studies have proven iPBS-retrotransposon a highly reproducible, robust and trustable marker system [22,23,40,41]. Our earlier studies in wild emmer [8], pea [24], tobacco [25], common bean [48], pepper [50] and safflower [51] also found this marker system highly robust, and trustable, which can be used for the investigation of genetic diversity in any crop. Similarly, Mehmood et al. [41] also found this marker system highly reproducible, robust and trustable. Moreover, trustbility of this marker compared to another marker system has been proven by earlier studies as well. Cömertpay et al. [52] compared iPBS retrotransposons with SSR markers for rice germplasm and found parallel results with both marker system, which confirm trustbility of this marker system. Andeden et al. [42] used iPBS-retrotransposon and an ISSR marker system for the taxonomic evaluation of seven Cicer species. They found similar clustering for species and confirmed the trustbility and robustness of this marker system. Yildiz et al. [53] also checked the reproducibility of this marker system in okra by repeating PCR two times and stated that band profiles remains unchanged after PCR amplification. In this study, we also checked the reproducibility of the iPBS-retrotransposon marker system by repeating PCR two times for two primers. We found that band profiles remain unchanged after repeating the PCR amplification for both primers. Total and polymorphic numbers of bands remained unchanged (Figure S2) and these results were found similar to the previous studies [41,53]. These results are confirming the higher reproducibility of this marker system. Therefore, this marker system can be suggested as a marker of choice for the investigation of genetic diversity due to its highly reproducibility, robustness and lower cost.
4.2. Genetic Diversity and Population Evaluation for Turkish Laurel Germplasm
Various diversity indices were calculated to elucidate the genetic diversity in Turkish laurel germplasm (Table 2). The maximum number of effective alleles is always desirable because they show the existence of greater genetic variations. Mean effective number of alleles (1.36) found in this study were higher than the reported by previous studies in various crops using iPBS-retrotransposons markers [25,50]. Mean gene diversity (0.22) resulted in this study was found higher than the reported by earlier studies [25,48,50] and thus explains the presence of higher diversity in studied germplasm. Shannon’s information index is an important criterion to understand the variation as it distinguishes the genetic diversity in a population combining abundance and evenness [51]. Mean Shannon’s information index (0.35) was found greater than the reported by earlier studies using the same molecular marker [25,50]. Genetic distance was calculated among the 94 Turkish laurel genotypes and mean genetic distance was 0.228. Bartin3 and Zonguldak3 genotypes showed a higher level of similarity as they accounted for the minimum (0.089) genetic distance. Canakkale1 and Mersin1 genotypes were found genetically to be the most distinct genotypes as the maximum genetic distance (0.607) were present between these genotypes. Development of breeding material reflecting desirable attributes remained the central focus of the breeding community and Arystanbekkyzy et al. [8] stated that genetically distinct genotypes always acts as a source of breeding material. Therefore, Canakkale1 and Mersin1 genotypes can be suggested as candidate parents for the various laurel breeding activities.
To elucidate the genetic variations in Turkish laurel germplasm more comprehensively, various diversity indices were also calculated upon their collection regions (Table 3). Genotypes from the Mediterranean region showed a higher level of diversity as compared to the rest of the regions and the Marmara region was found to be the least diverse region. Cuttelod et al. [54] stated that the Mediterranean basin is one of the most diverse places on this planet and Nadeem et al. [9] stated that laurel is an important plant of this region and shows a good level of diversity in its phenotype and genotype. Hatay2 and Antalya5 were found to be the most diverse genotypes in the Mediterranean region, while Muğla1 and İzmir5 were distinct genotypes for the Aegean region. In the Black Sea region, Giresun2 and Sinop5 were distinct genotypes, while Sakarya1 and Bursa1 were distinct genotypes for the Marmara region. Therefore, these diverse genotypes can act as genetic stock for the breeding activities in laurel.
Analysis of molecular variance (AMOVA) revealed the existence of higher variations within the laurel genotypes and the percentage of the total variance was 85% (Table 4). Pour et al. [49] stated that higher variations within genotypes might be due to selection, adaptation, gene flow, genetic drift, variation in ecotypes and the pollination method. Moreover, human activities and environmental fluctuations over time might be responsible for higher variations [55]. Our findings were in line with Gramazio et al. [56], as they also found higher genetic variation within the population of Larix decidua using SSR markers. However, this factor should be accounted for that iPBS-retrotransposon is a dominant marker system. A marker of this nature may not provide the credible information about the selection and adaptation phenomenon that occurs in a plant species. Therefore, a co-dominant marker system should be used in future studies in laurel germplasm to evaluate the selection and adaptation factors. There were no statistically significant variations among the population (only 15%), therefore, most of the variation in Turkish laurel germplasm are due to differences within populations. Laurel male flower matures early when compared to the female flower and this condition is known as protandry, which ultimately ensures there is no self-pollination in this plant. This cross-fertilization ultimately is responsible for higher genetic diversity within the population [57].
The neighbor-joining analysis was performed to understand the relationship among 94 Turkish laurel genotypes, which grouped the germplasm according to their geographic regions and provinces (Figure 2). Genotypes belonging to the same region were present together and the same was the case for the genotypes belonging to their respective province. However, it was also observable that genotypes belonging to the Aegean region were also grouped with the rest of the provinces. Laurel is native to Mediterranean countries like Italy, Spain, Israel, France Corsica Island and North Africa [13]. Hatay, Kahramanmaraş, Adana, Mersin and Antalya provinces are present in the Mediterranean region of Turkey and native to this region [9]. However, due to its medicinal and essential oil importance, it gains the attention of Turkish farmers and spread to Black Sea and Marmara regions. Later, it was introduced into the Aegean region from all three regions. It is believed that a lot of seed mixing happens during laurel introduction to the Aegean region. Results of this study are also supporting this hypothesis because genotypes from the Aegean region are showing similarity to all provinces. There is a possibility of seed mixing, human selection and hybridization process when the seed was introduced to the Aegean region, which ultimately resulted in higher gene flow in the genotypes of this region. Bulut et al. [38] used an SSR marker to characterize Turkish laurel germplasm and they also found admixture of genotypes and concluded this possibly due to natural gene flow in genotypes reflecting similarity with other cluster genotypes. Principal coordinate analysis (PCoA) was also performed, which confirmed the result obtained by the neighbor-joining analysis (Figure 3). The principal coordinate analysis also divided the genotypes upon their collection regions. Similar to the neighbor-joining analysis, genotypes from Aegean regions were mixed with all three regions and supported the hypothesis of gene flow in this region.
The model-based structure algorithm has been found more informative and precise as compared to other clustering algorithms [7,51]. Therefore, the structure was taken as a clustering benchmark in this study. Using this algorithm, 94 Turkish laurel germplasms were partitioned into two populations, largely upon the basis of their provinces and respective region (Figure 4). Population A was found admixture of genotypes from all four regions. All genotypes from the Marmara and Black Sea region were present in population A. All genotypes from the Aegean region were present in population A except three genotypes (Mugla1, Mugla2 and Izmir5), which were present in population B on the basis of membership coefficients. Population B largely comprised of genotypes from the Mediterranean region. Three genotypes from the Mediterranean region were clustered in population A with genotypes from the Aegean region. The Aegean region is close to the Mediterranean region. Izmir and Mugla provinces are close to the Mediterranean region and there is a possibility of a seed exchange and hybridization event. Genetic diversity assessment is an important step towards the breeding activities as it provides a novel source of variations [58,59]. Various genetic diversity indices in this study revealed the existence of higher diversity in studied germplasm, which can be utilized for future laurel breeding activities. Similarly, various diversity indices confirmed the existence of higher genetic diversity in the genotypes from the Mediterranean region and it is believed that laurel was native to this region. Later, it was introduced to other regions of Turkey. Seed mixing and selection of germplasm for favorable traits lowers the genetic diversity in the Black Sea, Aegean and Marmara region. Therefore, genotypes from these regions reflected higher genetic similarity with each other and were grouped in population A.
5. Conclusions
The present study comprehensively explored the genetic diversity and population structure of Turkish laurel germplasm. This study confirmed iPBS-retrotransposon as a highly reproducible, trustable and robust marker system. An analysis of molecular variance (AMOVA) revealed that most variations in the studied germplasm are explained by the differences that existed within the populations. Neighbor joining analysis and PCoA analysis clustered the 94 Turkish laurel genotypes largely according to their collection provinces and regions. Genotypes from the Mediterranean region showed more diversity compared to the Black Sea, Aegean and Marmara region. We are confident that information derived from this study can be used for the deeper understanding of the genetic relationships in laurel germplasm, establishment of a reference collection and in the determination of appropriate breeding and conservation strategies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/9/10/647/s1. Figure S1. A representative gel imaging picture revealing genetic diversity among 94 Turkish laurel genotypes using 13 iPBS-retrotransposon primers; Figure S2. Confirmation of reproducibility of DNA profiles for iPBS-retrotransposon marker system; Table S1. Passport data of 94 Turkish laurel genotypes.
Author Contributions
Conceptualization, F.S.B. and Ü.K.; methodology, M.A.N.; software, M.A.N.; validation, F.S.B., and, M.A.N.; formal analysis, M.A.N.; investigation, M.A.N., A.Y.; resources, Ü.K., G.C.; data curation, M.A.N., and A.Y.; writing—original draft preparation, M.A.N.; writing—review and editing, Ü.K., S.E., E.H., M.Y., S.H.Y.; visualization, G.C.; supervision, F.S.B.
Funding
This research was funded by Scientific research unit, Bolu Abant Izzet Baysal University, Bolu, Turkey (BAP project: 2018.10.01.1305 to FSB).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, S.L.; Yu, H.; Luo, H.M.; Wu, Q.; Li, C.F.; Steinmetz, A. Conservation and sustainable use of medicinal plants: Problems, progress, and prospects. Chin. Med. 2016, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Van de Wouw, M.; Kik, C.; van Hintum, T.; van Treuren, R.; Visser, B. Genetic erosion in crops: Concept, research results and challenges. Plant Genet. Resour. 2010, 8, 1–15. [Google Scholar] [CrossRef]
- Khoury, C.K.; Castañeda-Alvarez, N.P.; Achicanoy, H.A.; Sosa, C.C.; Bernau, V.; Kassa, M.T.; Norton, S.L.; van der Maesen, L.J.G.; Upadhyaya, H.D.; Ramírez-Villegas, J.; et al. Crop wild relatives of pigeonpea [Cajanus cajan (L.) Millsp.]: Distributions, ex situ conservation status, and potential genetic resources for abiotic stress tolerance. Biol. Conserv. 2015, 184, 259–270. [Google Scholar] [CrossRef]
- Roederer, C.; Nugent, R.; Wilson, P. Economic Impacts of Genetically Modified Crops on the Agrifood Sector: A Synthesis. Working Document, Directorate-General for Agriculture; EU Commission Working Document; EU Commission: Brussels, Belgium, 2000. [Google Scholar]
- Engels, J.M.M. A Guide to Effective Management of Germplasm Collections; Visser, L., Ed.; IPGRI: Rome, Italy, 2003; pp. 157–161. [Google Scholar]
- Dawson, I.; Were, J. Collecting germplasm from trees-some guidelines. Agrofor. Today 1997, 9, 6–9. [Google Scholar]
- Nadeem, M.A.; Habyarimana, E.; Çiftçi, V.; Nawaz, M.A.; Karaköy, T.; Comertpay, G.; Shahid, M.Q.; Hatipoğlu, R.; Yeken, M.Z.; Ali, F.; et al. Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseq-generated silicoDArT marker information. PLoS ONE 2018, 13, e0205363. [Google Scholar] [CrossRef] [PubMed]
- Arystanbekkyzy, M.; Nadeem, M.A.; Aktas, H.; Yeken, M.Z.; Zencirci, N.; Nawaz, M.A.; Ali, F.; Haider, M.S.; Tunc, K.; Chung, G.; et al. Phylogenetic and taxonomic relationship of turkish wild and cultivated emmer (triticum turgidum ssp. dicoccoides) revealed by iPBSretrotransposons markers. Int. J. Agric. Biol. 2019, 21, 155–163. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Aasim, M.; Kırıcı, S.; Karık, Ü.; Nawaz, M.A.; Yılmaz, A.; Maral, H.; Khawar, K.M.; Baloch, F.S. Laurel (Laurus nobilis L.): A Less-Known Medicinal Plant to the World with Diffusion, Genomics, Phenomics, and Metabolomics for Genetic Improvement. In Biotechnological Approaches for Medicinal and Aromatic Plants; Springer: Singapore, 2018; pp. 631–653. [Google Scholar]
- Vital, P.G.; Rivera, W.L. Antimicrobial activity and cytotoxicity of Chromolaena odorata (L. f.) King and Robinson and Uncaria perrottetii (A. Rich) Merr. Extracts. J. Med. Plant Res. 2009, 3, 511–518. [Google Scholar]
- Balunas, M.J.; Kinghorn, A.D. Drug discovery from medicinal plants. Life Sci. 2005, 78, 431–441. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef]
- Leung, A.Y.; Foster, S. Encyclopedia of Common Natural Ingredients Used in Food, Drugs and Cosmetics, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 2003; pp. 69–71. [Google Scholar]
- Aqili Khorasani, M.H. Collection of Drugs (Materia Media); Enqelab-e-Eslami Publishing and Educational Organization: Tehran, Iran, 1991; pp. 388–389. [Google Scholar]
- El, S.N.; Karagozlu, N.; Karakaya, S.; Sahın, S. Antioxidant and antimicrobial activities of essential oils extracted from Laurus nobilis L. leaves by using solvent-free microwave and hydrodistillation. FNS 2014, 5, 97. [Google Scholar] [CrossRef]
- Cherrat, L.; Espina, L.; Bakkali, M.; García-Gonzalo, D.; Pagán, R.; Laglaoui, A. Chemical composition and antioxidant properties of Laurus nobilis L. and Myrtus communis L. essential oils from Morocco and evaluation of their antimicrobial activity acting alone or in combined processes for food preservation. J. Sci. Food Agric. 2014, 94, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Baloch, F.S.; Alsaleh, A.; Shahid, M.Q.; Çiftçi, V.; de Miera, L.E.S.; Aasim, M.; Nadeem, M.A.; Aktaş, H.; Özkan, H.; Hatipoğlu, R. A whole genome DArTseq and SNP analysis for genetic diversity assessment in durum wheat from central fertile crescent. PLoS ONE 2017, 12, e0167821. [Google Scholar] [CrossRef] [PubMed]
- Boza, A.; Hepaksoy, S. Some leaf properties of natural Laurus nobilis L. population in Karaburun Peninsula (Izmir/Turkey). In Proceedings of the VII International Scientific Agriculture Symposium (Agrosym 2016), Jahorina, Bosnia and Herzegovina, 6–9 October 2016; University of East Sarajevo, Faculty of Agriculture: Lukavica, Bosnia and Herzegovina, 2016; pp. 717–722. [Google Scholar]
- Dadalioǧlu, I.; Evrendilek, G.A. Chemical compositions and antibacterial effects of essential oils of Turkish oregano (Origanum minutiflorum), bay laurel (Laurus nobilis), Spanish lavender (Lavandula stoechas L.), and fennel (Foeniculum vulgare) on common foodborne pathogens. J. Agric. Food Chem. 2004, 52, 8255–8260. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Guo, D.L.; Guo, M.X.; Hou, X.G.; Zhang, G.H. Molecular diversity analysis of grape varieties based on iPBS markers. Biochem. Syst. Ecol. 2014, 52, 27–32. [Google Scholar] [CrossRef]
- Baloch, F.S.; Alsaleh, A.; de Miera, L.E.S.; Hatipoğlu, R.; Çiftçi, V.; Karaköy, T.; Yıldız, M.; Özkan, H. DNA based iPBS-retrotransposon markers for investigating the population structure of pea (Pisum sativum) germplasm from Turkey. Biochem. Syst. Ecol. 2015, 61, 244–252. [Google Scholar] [CrossRef]
- Yaldiz, G.; Camlica, M.; Nadeem, M.A.; Nawaz, M.A.; Baloch, F.S. Genetic diversity assessment in Nicotiana tabacum L. with iPBS-retrotransposons. Turk. J. Agric. For. 2018, 42, 154–164. [Google Scholar] [CrossRef]
- Szmidt, A.E.; Wang, X.R.; Lu, M.Z. Empirical assessment of allozyme and RAPD variation in Pinus sylvestris (L.) using haploid tissue analysis. Heredity 1996, 76, 412. [Google Scholar] [CrossRef]
- Fan, X.X.; Shen, L.; Zhang, X.; Chen, X.Y.; Fu, C.X. Assessing genetic diversity of Ginkgo biloba L. (Ginkgoaceae) populations from China by RAPD markers. Biochem. Genet. 2004, 42, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Yalçın, H.; Anık, M.; Şanda, M.A.; Çakır, A. Gas chromatography/mass spectrometry analysis of Laurus nobilis essential oil composition of northern Cyprus. J. Med. Food 2007, 10, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Caputo, L.; Nazzaro, F.; Souza, L.F.; Aliberti, L.; De Martino, L.; Fratianni, F.; Coppola, R.; De Feo, V. Laurus nobilis: Composition of essential oil and its biological activities. Molecules 2017, 22, 930. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-García, R.; Martínez-Zapater, J.M.; Prieto, J.F.; Álvarez-Arbesú, R. AFLP evaluation of genetic similarity among laurel populations (Laurus L.). Euphytica 2001, 122, 155–164. [Google Scholar]
- Arroyo, J.M.; Rigueiro, C.; Rodríguez, R.; Hampe, A.; Valido, A.; Rodríguez-Sánchez, F.; Jordano, P. Isolation and characterization of 20 microsatellite loci for laurel species (Laurus, Lauraceae). Am. J. Bot. 2010, 97, e26–e30. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, F.; Guzmán, B.; Valido, A.; Vargas, P.; Arroyo, J. Late Neogene history of the laurel tree (Laurus L.; Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. J. Biogeogr. 2009, 36, 1270–1281. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Baloch, F.S.; Alsaleh, A.; Andeden, E.E.; Hatipoğlu, R.; Nachit, M.; Özkan, H. High levels of segregation distortion in the molecular linkage map of bread wheat representing the West Asia and North Africa region. Turk. J. Agric. For. 2016, 40, 352–364. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.; Boyle, T.J.; Ye, Z.; Xiyan, J.M. PopGene32, Microsoft Windows-Based Freeware for Population Genetic Analysis, Version 1.32. Mol. Biology and Biotechnology Centre; University of Alberta: Edmonton, AB, Canada, 2000. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Bulut, M.Ç.; Özmen, C.Y.; Ergül, A.; Ayanoğlu, F. Genetic Characterization of Bay Laurel (Laurus nobilis L.) Populations Using Microsatellite Markers and Flow Cytometry. MKUJAS 2018, 23, 242–253. [Google Scholar]
- Baránek, M.; Meszáros, M.; Sochorová, J.; Čechová, J.; Raddová, J. Utility of retrotransposon-derived marker systems for differentiation of presumed clones of the apricot cultivar Velkopavlovická. Sci. Hortic. 2012, 143, 1–6. [Google Scholar] [CrossRef]
- Nemli, S.; Kianoosh, T.; Tanyolac, M.B. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based interprimer binding sites (iPBSs) markers. Turk. J. Agric. For. 2015, 39, 940–948. [Google Scholar] [CrossRef]
- Mehmood, A.; Luo, S.; Ahmad, N.M.; Dong, C.; Mahmood, T.; Sajjad, Y.; Jaskani, M.J.; Sharp, P. Molecular variability and phylogenetic relationships of guava (Psidium guajava L.) cultivars using inter-primer binding site (iPBS) and microsatellite (SSR) markers. Genet. Resour. Crop Evol. 2016, 63, 1345–1361. [Google Scholar] [CrossRef]
- Andeden, E.E.; Baloch, F.S.; Derya, M.; Kilian, B.; Özkan, H. iPBS-Retrotransposons-based genetic diversity and relationship among wild annual Cicer species. J. Plant Biochem. Biotechnol. 2013, 22, 453–466. [Google Scholar] [CrossRef]
- Gedik, A.; Duygu, A.T.E.S.; Erdogmus, S.; Comertpay, G.; Tanyolac, M.B.; Ozkan, H. Genetic diversity of Crocus sativus and its close relative species analyzed by iPBS-retrotransposons. Turk. J. Agric. For. 2017, 22, 243–252. [Google Scholar] [CrossRef]
- Boronnikova, S.V.; Kalendar, R.N. Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants. Russ. J. Genet. 2010, 46, 36–42. [Google Scholar] [CrossRef]
- Mir, J.I.; Ahmed, N.; Khan, M.H.; Mokhdomi, T.A.; Wani, S.H.; Bukhari, S.; Asif, A.M.I.N.; Qadri, R.A. Molecular characterization of saffron-potential candidates for crop improvement. Not. Sci. Biol. 2015, 7, 81–89. [Google Scholar] [CrossRef][Green Version]
- Babaei, S.; Talebi, M.; Bahar, M.; Zeinali, H. Analysis of genetic diversity among saffron (Crocus sativus) accessions from different regions of Iran as revealed by SRAP markers. Sci. Hortic. 2014, 171, 27–31. [Google Scholar] [CrossRef]
- Sevindik, E. Molecular Genetic Diversity of Some Laurus nobilis L. (Lauraceae) Populations Grown in The Aegean Region/Turkey. AFS 2019, 41, 28–31. [Google Scholar]
- Aydın, M.F.; Baloch, F.S. Exploring the genetic diversity and population structure of Turkish common bean germplasm by the iPBS-retrotransposons markers. Legume Res. 2018, LR-423, 1–7. [Google Scholar]
- Pour, A.H.; Karahan, F.; İlhan, E.; İlçim, A.; Haliloğlu, K. Genetic structure and diversity of Adonis L. (Ranunculaceae) populations collected from Turkey by inter-primer binding site (iPBS) retrotransposon markers. Turk. J. Bot. 2019, 43, 585–596. [Google Scholar] [CrossRef]
- Yildiz, M.; Koçak, M.; Nadeem, M.A.; Cavagnaro, P.; Barboza, K.; Baloch, F.S.; Argün, D.; Keleş, D. Genetic diversity analysis in the Turkish pepper germplasm using iPBS retrotransposon-based markers. Turk. J. Agric. For. 2019, 43. [Google Scholar] [CrossRef]
- Ali, F.; Yılmaz, A.; Nadeem, M.A.; Habyarimana, E.; Subaşı, I.; Nawaz, M.A.; Chaudhary, H.J.; Shahid, M.Q.; Ercişli, S.; Zia, M.A.B.; et al. Mobile genomic element diversity in world collection of safflower (Carthamus tinctorius L.) panel using iPBS-retrotransposon markers. PLoS ONE 2019, 14, e0211985. [Google Scholar] [CrossRef]
- Cömertpay, G.; Baloch, F.S.; Derya, M.; Andeden, E.E.; Alsaleh, A.; Sürek, H.; Özkan, H. Population structure of rice varieties used in Turkish rice breeding programs determined using simple-sequence repeat and inter-primer binding site-retrotransposon data. Genet. Mol. Res. 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Yildiz, M.; Koçak, M.; Baloch, F.S. Genetic bottlenecks in Turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment. Genet. Mol. Res. 2015, 14, 10588–10602. [Google Scholar] [CrossRef]
- Cuttelod, A.; García, N.; Malak, D.A.; Temple, H.J.; Katariya, V. The Mediterranean: A biodiversity hotspot under threat. In Wildlife in a Changing World–an analysis of the 2008 IUCN Red List of Threatened Species; Jean-Christophe, V., Craig, H.-T., Stuart, S.N., Eds.; IUCN: Gland, Switzerland, 2009; p. 89. [Google Scholar]
- Solouki, M.; Mehdikhani, H.; Zeinali, H.; Emamjomeh, A.A. Study of genetic diversity in Chamomile (Matricaria chamomilla) based on morphological traits and molecular markers. Sci. Hortic. 2008, 117, 281–287. [Google Scholar] [CrossRef]
- Gramazio, P.; Plesa, I.M.; Truta, A.M.; Sestras, A.F.; Vilanova, S.; Plazas, M.; Vicente, O.; Boscaiu, M.; Prohens, J.; Sestras, R.E. Highly informative SSR genotyping reveals large genetic diversity and limited differentiation in European larch (Larix decidua) populations from Romania. Turk. J. Agric. For. 2018, 42, 165–175. [Google Scholar] [CrossRef]
- Denisow, B.; Wrzesien, M.; Cwener, A. Pollination and floral biology of Adonis vernalis L. (Ranunculaceae)-a case study of threatened species. Acta Soc. Bot. Pol. 2014, 83, 29–37. [Google Scholar] [CrossRef]
- Guliyev, N.; Sharifova, S.; Ojaghi, J.; Abbasov, M.; Akparov, Z. Genetic diversity among melon (Cucumis melo L.) accessions revealed by morphological traits and ISSR markers. Turk. J. Agric. For. 2018, 42, 393–401. [Google Scholar] [CrossRef]
- Eser, E.; Topcu, H.; Kefayati, S.; Sütyemez, M.; Islam, M.R.; Kafkas, S. Highly polymorphic novel simple sequence repeat markers from Class I repeats in walnut (Juglans regia L.). Turk. J. Agric. For. 2019, 43, 174–183. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).