Liming Positively Modulates Microbial Community Composition and Function of Sugarcane Fields
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Measurement of Sucrose Content and Theoretical Yield
- (a)
- Single stalk weight (kg) = (stalk diameter (cm))2 × (stalk height (cm) − 30) × 1 (g/cm3) × 0.7854/1000.
- (b)
- Theoretical production (kg/hm2) = single stalk weight (kg) × productive stem numbers (hm2).
2.3. Measurement of Soil Chemical Properties
2.4. Soil DNA Extraction and PCR Amplification
2.5. Illumina MiSeq Sequencing
2.6. Processing and Analyzing of Sequencing Data
3. Results
3.1. Sugarcane Agronomic Properties and Production
3.2. Soil Physiochemical Properties
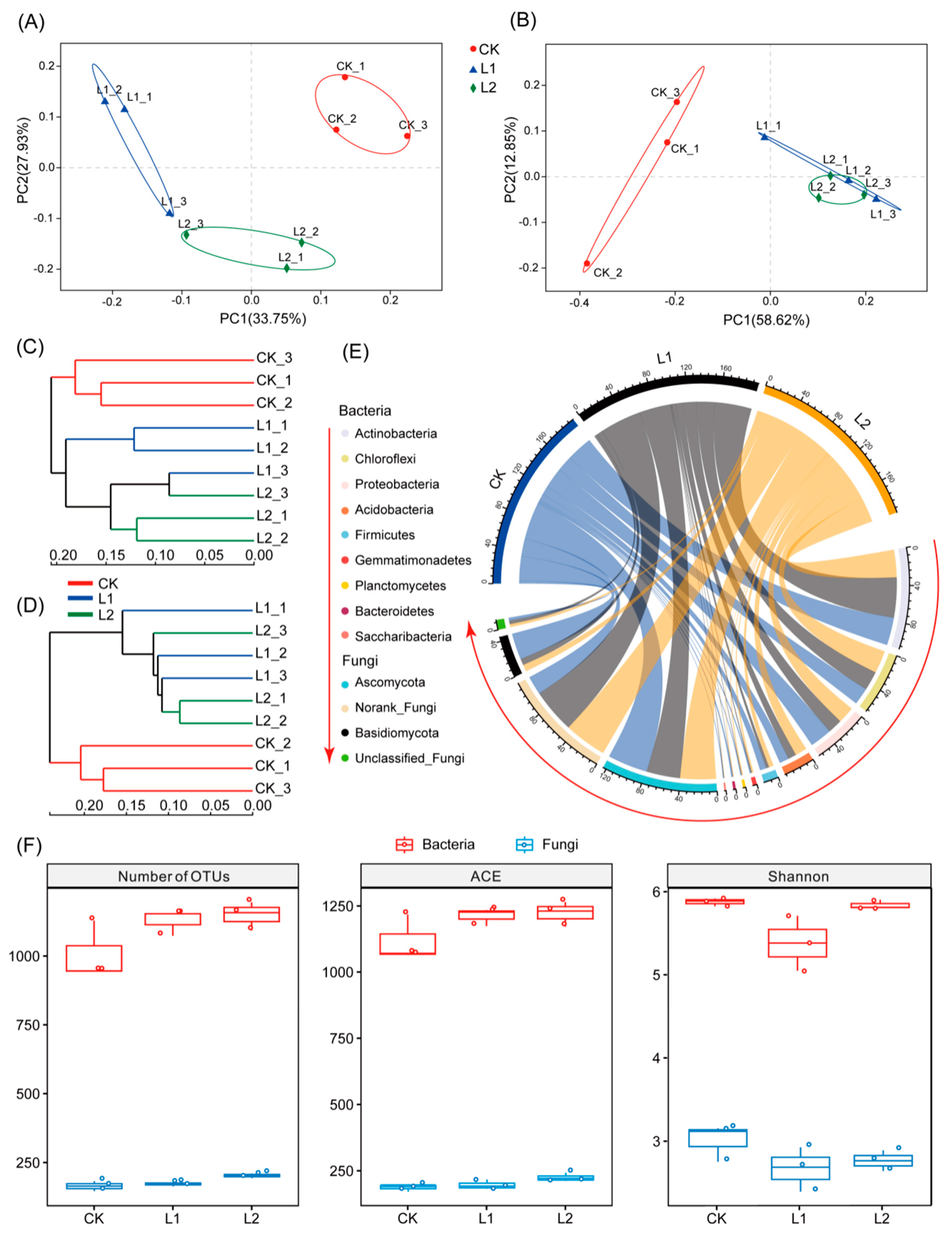
3.3. Microbial Alpha Diversity
3.4. Microbial Beta Diversity
3.5. Microbial Community Composition
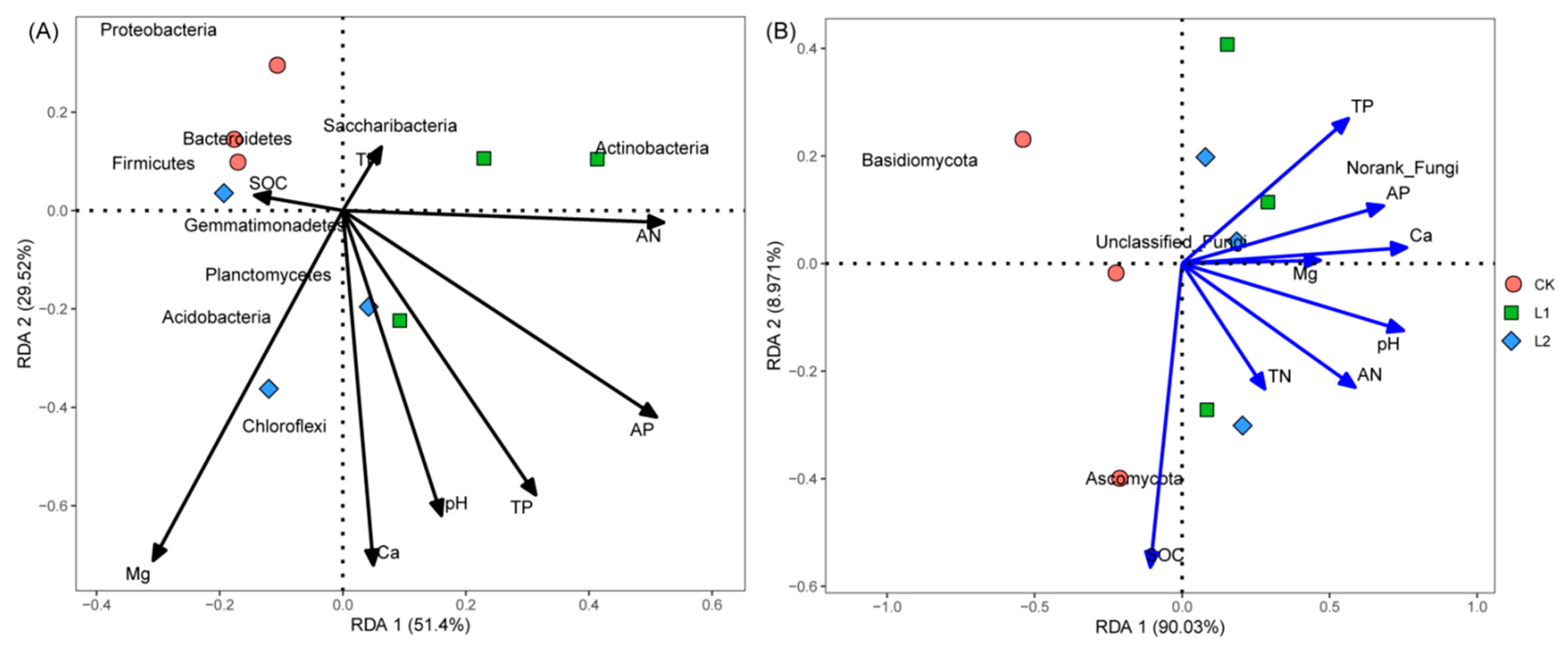
3.6. Correlation among Soil Chemical Properties and Microbial Community
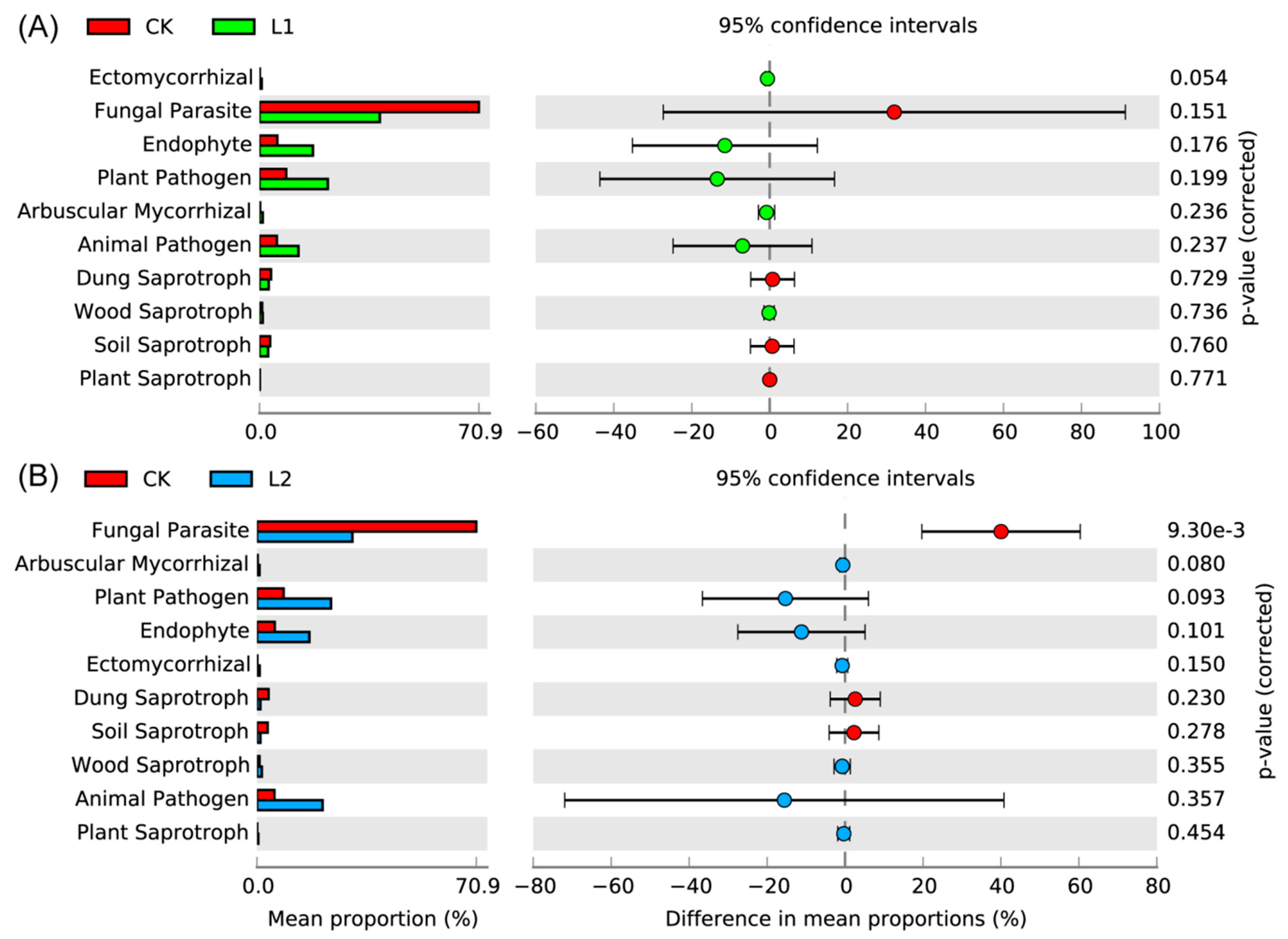
3.7. Bacterial Function Analysis
3.8. Fungal Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khalil, F.; Yueyu, X.; Naiyan, X.; Di, L.; Tayyab, M.; Hengbo, W.; Islam, W.; Rauf, S.; Pinghua, C. Genome characterization of Sugarcane Yellow Leaf Virus with special reference to RNAi based molecular breeding. Microb. Pathog. 2018, 120, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Eggleston, G.; Legendre, B.; Godshall, M.A. Sugar and Other Sweeteners. In Handbook of Industrial Chemistry and Biotechnology; Springer: Berlin, Germany, 2017; pp. 933–978. [Google Scholar]
- Chandel, A.K.; da Silva, S.S.; Carvalho, W.; Singh, O. V Sugarcane bagasse and leaves: Foreseeable biomass of biofuel and bio-products. J. Chem. Technol. Biotechnol. 2012, 87, 11–20. [Google Scholar] [CrossRef]
- Zhang, M.; Govindaraju, M. Sugarcane Production in China; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
- Tayyab, M.; Islam, W.; Arafat, Y.; Pang, Z.; Zhang, C.; Lin, Y.; Waqas, M.; Lin, S.; Lin, W.; Zhang, H. Effect of Sugarcane Straw and Goat Manure on Soil Nutrient Transformation and Bacterial Communities. Sustainability 2018, 10, 2361. [Google Scholar] [CrossRef]
- Das, S.; Adhya, T.K. Effect of combine application of organic manure and inorganic fertilizer on methane and nitrous oxide emissions from a tropical flooded soil planted to rice. Geoderma 2014, 213, 185–192. [Google Scholar] [CrossRef]
- Thorburn, P.J.; Biggs, J.S.; Palmer, J.; Meier, E.A.; Verburg, K.; Skocaj, D.M. Prioritizing Crop Management to Increase Nitrogen Use Efficiency in Australian Sugarcane Crops. Front. Plant Sci. 2017, 8, 1504. [Google Scholar] [CrossRef] [PubMed]
- De Castro, S.G.Q.; Franco, H.C.J. N-Fertilization Adjustment in Sugarcane Crop Cultivated in Intensive Mechanization. In Nitrogen in Agricultural Systems; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Hartemink, A.E.; Wood, A.W. Sustainable land management in the tropics: The case of sugarcane plantations. In Proceedings of the 16th World Congress of Soil Science, Montpellier, France, 20–26 August 1998; p. 7. [Google Scholar]
- Paungfoo-Lonhienne, C.; Yeoh, Y.K.; Kasinadhuni, N.R.P.; Lonhienne, T.G.A.; Robinson, N.; Hugenholtz, P.; Ragan, M.A.; Schmidt, S. Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 2015, 5, 8678. [Google Scholar] [CrossRef] [PubMed]
- Otto, R.; Castro, S.A.Q.; Mariano, E.; Castro, S.G.Q.; Franco, H.C.J.; Trivelin, P.C.O. Nitrogen Use Efficiency for Sugarcane-Biofuel Production: What Is Next? Bioenergy Res. 2016, 9, 1272–1289. [Google Scholar] [CrossRef]
- De Oliveira, M.W.; Macêdo, G.A.R.; Martins, J.A.; da Silva, V.S.G.; de Oliveira, A.B. Mineral nutrition and fertilization of sugarcane. In Sugarcane-Technology and Research; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar]
- Liu, X.Y.; Rezaei Rashti, M.; Esfandbod, M.; Powell, B.; Chen, C.R. Liming improves soil microbial growth, but trash blanket placement increases labile carbon and nitrogen availability in a sugarcane soil of subtropical Australia. Soil Res. 2018, 56, 235–243. [Google Scholar] [CrossRef]
- Singha, D.D. Effect of liming on the sugarcane (Saccharum officinarum) yield, juice quality and available soil nutrients in acid soil. Indian J. Agric. Sci. 2006, 76, 203–204. [Google Scholar]
- Daniel, J.L.P.; Santos, M.C.; Zopollatto, M.; Huhtanen, P.; Nussio, L.G. A data-analysis of lime addition on the nutritive value of sugarcane in Brazil. Anim. Feed Sci. Technol. 2013, 184, 17–23. [Google Scholar] [CrossRef]
- Lourenço, K.S.; Suleiman, A.K.A.; Pijl, A.; van Veen, J.A.; Cantarella, H.; Kuramae, E.E. Resilience of the resident soil microbiome to organic and inorganic amendment disturbances and to temporary bacterial invasion. Microbiome 2018, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- Altieri, M.A. The ecological role of biodiversity in agroecosystems. Agric. Ecosyst. Environ. 1999, 74, 19–31. [Google Scholar] [CrossRef]
- Xun, W.; Xiong, W.; Huang, T.; Ran, W.; Li, D.; Shen, Q.; Li, Q.; Zhang, R. Swine manure and quicklime have different impacts on chemical properties and composition of bacterial communities of an acidic soil. Appl. Soil Ecol. 2016, 100, 38–44. [Google Scholar] [CrossRef]
- Narendrula-Kotha, R.; Nkongolo, K.K. Microbial response to soil liming of damaged ecosystems revealed by pyrosequencing and phospholipid fatty acid analyses. PLoS ONE 2017, 12, e0168497. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, N.; Brodie, E.; Connolly, J.; Clipson, N. Impact of lime, nitrogen and plant species on bacterial community structure in grassland microcosms. Environ. Microbiol. 2004, 6, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Singh, B.; Dijkstra, F.A.; Dalal, R.C. Inorganic and organic carbon dynamics in a limed acid soil are mediated by plants. Soil Biol. Biochem. 2013, 57, 549–555. [Google Scholar] [CrossRef]
- Jones, J.P.; Woltz, S.S.; Everett, P.H. Effect of liming and nitrogen source on Fusarium wilt of cucumber and watermelon. In Proceedings of the Florida State Horticultural Society; 1975; Volume 88, pp. 200–203. [Google Scholar]
- Gatch, E.W.; du Toit, L.J. Limestone-Mediated Suppression of Fusarium Wilt in Spinach Seed Crops. Plant Dis. 2017, 101, 81–94. [Google Scholar] [CrossRef]
- Jones, J.P.; Woltz, S.S. Effect of soil pH and micronutrient amendments on Verticillium and Fusarium wilt of tomato. Plant Dis. Report. 1972, 56, 151. [Google Scholar]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Toju, H.; Kishida, O.; Katayama, N.; Takagi, K. Networks depicting the fine-scale co-occurrences of fungi in soil horizons. PLoS ONE 2016, 11, e0165987. [Google Scholar] [CrossRef]
- Glymenaki, M.; Barnes, A.; O’Hagan, S.; Warhurst, G.; McBain, A.J.; Wilson, I.D.; Kell, D.B.; Else, K.J.; Cruickshank, S.M. Stability in metabolic phenotypes and inferred metagenome profiles before the onset of colitis-induced inflammation. Sci. Rep. 2017, 7, 8836. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Wang, B.; Kaliannan, K.; Wang, X.; Lang, H.; Hui, S.; Huang, L.; Zhang, Y.; Zhou, M.; Chen, M.; et al. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio 2017, 8, e00417–e00470. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Huang, Y.; Zhang, M.; Yu, Y.; Lu, Y.; Kong, F. Bacterial community dynamics and functional variation during the long-term decomposition of cyanobacterial blooms in-vitro. Sci. Total Environ. 2017, 598, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, Q.; Zhang, H.; Bao, J. Inhibitor degradation and lipid accumulation potentials of oleaginous yeast Trichosporon cutaneum using lignocellulose feedstock. Bioresour. Technol. 2016, 218, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Tayyab, M.; Islam, W.; Lee, C.; Pang, Z.; Khalil, F.; Lin, S.; Lin, W.; Zhang, H. Short-Term Effects of Different Organic Amendments on Soil Fungal Composition. Sustainability 2019, 11, 198. [Google Scholar] [CrossRef]
- Farid, R.; Khordadpour, G. The Effect of Lime on Controlling Manganese Toxicity, Tea Yield and Quality in Sprinkler Condition. 2008. Available online: http://agris.fao.org/agris-search/search.do?recordID=IR2012049013 (accessed on 26 November 2019).
- Zhang, Y.; He, X.; Liang, H.; Zhao, J.; Zhang, Y.; Xu, C.; Shi, X. Long-term tobacco plantation induces soil acidification and soil base cation loss. Environ. Sci. Pollut. Res. 2016, 23, 5442–5450. [Google Scholar] [CrossRef]
- Lin, W.; Wu, L.; Lin, S.; Zhang, A.; Zhou, M.; Lin, R.; Wang, H.; Chen, J.; Zhang, Z.; Lin, R. Metaproteomic analysis of ratoon sugarcane rhizospheric soil. BMC Microbiol. 2013, 13, 135. [Google Scholar] [CrossRef]
- Bao, S.D. Methods of Soil Agricultural Chemistry Analysis; Agriculture Public House: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; U.S. Department of Agriculture: Washington, DC, USA, 1954; Volume 939, pp. 1–19.
- Mulvaney, R.L.; Khan, S.A. Diffusion methods to determine different forms of nitrogen in soil hydrolysates. Soil Sci. Soc. Am. J. 2001, 65, 1284–1292. [Google Scholar] [CrossRef]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer Science & Business Media: New York, NY, USA, 2007; ISBN 3540312110. [Google Scholar]
- Chen, C.; Zhang, J.; Lu, M.; Qin, C.; Chen, Y.; Yang, L.; Huang, Q.; Wang, J.; Shen, Z.; Shen, Q. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol. Fertil. Soils 2016, 52, 455–467. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Green, S.; Dangl, J.L. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Song, Z.; Yang, X.; Mao, Z.; Nie, X.; Guo, H.; Peng, X. Microbial community analysis of apple rhizosphere around Bohai Gulf. Sci. Rep. 2017, 7, 8918. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA\nsequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, A. Non-parametric estimation of the number of classes in a population. Ann. Math. Stat. 1984, 20, 265–270. [Google Scholar]
- Chao, A.; Lee, S.M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Keylock, C.J. Simpson diversity and the Shannon—Wiener index as special cases of a generalized entropy. Oikos 2005, 109, 203–207. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Bal, S.C.; Nayak, R.K.; Sahu, S.K. Effect of major nutrients (NPK) and lime on growth, yield and quality of mango cv Latsundari grown in acid lateritc soil of Bhubaneswar, Orissa. Environ. Ecol. 2009, 27, 1175–1177. [Google Scholar]
- Njoku, B.O.; Enwezor, W.O. Differential response of four cassava cultivars (Manihot esculenta) to liming of two acid soils in pot and field experiments. Field Crop. Res. 1991, 28, 163–172. [Google Scholar] [CrossRef]
- Chan, K.Y.; Oates, A.; Liu, D.L.; Li, G.D.; Prangnell, R.; Poile, G.J.; Conyers, M.K. A Farmer’s Guide to Increasing Soil Organic Carbon under Pastures; Industry & Investment NSW: Narrabri, Australia, 2010. [Google Scholar]
- Pham, T.G.; Nguyen, H.T.; Kappas, M. Assessment of soil quality indicators under different agricultural land uses and topographic aspects in Central Vietnam. Int. Soil Water Conserv. Res. 2018, 6, 280–288. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Abundance and Diversity of Bacterial, Archaeal, and Fungal Communities Along an Altitudinal Gradient in Alpine Forest Soils: What Are the Driving Factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F.; Ma, P. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Bei, S.; Li, B.; Zhang, J.; Christie, P.; Li, X. Organic fertilizer, but not heavy liming, enhances banana biomass, increases soil organic carbon and modifies soil microbiota. Appl. Soil Ecol. 2019, 136, 67–79. [Google Scholar] [CrossRef]
- Arafat, Y.; Wei, X.; Jiang, Y.; Chen, T.; Saqib, H.S.A.; Lin, S.; Lin, W. Spatial distribution patterns of root-associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 2017, 18, 1727. [Google Scholar] [CrossRef] [Green Version]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02717–e02738. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-soud, W.A.; Sørensen, S.J.; He, Z.; White, D.; Sinclair, A.; et al. Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ. Microbiol. 2014, 16, 2408–2420. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Hu, X.; Li, X.; Zhang, Y.; Jiang, L.; Li, J.; Guan, Z.; Cai, Y.; Liao, X. The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PLoS ONE 2017, 12, e0174411. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Ruan, Y.; Xue, C.; Zhong, S.; Li, R.; Shen, Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 2015, 393, 21–33. [Google Scholar] [CrossRef]
- Bryant, D.A.; Frigaard, N.U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006, 14, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Lücker, S.; Vejmelkova, D.; Kostrikina, N.A.; Kleerebezem, R.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Le Paslier, D.; Muyzer, G.; Wagner, M.; et al. Nitrification expanded: Discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012, 6, 2245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, K.; Kamagata, Y. The challenges of studying the anaerobic microbial world. Microbes Environ. 2014, 29, 335–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Gu, H.; Wang, H.; An, Q.; Xu, J. Complete genome sequence of Massilia sp. WG5, an efficient phenanthrene-degrading bacterium from soil. J. Biotechnol. 2016, 218, 49–50. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, H.; Yang, R.; Li, S.; Zhou, M.; Gao, T.; An, L.; Chen, X.; Dyson, P. Complete genome sequence of a psychotrophic Pseudarthrobacter sulfonivorans strain Ar51 (CGMCC 4.7316), a novel crude oil and multi benzene compounds degradation strain. J. Biotechnol. 2016, 231, 81–82. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.H.; Yokota, A. Sphingomonas azotifigens sp. nov., a nitrogen-fixing bacterium isolated from the roots of Oryza sativa. Int. J. Syst. Evol. Microbiol. 2006, 56, 889–893. [Google Scholar] [CrossRef]
- Lewis, R.; Barth, V.; Coffey, T.; McFarland, C.; Huggins, D.; Sullivan, T. Altered Bacterial Communities in Long-Term No-Till Soils Associated with Stratification of Soluble Aluminum and Soil pH. Soil Syst. 2018, 2, 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Song, Y.; Ma, T.; Raza, W.; Li, J.; Howland, J.G.; Huang, Q.; Shen, Q. Impacts of inorganic and organic fertilization treatments on bacterial and fungal communities in a paddy soil. Appl. Soil Ecol. 2017, 112, 42–50. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Townsend, A.R.; Sattin, S.R.; Freeman, K.R.; Fierer, N.; Neff, J.C.; Bowman, W.D.; Schadt, C.W.; Weintraub, M.N.; Schmidt, S.K. The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: Implications for carbon and nitrogen cycling. Environ. Microbiol. 2008, 10, 3093–3105. [Google Scholar] [CrossRef] [PubMed]
- Igiehon, N.O.; Babalola, O.O. Biofertilizers and sustainable agriculture: Exploring arbuscular mycorrhizal fungi. Appl. Microbiol. Biotechnol. 2017, 101, 4871–4881. [Google Scholar] [CrossRef]
- Rouphael, Y.; Franken, P.; Schneider, C.; Schwarz, D.; Giovannetti, M.; Agnolucci, M.; De Pascale, S.; Bonini, P.; Colla, G. Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Sci. Hortic. 2015, 196, 91–108. [Google Scholar] [CrossRef]



| Treatment | 2016 | 2017 | ||||||
|---|---|---|---|---|---|---|---|---|
| Urea | Calcium Superphosphate | Potassium Chloride | Quicklime | Urea | Calcium Superphosphate | Potassium Chloride | Quicklime | |
| CK | 675 | 1500 | 525 | 0 | 675 | 1500 | 525 | 0 |
| L1 | 675 | 1500 | 525 | 0 | 675 | 1500 | 525 | 1500 |
| L2 | 675 | 1500 | 525 | 1500 | 675 | 1500 | 525 | 1500 |
| Treatment | Sucrose Content (%) | Available Stalk Number (hm−2) | Stalk Height (cm) | Stalk Diameter (cm) | Single Stalk Weight (kg) | Theoretical Production (kg/hm2) |
|---|---|---|---|---|---|---|
| CK | 12.03 ± 0.54 a | 52,778 ± 1604 a | 300.7 ± 6.2 b | 2.56 ± 0.03 a | 1.40 ± 0.06 b | 73,716 ± 2916 a |
| L1 | 12.23 ± 0.38 a | 50,926 ± 2450 a | 321.0 ± 4.6 a | 2.61 ± 0.02 a | 1.55 ± 0.04 ab | 78,974 ± 2755 a |
| L2 | 12.48 ± 0.23 a | 51,852 ± 4036 a | 329.0 ± 2.3 a | 2.61 ± 0.03 a | 1.60 ± 0.03 a | 83,269 ± 7198 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Z.; Tayyab, M.; Kong, C.; Hu, C.; Zhu, Z.; Wei, X.; Yuan, Z. Liming Positively Modulates Microbial Community Composition and Function of Sugarcane Fields. Agronomy 2019, 9, 808. https://doi.org/10.3390/agronomy9120808
Pang Z, Tayyab M, Kong C, Hu C, Zhu Z, Wei X, Yuan Z. Liming Positively Modulates Microbial Community Composition and Function of Sugarcane Fields. Agronomy. 2019; 9(12):808. https://doi.org/10.3390/agronomy9120808
Chicago/Turabian StylePang, Ziqin, Muhammad Tayyab, Chuibao Kong, Chaohua Hu, Zhisheng Zhu, Xin Wei, and Zhaonian Yuan. 2019. "Liming Positively Modulates Microbial Community Composition and Function of Sugarcane Fields" Agronomy 9, no. 12: 808. https://doi.org/10.3390/agronomy9120808





