Hydrangea DNA Methylation Caused by pH Substrate Changes to Modify Sepal Colour is Detected by MSAP and ISSR Markers
Abstract
:1. Introduction
2. Materials and Methods
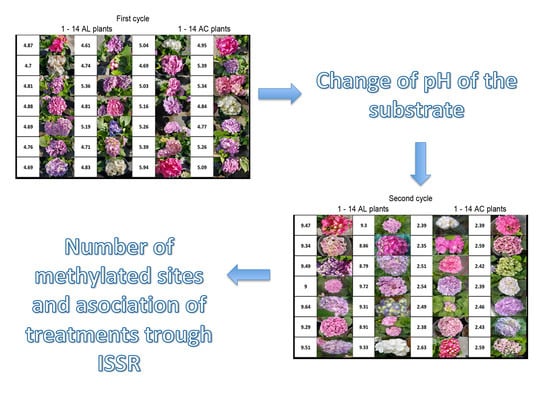
2.1. Plant Material
2.2. Treatments
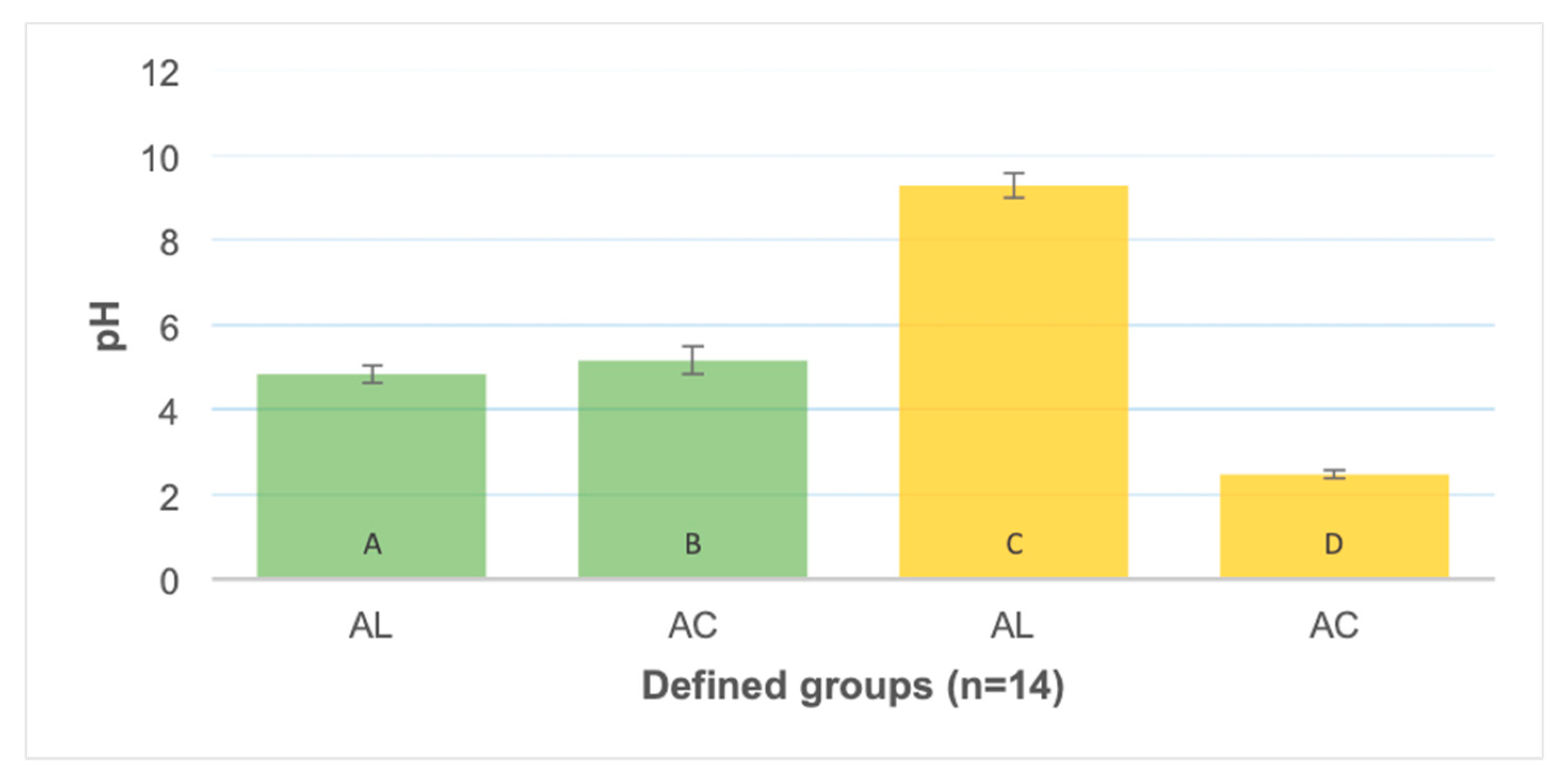
2.3. pH and Colour Registration
2.4. DNA Extraction
2.5. Methylation Sensitive Amplification Polymorphism (MSAP)
2.6. Inter-Simple Sequence Repeat
2.7. Statistical Analysis
3. Results
3.1. pH and Colour Registration
3.2. MSAP
3.3. ISSR
3.4. Mantel Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McClintock, E. A monograph of the genus Hydrangea. Proc. Calif. Acad. Sci. USA 1957, 29, 147–256. [Google Scholar]
- Cerbah, M.; Mortreau, E.; Brown, S.; Siljak-Yakovlev, S.; Bertrand, H.; Lambert, C. Genome size variation and species relationships in the genus Hydrangea. Theor. Appl. Genet. 2009, 103, 45–51. [Google Scholar] [CrossRef]
- Ball, V. Ball RedBook, 16th ed.; Ball Publishing: Batavia, IL, USA, 1998; p. 802. [Google Scholar]
- Durán-Espinosa, C. Hydrangeaceae. In Flora de Veracruz; Instituto Nacional de Investigaciones sobre Recursos Bióticos: Xalapa, Veracruz, 1999; pp. 1–22. [Google Scholar]
- Kesumawati, E.; Kimata, T.; Uemachi, T.; Hosokawa, M.; Yazawa, S. Correlation of phytoplasma concentration in Hydrangea macrophylla with green-flowering stability. Sci. Hort. 2006, 108, 74–78. [Google Scholar] [CrossRef]
- Galopin, G.; Codarin, S.; Viemont, J.D.; Morel, P. Architectural development of inflorescence in Hydrangea macrophylla cv. Hermann Dienemann. HortScience 2008, 43, 361–365. [Google Scholar] [CrossRef] [Green Version]
- Owen, J.S.; Fulcher, A.F.; Lebude, A.; Chappell, M. Hydrangea Production: Cultivar Selection and General Practices to Consider When Propagating and Growing Hydrangea. Available online: https://extension.tennessee.edu/publications/Documents/PB1840-A.pdf (accessed on 24 September 2019).
- Uemachi, T.; Mizuhara, Y.; Deguchi, K.; Shinjo, Y.; Kajino, E.; Ohba, H. Phylogenetic relationship of Hydrangea macrophylla (Thunb.) Ser. And, H. serrata (Thunb.) Ser. evaluated Using RAPD markers and plastid DNA Sequences. J. Jpn. Soc. Hortic. Sci. 2014, 83, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Allen, R.C. Influence of aluminium on the flower colour of Hydrangea macrophylla DC. Contr. Boyce Thompson Inst. 1943, 13, 221–242. [Google Scholar]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Rausher, M.D. Evolutionary transitions in floral color. Int. J. Plant Sci. 2008, 169, 7–21. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Brugliera, F.; Chandler, S. Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 2009, 10, 5350–5369. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Tuan, P.A.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 2016, 1–14. [Google Scholar] [CrossRef]
- Takeda, K.; Kariuda, M.; Itoi, H. Blueing of sepal colour of Hydrangea macrophylla. Phytochemistry 1985, 24, 2251–2254. [Google Scholar] [CrossRef]
- Takeda, K.; Yamashita, T.; Takahashi, A.; Timberlake, C.F. Stable blue complexes of anthocyanin-aluminium-3-p-coumaroyl-or 3-caffeoyl-quinic acid involved in the blueing of Hydrangea flower. Phytochemistry 1990, 29, 1089–1091. [Google Scholar] [CrossRef]
- Yoshida, K.; Toyama-Kato, Y.; Kameda, K.; Kondo, T. Sepal color variation of Hydrangea macrophylla and vacuolar pH measured with a proton-selective microelectrode. Plant Cell Physiol. 2003, 44, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Ito, D.; Shinkai, Y.; Kondo, T. Change of color and components in sepals of chameleon hydrangea during maturation and senescence. Phytochemistry 2008, 69, 3159–3165. [Google Scholar] [CrossRef]
- Ito, D.; Shinkai, Y.; Kato, Y.; Kondo, T.; Yoshida, K. Chemical studies on different color development in blue-and red-colored sepal cells of Hydrangea macrophylla. Biosci. Biotechnol. Biochem. 2009, 73, 1054–1059. [Google Scholar] [CrossRef] [Green Version]
- Ezaki, B.; Higashi, A.; Nanba, N.; Nishiuchi, T. An S-adenosyl Methionine Synthetase (SAMS) gene from Andropogon virginicus L. confers aluminum stress tolerance and facilitates epigenetic gene regulation in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1627. [Google Scholar] [CrossRef] [Green Version]
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast- and plasma membrane- localized aquaporin-Family transporters in blue hydrangea sepals of Aluminum hyperaccumulating plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef] [Green Version]
- Negishi, T.; Oshima, K.; Hattori, M.; Yoshida, K. Plasma membrane-localized Al-transporter from blue hydrangea sepals is a member of the anion permease family. Genes Cells 2013, 18, 341–352. [Google Scholar] [CrossRef]
- Chen, H.; Lu, C.; Jiang, H.; Peng, J. Global transcriptome analysis reveals distinct aluminum- tolerance pathways in the Al-accumulating species Hydrangea macrophylla and marker identification. PLoS ONE 2015, 10, e0144927. [Google Scholar] [CrossRef] [Green Version]
- Cocciolone, S.M.; Cone, K.C. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 1993, 135, 575–588. [Google Scholar]
- Sekhon, R.S.; Chopra, S. Progressive loss of DNA methylation releases epigenetic gene silencing from a tandemly repeated maize Myb gene. Genetics 2009, 181, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Fu, Z.; Chen, S.; Damaris, R.N.; Wang, K.; Li, T.; Yang, P. Proteomic and epigenetic analyses of lotus (Nelumbo nucifera) petals between red and white cultivars. Plant Cell Physiol. 2015, 56, 1546–1555. [Google Scholar] [CrossRef] [Green Version]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.Z.; Xu, C.G.; Maroof, M.S.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 1999, 261, 439–446. [Google Scholar] [CrossRef]
- Sanchez-Muñoz, R.; Moyano, E.; Khojasteh, A.; Bonfill, M.; Cusido, R.M.; Palazon, J. Genomic methylation in plant cell cultures: A barrier to the development of commercial long-term biofactories. Eng. Life Sci. 2019. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Li, X.; Korban, S.S. AFLP-based detection of DNA methylation. Plant Mol. Biol. Rep. 2000, 18, 361–368. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Eguiarte, L.E.; Souza, V.; Aguirre, X. Ecología Molecular; Instituto Nacional de Ecología, Semarnat: Ciudad de Mexico, México, 2007; p. 592. [Google Scholar]
- Heubl, G. DNA-based authentication of TCM-plants: Current progress and future perspectives. In Evidence and Rational Based Research on Chinese Drugs; Ulrich-Merzenich, G., Wagner, H., Eds.; Springer: Vienna, Austria, 2013; pp. 27–85. [Google Scholar] [CrossRef]
- Chandrika, M.; Rai, V.R. Genetic fidelity in micropropagated plantlets of Ochreinauclea missionis an endemic, threatened and medicinal tree using ISSR markers. Afr. J. Biotechnol. 2009, 8, 13. [Google Scholar]
- Lata, H.; Chandra, S.; Techen, N.; Wang, Y.H.; Khan, I.A. Molecular analysis of genetic fidelity in micropropagated plants of Stevia rebaudiana Bert. using ISSR marker. Am. J. Plant Sci. 2013, 4, 964–971. [Google Scholar] [CrossRef] [Green Version]
- Torres, A.; Camberato, D.; López, R.G.; Mickelbart, M. Medición de pH y Conductividad Eléctrica en Sustratos. Available online: https: //www.extension.purdue.edu/extmedia/HO/HO-237-SW.pdf (accessed on 24 September 2019).
- Baurens, F.C.; Causse, S.; Legavre, T. Methylation-sensitive amplification polymorphism (MSAP) protocol to assess CpG and CpNpG methylation in the banana genome. Fruits 2008, 63, 117–123. [Google Scholar] [CrossRef]
- Sanguinetti, C.J.; Neto, E.D.; Simpson, A.J. RAPD silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 1994, 17, 914–921. [Google Scholar] [PubMed]
- Pérez-Figueroa, A. MSAP: A tool for the statistical analysis of methylation-sensitive amplified polymorphism data. Mol. Ecol. Res. 2013, 13, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Reif, J.C.; Melchinger, A.E.; Frisch, M. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop. Sci. 2005, 45, 1–7. [Google Scholar] [CrossRef]
- Fulneček, J.; Kovařík, A. How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? Genetics 2014, 15, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Goyal, A.K.; Pradhan, S.; Basistha, B.C.; Sen, A. Micropropagation and assessment of genetic fidelity of Dendrocalamus strictus (Roxb.) nees using RAPD and ISSR markers. Biotech 2015, 5, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeats (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef]
- Nishijima, T.; Niki, T.; Niki, T. A Novel “Petaloid” mutant of Torenia (Torenia fournieri Lind. ex Fourn.) bears double flowers through insertion of the DNA transposon Ttf1 into a C-class floral homeotic gene. Horticul. J. 2016, 85, 272–283. [Google Scholar] [CrossRef] [Green Version]




| Name | Sequence | Technique |
|---|---|---|
| ELINK1 | CTCGTAGACTGCGTACC | MSAP adapter |
| ELINK2 | AATTGGTACGCAGTCTAC | MSAP adapter |
| HLINK1 | GATCATGAGTCCTGCT | MSAP adapter |
| HLINK2 | CGAGCAGGACTCATGA | MSAP adapter |
| HPA + A | ATCATGAGTCCTGCTCGGA | MSAP primer pre-selective PCR |
| ECO + A | GACTGCGTACCAATTCA | MSAP primer pre-selective PCR |
| ECO + AC | GACTGCGTACCAATTCAC | MSAP primer selective PCR |
| HPA2ATG | ATCATGAGTCCTGCTCGGATG | MSAP primer selective PCR |
| MAO | CTCCTCCTCCTCRC | ISSR primer |
| OMAR | GAGGAGGAGGAGRC | ISSR primer |
| AW3 | GTGTGTGTGTGTRG | ISSR primer |
| 898 | CACACACACACARY | ISSR primer |
| BECKY | CACACACACACACAYC | ISSR primer |
| 844 | CTCTCTCTCTCTCTCTRC | ISSR primer |
| Methylation Levels | Proportion | |||
|---|---|---|---|---|
| AL 1st Cycle | AC 1st Cycle | AL 2nd Cycle | AC 2nd Cycle | |
| HpaII+/MspI+ (Unmethylated) | 0.47727 | 0.5292 | 0.3312 | 0.2305 |
| HpaII+/MspI− (Hemimethylated *) | 0.08442 | 0.1136 | 0.1818 | 0.2565 |
| HpaII−/MspI+ (Internal cytosine methylation *) | 0.24351 | 0.1753 | 0.3182 | 0.3539 |
| HpaII−/MspI− (Full methylation or absence of target **) | 0.19481 | 0.1818 | 0.1688 | 0.1591 |
| ISSR 1st Cycle | ISSR 2nd Cycle | HpaII+/MspI− 1st Cycle | HpaII−/MspI+ 1st Cycle | HpaII+/MspI− 2nd Cycle | HpaII−/MspI+ 2nd Cycle | |
|---|---|---|---|---|---|---|
| ISSR 1st cycle | 0 | |||||
| ISSR 2nd cycle | 0.283 | 0 | ||||
| HpaII+/MspI− 1st cycle | 0.359 | 0.367 | 0 | |||
| HpaII−/MspI+ 1st cycle | 0.042 | 0.133 | 0.089 | 0 | ||
| HpaII+/MspI− 2nd cycle | 0.460 | 0.408 | 0.689 | 0.123 | 0 | |
| HpaII−/MspI+ 2nd cycle | 0.233 | −0.0221 | 0.609 | 0.029 | 0.121 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anaya-Covarrubias, J.Y.; Larranaga, N.; Almaráz-Abarca, N.; Escoto-Delgadillo, M.; Rodríguez-Macías, R.; Torres-Morán, M.I. Hydrangea DNA Methylation Caused by pH Substrate Changes to Modify Sepal Colour is Detected by MSAP and ISSR Markers. Agronomy 2019, 9, 871. https://doi.org/10.3390/agronomy9120871
Anaya-Covarrubias JY, Larranaga N, Almaráz-Abarca N, Escoto-Delgadillo M, Rodríguez-Macías R, Torres-Morán MI. Hydrangea DNA Methylation Caused by pH Substrate Changes to Modify Sepal Colour is Detected by MSAP and ISSR Markers. Agronomy. 2019; 9(12):871. https://doi.org/10.3390/agronomy9120871
Chicago/Turabian StyleAnaya-Covarrubias, Julio Y., Nerea Larranaga, Norma Almaráz-Abarca, Martha Escoto-Delgadillo, Ramón Rodríguez-Macías, and Martha I. Torres-Morán. 2019. "Hydrangea DNA Methylation Caused by pH Substrate Changes to Modify Sepal Colour is Detected by MSAP and ISSR Markers" Agronomy 9, no. 12: 871. https://doi.org/10.3390/agronomy9120871
APA StyleAnaya-Covarrubias, J. Y., Larranaga, N., Almaráz-Abarca, N., Escoto-Delgadillo, M., Rodríguez-Macías, R., & Torres-Morán, M. I. (2019). Hydrangea DNA Methylation Caused by pH Substrate Changes to Modify Sepal Colour is Detected by MSAP and ISSR Markers. Agronomy, 9(12), 871. https://doi.org/10.3390/agronomy9120871