1. Introduction
Rapeseed (
Brassica naups L.) is a major oil crop globally. It is an allotetraploid (AACC, 2
n = 38) with a complex genome. Heterosis is used widely in rapeseed breeding, which has resulted in plants that are stronger and taller [
1] at the vegetative stage. Plant height is an important yield-related trait; taller plants are more prone to lodging, which results in decreased yield due to grain loss and spoilage, and also makes mechanized harvesting difficult [
2]. In some crops, such as wheat (
Triticum sp.) and rice (
Oryza sativa), dwarf or semi-dwarf plant types were introduced in the 1960s and 1970s, resulting in increased yields and the so-called “green revolution” [
2]. Therefore, breeding rapeseed for dwarfism or semi-dwarfism plant stature could increase lodging resistance, stabilizing production and facilitating mechanized harvesting.
Plant height in crops is generally considered to be a quantitative trait controlled by multiple loci and significantly affected by environmental factors [
3]. Li et al. [
4] used 472 rapeseed accessions to conduct a genome-wide association study (GWAS) and identified eight quantitative trait loci (QTLs) for plant height on chromosomes A03, A05, A07, and C07. Mei et al. [
5] used a segregating population of rapeseed with 145 F
2:3 lines, which was obtained by crossing a tall/later flowering line with a dwarf/early flowering line, and found seven QTLs related to plant height. These QTLs accounted for 8.5–28.6% of the phenotypic variation, and two of them were identified in both years. Shi et al. [
6] used a doubled-haploid population with 188 lines and found 21 QTLs related to plant height under phosphorus deficiency conditions, which decreased plant height.
In other instances where the dwarfism trait was obtained from mutation, dwarfism was found to be controlled by one major gene. Wang et al. [
7] fine-mapped an ethyl methane sulfonate (EMS) mutagenized line with a dominant locus controlling dwarfism to linkage group A09 using single-nucleotide polymorphism (SNP) markers. Liu et al. [
8] investigated a semi-dwarf mutant phenotype and found that it was caused by a missense mutation in a DELLA protein. Wang et al. [
9] and Li et al. [
10] studied a dwarf rapeseed mutant ‘NDF-1′, in which dwarfism is controlled by a major gene with a mainly additive and non-significant dominance effect, and revealed that three bases mutated in the pyrimidine box (P-box) of the
Gibberellin Insensitive Dwarf 1 (
BnGID1) promoter changed its expression. To date, many height-related loci/genes involved in hormone biosynthesis or signal transduction in crops have been identified, especially genes involved in gibberellin (GA) signal transduction:
Reduced Height (
Rht) [
11] in wheat, orthologs of
Arabidopsis thaliana GIBBERELLIN INSENSITIVE (
GAI);
Semidwarfing Gene (
sd1) in rice, which encodes a mutant enzyme involved in gibberellin synthesis [
12,
13,
14]; and
ZmGA3ox2 in maize (
Zea mays) [
15], an ortholog of
OsGA3ox2, which encodes a GA3 β-hydroxylase.
Bulked segregant analysis (BSA) [
16] was traditionally combined with markers such as sequence-related amplified polymorphism (SRAP) [
17], simple sequence repeats (SSR) [
18], and random amplified polymorphic DNA (RAPD) [
19] to locate the trait-related regions on chromosomes. More recently, next-generation sequencing technologies, such as whole-genome re-sequencing (WGR), have been commonly used to identify key loci related to agronomic traits. The sequencing of the
B. napus genome [
20] facilitated great advances in the identification of key genes related to branch angle [
21], seed weight [
22], and petal color [
23], among others, in rapeseed.
In this study, a dwarfism trait in the dwarf rapeseed line YA2016-12, which was first discovered in B. napus × B. rapa hybrid offspring, was crossed with the tall rapeseed line G184-189. Using plants from the F2 population, we constructed two DNA pools (tall pool and dwarf pool) by the BSA method and performed WGR. A sliding window analysis was used to identify the trait-associated regions from the SNP and insertion/deletion (InDel) data. Genes within the associated region were then aligned with known plant height-related genes in Arabidopsis, and their annotations were also analyzed. Finally, we selected nine potential candidate genes for this dwarfism trait in rapeseed.
4. Discussion
Heterosis is extensively utilized in rapeseed breeding in China. Heterosis has resulted in taller rapeseed plants with more biomass, but this increase in plant height has had the undesired effect of enhanced risk of lodging. Lodging can reduce the seed yield [
36] and significantly reduce the oil content of the seed, as well as the seed number per pod, seed weight, and other yield-related traits. Moreover, lodging increases the difficulty of mechanized harvesting and decreases the harvesting index [
37]. Therefore, dwarf or semi-dwarf rapeseed varieties are considered ideal to increase and stabilize the yield and harvesting index—especially via use of the dwarfism genes produced by mutation, as these genes are known to confer lodging resistance and high yield [
9]. The ideal height for mechanized harvesting is semi-dwarf.
The dwarfism trait investigated in this study was originally discovered in B. napus × B. rapa hybrid offspring. The homozygous line YA2016-12 was obtained after self-pollination for several generations. At the seedling stage, unlike the other plants, the leaves of YA2016-12 were dark green and wrinkled. When the other genotypes (including G184-189) started bolting, YA2016-12 had only procumbent leaves and a shorter stem, showing obviously stunted growth. At the final-flowering stage, YA2016-12 was approximately 50% of the height of G184-189, as well as some other rapeseed materials observed in this study. These phenomena could be due to an endogenous hormone deficiency in the plant.
G184-189 has a tall stature, which made it suitable as the other parent for a cross to study the segregation of the dwarfism trait. All of the F
1 individuals showed a semi-dwarf plant stature, which implied that the trait was semi-dominant. Variations in plant height were observed in the F
2 population: the number of tall, semi-dwarf, and dwarf individuals satisfied the 1:2:1 segregation ratio, further supporting that the dwarfism trait is controlled by a single semi-dominant allele. Most reported natural dwarfism traits are controlled by multiple minor loci, which reflect continuous variation in a population [
30,
38,
39,
40]. However, other dwarfism traits were found to be controlled by a single allele, especially those discovered from artificial mutation. Foisset et al. [
1] used EMS to mutagenize rapeseed and discovered a dwarf line controlled by a recessive allele (bzh/bzh). Zeng et al. [
41] identified a recessive gene in a dwarf mutant (
bnaC.dwf) mutagenized by EMS that was responsible for the dwarfism.
The correlation analysis indicated that plant height was significantly positively correlated with branch segment length (BSL) but not with main inflorescence length or first primary branch height (FPBH), i.e., the dwarfism trait in YA2016-12 is mainly due to the shortened BSL. Here, one advantage of a semi-dominant dwarfism allele was that the hybrid (F1) plant was semi-dwarf, an ideal height for mechanized harvesting. A too low plant height or FPBH would hinder mechanized harvesting; instead, with this dwarfism trait, the BSL is shorter, which results in a more compact plant type. In addition, because this trait is conferred by a single allele, it would be easier to use in breeding.
BSA, also known as linkage disequilibrium (LD) mapping, provided a convenient but effective strategy for identifying molecular markers linked to target genes by genotyping bulked DNA samples from two populations with distinct phenotypes [
16]. In this study, instead of traditional markers like SRAP [
17], SSRs [
18], or RAPD [
19], we used SNPs/InDels detected by WGR. SNPs occur at high frequencies in plant genomes, offering larger quantities of data, and are suitable for the construction of high-density genetic linkage maps for crops with large genomes [
42,
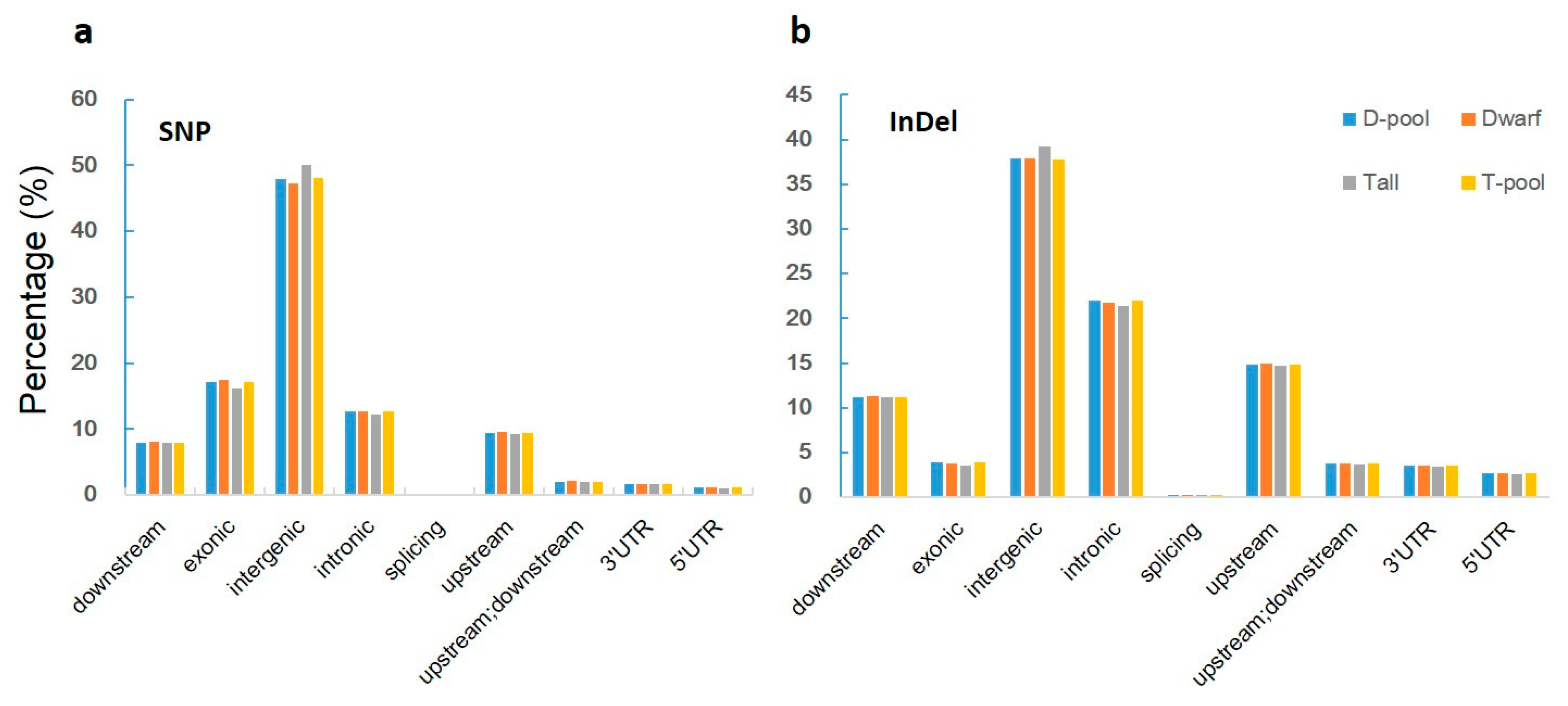
43]. Moreover, compared to reduced-representation sequencing (RRS), WGR produced more data and covered more regions on the reference genome with more sequencing depth. We obtained 225 Gbp of raw data, approximately 339 M HQ clean reads for each sample after filtration, with the average sequencing depth of 37.5×. After screening, 897,550 SNPs qualified for association analysis by the SNP-index method, which provided higher density.
According to the Δ(SNP-index) data, the peak regions above the threshold line revealed a candidate association region on ChrA03 with a length of 12.4 Mbp. Although this region was larger than expected, its segregation fitted the 1:2:1 ratio. Li et al. [
4] also identified a locus on ChrA03, but its position was different from the one identified here. Further analysis revealed that there are 1225 genes within our candidate association region and 811 of them contained sequence variations between YA2016-12 and G184-189.
We aligned these 811 genes to the known 143 plant height-related genes in
Arabidopsis gathered by Shi et al. [
30] and selected two important genes:
BnaA03g31770D (ortholog of AT3G11540) and
BnaA03g37960D (ortholog of AT5G35750). AT3G11540 [
34], known as
SPINDLY (
SPY) in
Arabidopsis, encodes an O-linked N-acetylglucosamine transferase, which is a negative regulator of gibberellin signaling and activates DELLA proteins [
44,
45,
46,
47], which are critical in the gibberellin signaling pathway. The
spy-4 mutant in
Arabidopsis has decreased plant height compared to the wild type [
47]. AT5G35750, known as
Arabidopsis Histidine Kinase 2 (
AHK2), encodes a histidine kinase, which acts as a cytokinin receptor. In
ahk2 ahk3 double mutants of
Arabidopsis, the shoot growth is reduced [
35,
48].
BnaA03g31770D and
BnaA03g37960D shared 91% and 76% amino acid identification with AT3G11540 and AT5G35750, respectively; therefore, they are presumed to perform similar functions and are considered important candidate genes.
YA2016-12 resembled some reported dwarf rapeseed plants resulting, to some extent, from GA deficiency, such as wrinkled, dark green leaves and sclerotiniose susceptibility; therefore, using the gene annotations, especially GO annotation and the KEGG pathway, we found seven additional important genes: (1)
BnaA03g24740D was described as a suppressor of the G2 allele of skp1. Skp1 is a subunit of the Skp1/cullin/F-box (SCF) protein complex, which interacts with and ubiquitylates DELLA proteins so that they can be degraded by the 26S proteasome. DELLA proteins are vital repressor proteins in the gibberellin signaling pathway [
8,
49,
50]. (2)
BnaA03g40550D was annotated to respond to gibberellin (GO: 0009739), cellular response to gibberellin stimulus (GO: 0071370), and the gibberellin-mediated signaling pathway (GO: 0010476). (3)
BnaA03g26120D was described as a cullin protein, which is another subunit of the SCF protein complex, combined with its annotations as ubiquitin ligase complex (GO: 0000151) and cullin-RING ubiquitin ligase complex (GO: 0031461). (4)
BnaA03g35130D is involved in the cytokinin metabolic process (GO: 0009690). (5)
BnaA03g42350D is related to plant hormone signal transduction (ko04075) and response to cytokinin (GO: 0009735). (6)
BnaA03g25610D and (7)
BnaA03g39850D are both related to the cullin-RING ubiquitin ligase complex (GO: 0031461). Although the last four genes need more investigation to support their role in plant height, they are also considered potential candidate genes based on our current information.