Phylogenomic Analysis of the PEBP Gene Family from Kalanchoë
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of PEBP Sequences
2.2. Phylogenetic Analysis
2.3. Protein Characterization
2.4. Gene Structure Analysis
2.5. Gene Duplication Analysis
3. Results
3.1. Identification of PEBP Sequences
3.2. Comparative Phylogenetic Analysis
3.3. Kalanchoë Protein Characterization
3.4. Gene Structure
3.5. Gene Duplication Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zheng, X.M.; Wu, F.Q.; Zhang, X.; Lin, Q.B.; Wang, J.; Guo, X.P.; Lei, C.L.; Cheng, Z.J.; Zou, C.; Wan, J.M. Evolution of the PEBP gene family and selective signature on FT-like clade. J. Syst. Evol. 2016, 54, 502–510. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Yang, K.Z.; Wei, X.X.; Wang, X.Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Zeevaart, J.A. Florigen Coming of Age after 70 Years. Plant Cell 2006, 18, 1783–1789. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.-I.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef]
- Gao, J.; Huang, B.-H.; Wan, Y.-T.; Chang, J.; Li, J.-Q.; Liao, P.-C. Functional divergence and intron variability during evolution of angiosperm TERMINAL FLOWER 1 (TFL1) genes. Sci. Rep. 2017, 7, 14830. [Google Scholar] [CrossRef] [PubMed]
- Rinne, P.L.; Welling, A.; Vahala, J.; Ripel, L.; Ruonala, R.; Kangasjärvi, J.; van der Schoot, C. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1, 3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 2011, 23, 130–146. [Google Scholar] [CrossRef]
- Koskela, E.A.; Mouhu, K.; Albani, M.C.; Kurokura, T.; Rantanen, M.; Sargent, D.J.; Battey, N.H.; Coupland, G.; Elomaa, P.; Hytönen, T. Mutation in TERMINAL FLOWER 1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 2012, 159, 1043–1054. [Google Scholar] [CrossRef]
- Randoux, M.; Davière, J.M.; Jeauffre, J.; Thouroude, T.; Pierre, S.; Toualbia, Y.; Perrotte, J.; Reynoird, J.P.; Jammes, M.J.; Oyant, H.S. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol. 2014, 202, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Narumi, T.; Oda, A.; Nakano, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 2013, 110, 17137–17142. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Blackman, B.K.; Strasburg, J.L.; Raduski, A.R.; Michaels, S.D.; Rieseberg, L.H. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 2010, 20, 629–635. [Google Scholar] [CrossRef]
- Pin, P.A.; Benlloch, R.; Bonnet, D.; Wremerth-Weich, E.; Kraft, T.; Gielen, J.J.; Nilsson, O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 2010, 330, 1397–1400. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Adams, J.P.; Kim, H.; No, K.; Ma, C.; Strauss, S.H.; Drnevich, J.; Vandervelde, L.; Ellis, J.D.; Rice, B.M. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 2011, 108, 10756–10761. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Baldwin, S.; Kenel, F.; McCallum, J.; Macknight, R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 2013, 4, 2884. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Nakazono, K.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef]
- Mackenzie, K.K.; Lütken, H.; Coelho, L.L.; Kaaber, M.D.; Hegelund, J.N.; Müller, R. Kalanchoë. In Ornamental Crops; Handbook of Plant Breeding; Van Huylenbroeck, J., Ed.; Springer: Cham, Switzerland, 2018; Volume 11, pp. 459–479. [Google Scholar]
- Kuligowska, K.; Lütken, H.; Christensen, B.; Müller, R. Quantitative and qualitative characterization of novel features of Kalanchoë interspecific hybrids. Euphytica 2015, 205, 927–940. [Google Scholar] [CrossRef]
- Currey, C.; Erwin, J. Variation among Kalanchoe species in their flowering responses to photoperiod and short-day cycle number. J. Hortic. Sci. Biotechnol. 2010, 85, 350–355. [Google Scholar] [CrossRef]
- Coelho, L.L.; Mackenzie, K.K.; Lütken, H.; Müller, R. Effect of cold night temperature on flowering of Kalanchoë species. Acta Sci. Pol. Hortorum Cultus 2018, 17, 121–125. [Google Scholar] [CrossRef]
- Coelho, L.L.; Fkiara, A.; Mackenzie, K.K.; Müller, R.; Lütken, H. Exogenous Application of Gibberellic Acid Improves Flowering in Kalanchoë. HortSciience 2018, 53, 342–346. [Google Scholar] [CrossRef]
- Sharma, G.J. Flower formation in Kalanchoe velutina induced by low night temperature. Southwest. Nat. 1973, 18, 331–334. [Google Scholar] [CrossRef]
- Mortensen, L.M. The effect of wide-range photosynthetic active radiations on photosynthesis, growth and flowering of Rosa sp. and Kalanchoe blossfeldiana. Am. J. Plant Sci. 2014, 5, 1489. [Google Scholar] [CrossRef]
- Wadhi, M.; Ram, H.M. Shortening the juvenile phase for flowering in Kalanchoe pinnata Pers. Planta 1967, 73, 28–36. [Google Scholar] [CrossRef]
- Zimmer, K. Untersuchungen zur Blühinduktion bei Kalanchoë marmorata Baker. KuaS 1996, 47, 188–191. [Google Scholar]
- Yang, X.; Hu, R.; Yin, H.; Jenkins, J.; Shu, S.; Tang, H.; Liu, D.; Weighill, D.A.; Yim, W.C.; Ha, J. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017, 8, 1899. [Google Scholar] [CrossRef] [PubMed]
- Winter, K.; Holtum, J.A. Facultative crassulacean acid metabolism (CAM) plants: Powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 2014, 65, 3425–3441. [Google Scholar] [CrossRef]
- Kolodziejczyk-Czepas, J.; Sieradzka, M.; Moniuszko-Szajwaj, B.; Pecio, Ł.; Ponczek, M.B.; Nowak, P.; Stochmal, A. Bufadienolides from Kalanchoe daigremontiana as thrombin inhibitors—In vitro and in silico study. Int. J. Biol. Macromol. 2017, 99, 141–150. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Schäffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct 2012, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–247. [Google Scholar] [CrossRef]
- Li, C.; Dubcovsky, J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008, 55, 543–554. [Google Scholar] [CrossRef]
- Leeggangers, H.A.; Rosilio-Brami, T.; Bigas-Nadal, J.; Rubin, N.; van Dijk, A.D.; Nunez de Caceres Gonzalez, F.F.; Saadon-Shitrit, S.; Nijveen, H.; Hilhorst, H.W.; Immink, R.G. Tulipa gesneriana and Lilium longiflorum PEBP genes and their putative roles in flowering time control. Plant Cell Physiol. 2018, 59, 90–106. [Google Scholar] [CrossRef]
- Mathieu, J.; Warthmann, N.; Küttner, F.; Schmid, M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 2007, 17, 1055–1060. [Google Scholar] [CrossRef]
- Li, D.; Lu, F.; Zhu, G.; Sun, Y.; Liu, H.; Liu, J.; Wang, Z. Molecular characterization and functional analysis of a Flowering locus T homolog gene from a Phalaenopsis orchid. Genet. Mol. Res. 2014, 13, 5982–5994. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Su, S.; Ma, L.; Wen, Y.; Wang, X. Molecular cloning and functional characterization of CoFT1, a homolog of FLOWERING LOCUS T (FT) from Camellia oleifera. Gene 2017, 626, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Zhang, C.; Zhang, H.; Li, X.; Wen, C.; Liang, Y. Molecular cloning, expression analysis, and subcellular localization of FLOWERING LOCUS T (FT) in carrot (Daucus carota L.). Mol. Breed. 2017, 37, 149. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 2007, 19, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Chu, C.-Y. The flower development and photoperiodism of native Kalanchoe spp. in Taiwan. Sci. Hortic. 2012, 146, 59–64. [Google Scholar] [CrossRef]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.-I. TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, R.K.; Han, J.S.H.; Vijayakumar, H.; Subramani, B.; Thamilarasan, S.K.; Park, J.-I.; Nou, I.-S. Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa. Molecules 2016, 21, 217. [Google Scholar] [CrossRef]
- Kotoda, N.; Hayashi, H.; Suzuki, M.; Igarashi, M.; Hatsuyama, Y.; Kidou, S.-i.; Igasaki, T.; Nishiguchi, M.; FYano, K.; Shimizu, T. Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus× domestica Borkh.). Plant Cell Physiol. 2010, 51, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Böhlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef]
- Mimida, N.; Kotoda, N.; Ueda, T.; Igarashi, M.; Hatsuyama, Y.; Iwanami, H.; Moriya, S.; Abe, K. Four TFL1/CEN-Like Genes on Distinct Linkage Groups Show Different Expression Patterns to Regulate Vegetative and Reproductive Development in Apple (Malus × domestica Borkh.). Plant Cell Physiol. 2009, 50, 394–412. [Google Scholar] [CrossRef]
- Mohamed, R.; Wang, C.-T.; Ma, C.; Shevchenko, O.; Dye, S.J.; Puzey, J.R.; Etherington, E.; Sheng, X.; Meilan, R.; Strauss, S.H.; et al. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 2010, 62, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Koskela, E.A.; Sønsteby, A.; Flachowsky, H.; Heide, O.M.; Hanke, M.V.; Elomaa, P.; Hytönen, T. TERMINAL FLOWER 1 is a breeding target for a novel everbearing trait and tailored flowering responses in cultivated strawberry (Fragaria× ananassa Duch.). Plant Biotechnol. J. 2016, 14, 1852–1861. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, W. Flower inhibition in Kalanchoe blossfeldiana. Bioassay of an endogenous long-day inhibitor and inhibition by (±) abscisic acid and xanthoxin. Planta 1972, 103, 18–23. [Google Scholar] [CrossRef]
- Schwabe, W.W. Evidence for a flowering inhibitor produced in long lays in Kalanchoe blossfeldiana. Ann. Bot. 1956, 20, 1–14. [Google Scholar] [CrossRef]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2011, 13, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Claros, M.G.; Bautista, R.; Guerrero-Fernández, D.; Benzerki, H.; Seoane, P.; Fernández-Pozo, N. Why Assembling Plant Genome Sequences Is So Challenging. Biology 2012, 1, 439–459. [Google Scholar] [CrossRef]
- Drabešová, J.; Černá, L.; Mašterová, H.; Koloušková, P.; Potocký, M.; Štorchová, H. The evolution of the FT/TFL1 genes in Amaranthaceae and their expression patterns in the course of vegetative growth and flowering in Chenopodium rubrum. G3: Genes Genomes Genet. 2016, 6, 3065–3076. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Koyanagi, K.O.; Itoh, T. Highly diversified molecular evolution of downstream transcription start sites in rice and Arabidopsis. Plant Physiol. 2009, 149, 1316–1324. [Google Scholar] [CrossRef]
- Tsaftaris, A.; Pasentsis, K.; Kalivas, A.; Michailidou, S.; Madesis, P.; Argiriou, A. Isolation of a CENTRORADIALIS/TERMINAL FLOWER1 homolog in saffron (Crocus sativus L.): Characterization and expression analysis. Mol. Biol. Rep. 2012, 39, 7899–7910. [Google Scholar] [CrossRef] [PubMed]
- McGarry, R.C.; Ayre, B.G. Manipulating plant architecture with members of the CETS gene family. Plant Sci. 2012, 188, 71–81. [Google Scholar] [CrossRef]
- Voogd, C.; Brian, L.A.; Wang, T.; Allan, A.C.; Varkonyi-Gasic, E. Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 2017, 68, 1479–1547. [Google Scholar] [CrossRef] [PubMed]
- Noy-Porat, T.; Cohen, D.; Mathew, D.; Eshel, A.; Kamenetsky, R.; Flaishman, M.A. Turned on by heat: Differential expression of FT and LFY-like genes in Narcissus tazetta during floral transition. J. Exp. Bot. 2013, 64, 3273–3284. [Google Scholar] [CrossRef]
- Wang, R.; Albani, M.C.; Vincent, C.; Bergonzi, S.; Luan, M.; Bai, Y.; Kiefer, C.; Castillo, R.; Coupland, G. Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 2011, 23, 1307–1321. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.S.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012, 69, 116–125. [Google Scholar] [CrossRef]

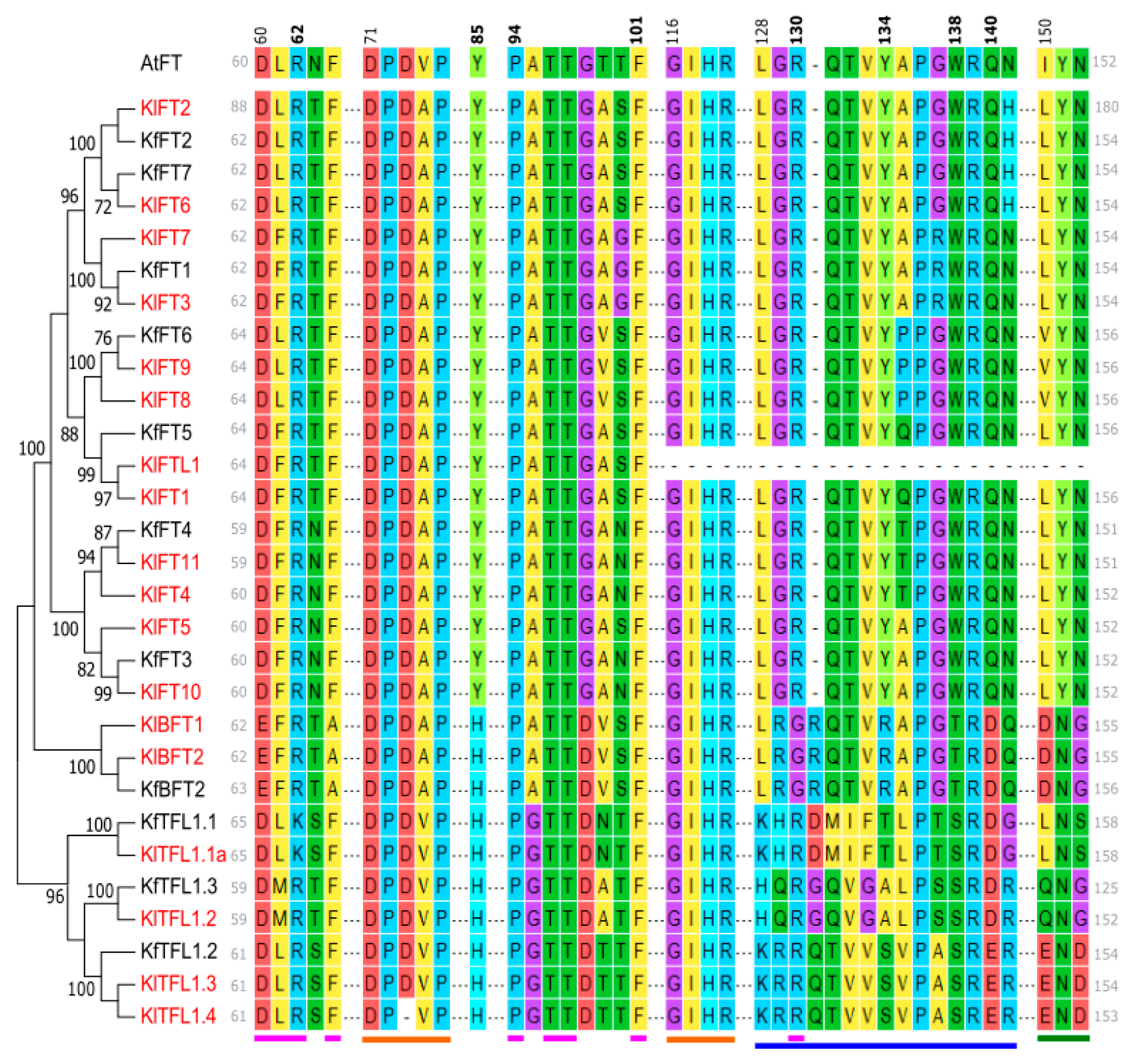
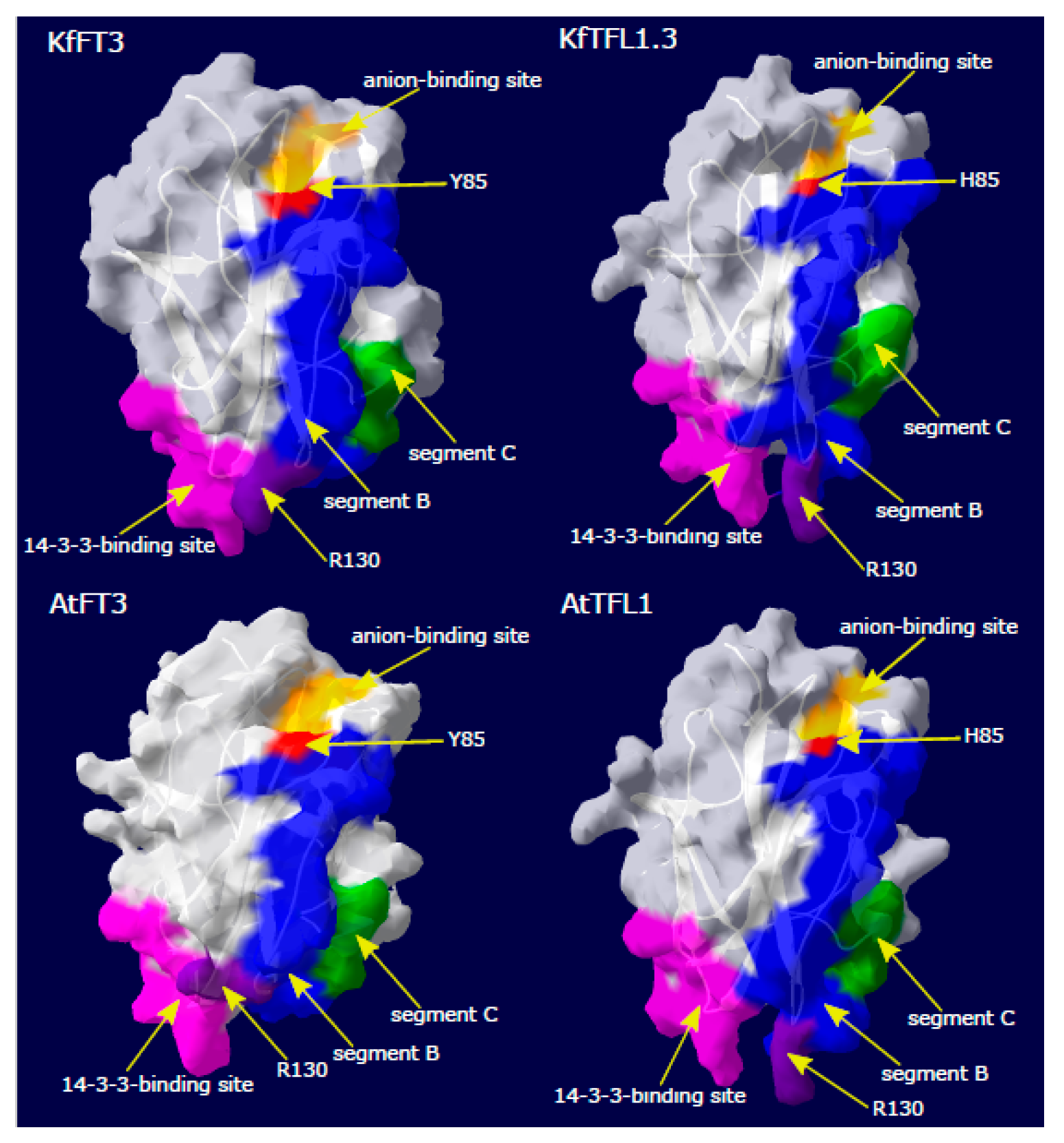

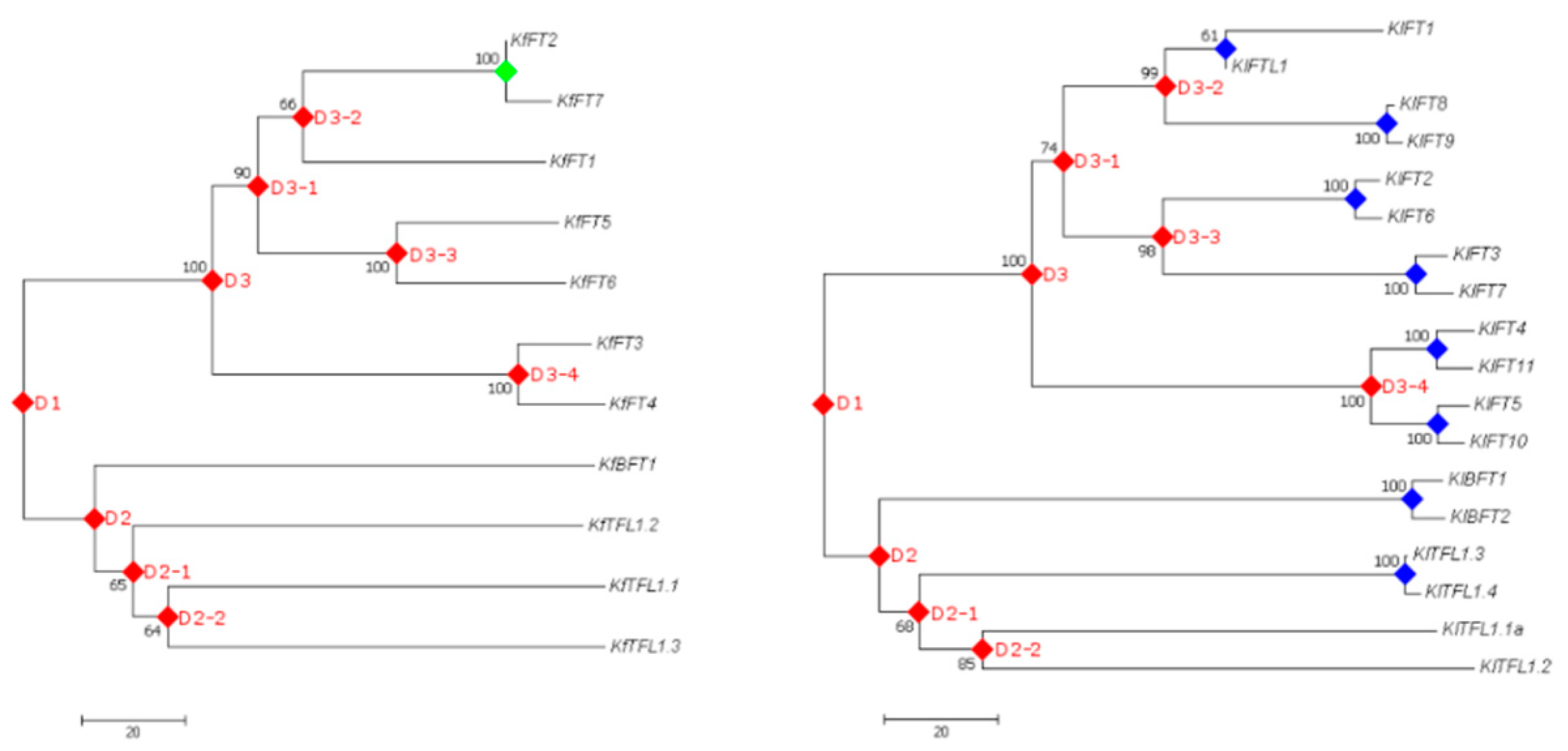

| No. | Accession IDs | Gene Name | No. of Transcripts | Protein Length (aa) | Molecular Weight (kDa) | pI | Instability Index |
|---|---|---|---|---|---|---|---|
| 1 | Kaladp0007s0057 | KfFT1 | 1 | 177 | 20.21 | 8.88 | 41.81; U 1 |
| 2 | Kaladp0031s0098 | KfFT2 | 1 | 181 | 20.31 | 8.75 | 44.11; U |
| 3 | Kaladp0055s0469 | KfFT3 | 1 | 178 | 20.21 | 9.09 | 38.93; S 2 |
| 4 | Kaladp0055s0470 | KfFT4 | 1 | 176 | 19.88 | 7.72 | 38.51, S |
| 5 | Kaladp0060s0252 | KfFT5 | 1 | 180 | 20.28 | 8.39 | 38.23; S |
| 6 | Kaladp0099s0141 | KfFT6 | 1 | 181 | 20.41 | 7.73 | 51.05; U |
| 7 | Kaladp1016s0006 | KfFT7 | 1 | 177 | 19.97 | 8.76 | 44.12; U |
| 8 | Kalax.0016s0040 | KlFT1 | 1 | 180 | 20.27 | 8.39 | 38.23; S |
| 9 | Kalax.0052s0013 | KlFT2 | 1 | 203 | 22.90 | 7.47 | 44.14; U |
| 10 | Kalax.0084s0028 | KlFT3 | 1 | 177 | 20.21 | 8.88 | 41.81; U |
| 11 | Kalax.0100s0001 | KlFT4 | 1 | 177 | 20.02 | 7.72 | 37.86; S |
| 12 | Kalax.0100s0002 | KlFT5 | 1 | 177 | 19.97 | 9.27 | 41.29; U |
| 13 | Kalax.0114s0057 | KlFT6 | 1 | 177 | 19.96 | 8.76 | 43.51; U |
| 14 | Kalax.0189s0069 | KlFT7 | 1 | 177 | 20.25 | 8.88 | 42.24; U |
| 15 | Kalax.0399s0008 | KlFT8 | 1 | 181 | 20.39 | 7.73 | 50.63; U |
| 16 | Kalax.0435s0026 | KlFT9 | 1 | 181 | 20.41 | 7.73 | 51.05; U |
| 17 | Kalax.0644s0009 | KlFT10 | 1 | 178 | 20.21 | 9.09 | 38.93; S |
| 18 | Kalax.0798s0001 | KlFT11 | 1 | 176 | 19.88 | 7.72 | 38.51; S |
| 19 | Kalax.0508s0010 | KlFTL1 | 1 | 112 | 12.28 | 5.24 | 32.41; S |
| 20 | Kaladp0011s0878 | KfTFL1.1 | 1 | 179 | 20.39 | 8.35 | 33.13; S |
| 21 | Kaladp0093s0128 | KfTFL1.2 | 1 | 175 | 19.89 | 8.73 | 54.72; U |
| 22 | Kaladp0095s0167 | KfTFL1.3 | 1 | 173 | 19.39 | 7.96 | 46.78; U |
| 23 | Kalax.0010s0108 | KlTFL1.1 | 2 | 179 | 20.36 | 8.35 | 27.75; U |
| 145 | 15.89 | 5.62 | 36.31; S | ||||
| 24 | Kalax.0018s0096 | KlTFL1.2 | 1 | 173 | 19.39 | 7.96 | 46.78; U |
| 25 | Kalax.0073s0028 | KlTFL1.3 | 1 | 175 | 19.88 | 8.73 | 54.72; U |
| 26 | Kalax.0154s0031 | KlTFL1.4 | 1 | 175 | 19.88 | 8.73 | 54.72; U |
| 27 | Kaladp0096s0031 | KfBFT1 | 1 | 177 | 20.06 | 9.16 | 50.66; U |
| 28 | Kalax.0003s0185 | KlBFT1 | 1 | 176 | 19.88 | 7.93 | 49.42; U |
| 29 | Kalax.0635s0001 | KlBFT2 | 1 | 176 | 19.85 | 9.16 | 48.94; U |
| Gene 1 | Gene 2 | CDS Identity | Ks 1 | Mya 2 | Purifying Selection 3 |
|---|---|---|---|---|---|
| KfFT1 | KfFT2 | 83.2 | 0.461 | 15.4 | yes |
| KfFT1 | KfFT7 | 82.5 | 0.480 | 16.0 | yes |
| KfFT2 | KfFT7 | 99.0 | 0.027 | 0.9 | no |
| KfFT3 | KfFT4 | 94.2 | 0.166 | 5.5 | yes |
| KfFT5 | KfFT6 | 88.2 | 0.222 | 7.4 | yes |
| KfTFL1.1 | KfTFL1.2 | 65.3 | 0.727 | 24.2 | yes |
| KfTFL1.1 | KfTFL1.3 | 67.6 | 0.650 | 21.7 | yes |
| KfTFL1.2 | KfTFL1.3 | 66.7 | 0.763 | 25.4 | yes |
| KfTFL1.1 | KfBFT1 | 64.8 | 0.614 | 20.5 | yes |
| KfTFL1.2 | KfBFT1 | 62.1 | 0.811 | 27.0 | yes |
| KfTFL1.3 | KfBFT1 | 62.6 | 0.644 | 21.5 | yes |
| KlFT15 | KlFTL1 | 95.0 | 0.063 | 2.1 | no |
| KlFT2 | KlFT6 | 98.3 | 0.053 | 1.8 | no |
| KlFT3 | KlFT7 | 97.8 | 0.095 | 3.2 | yes |
| KlFT4 | KlFT11 | 97.6 | 0.053 | 1.8 | yes |
| KlFT5 | KlFT10 | 98.1 | 0.026 | 0.9 | no |
| KlFT8 | KlFT9 | 99.3 | 0.026 | 0.9 | yes |
| KlBFT1 | KlBFT2 | 97.9 | 0.096 | 3.2 | yes |
| KlTFL1.1a | KlTFL1.2 | 68.2 | 0.621 | 20.7 | yes |
| KlTFL1.3 | KlTFL1.4 | 99.4 | 0.041 | 1.4 | yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuligowska Mackenzie, K.; Lopes Coelho, L.; Lütken, H.; Müller, R. Phylogenomic Analysis of the PEBP Gene Family from Kalanchoë. Agronomy 2019, 9, 171. https://doi.org/10.3390/agronomy9040171
Kuligowska Mackenzie K, Lopes Coelho L, Lütken H, Müller R. Phylogenomic Analysis of the PEBP Gene Family from Kalanchoë. Agronomy. 2019; 9(4):171. https://doi.org/10.3390/agronomy9040171
Chicago/Turabian StyleKuligowska Mackenzie, Kathryn, Lívia Lopes Coelho, Henrik Lütken, and Renate Müller. 2019. "Phylogenomic Analysis of the PEBP Gene Family from Kalanchoë" Agronomy 9, no. 4: 171. https://doi.org/10.3390/agronomy9040171
APA StyleKuligowska Mackenzie, K., Lopes Coelho, L., Lütken, H., & Müller, R. (2019). Phylogenomic Analysis of the PEBP Gene Family from Kalanchoë. Agronomy, 9(4), 171. https://doi.org/10.3390/agronomy9040171





