Abstract
Realizing the yield potential of crop plants in the presence of shifting pathogen populations, soil quality, rainfall, and other agro-environmental variables remains a challenge for growers and breeders worldwide. In this review, we discuss current approaches for combatting the soilborne phytopathogenic nematodes, Pratylenchus and Heterodera of wheat and barley, and Meloidogyne graminicola Golden and Birchfield, 1965 of rice. The necrotrophic fungal pathogens, Rhizoctonia solani Kühn 1858 AG-8 and Fusarium spp. of wheat and barley, also are discussed. These pathogens constitute major causes of yield loss in small-grain cereals of the Pacific Northwest, USA and throughout the world. Current topics include new sources of genetic resistance, molecular leads from whole genome sequencing and genome-wide patterns of hosts, nematode or fungal gene expression during root-pathogen interactions, host-induced gene silencing, and building a molecular toolbox of genes and regulatory sequences for deployment of resistance genes. In conclusion, improvement of wheat, barley, and rice will require multiple approaches.
Keywords:
genetic resistance; root disease; wheat; rice; barley; nematodes; transcriptomics; HIGS; DORN1; nematode-inducible promoters 1. Introduction
Grain crops including rice (Oryza sativa L.), common or bread wheat (Triticum aestivum L.), durum wheat (Triticum turgidum L. ssp. durum), and corn (Zea mays L.) are preferred staple foods throughout many parts of the world. Yield enhancement of these crops poses a challenge for growers and breeders who seek to maintain production rates in accordance with consumer needs and in the face of environmental challenges. For instance, an approximate 1% increase in wheat yields per year is estimated to meet consumer demands for the next 5 to 15 years, whereas yield reductions of 6%–13% are estimated in the face of temperature shifts, and development of new adapted cultivars is accomplished in a span of 10 or more years [1,2]. In this review, we focus on prospects for improvement of wheat, barley (Hordeum vulgare L.), and rice with respect to resistance against the soilborne pathogens Rhizoctonia solani, Fusarium culmorum (W.G. Smith) Sacc., Pratylenchus neglectus (Rensch, 1924) Filijev and Stuurmans Stekhoven, 1941, P. thornei Scher and Allen, 1953, Hereodera avenae Wollenweber, 1924, and Meloidogyne graminicola. These pathogens are problems in the dryland production regions of the Pacific Northwest, USA (PNW), Australia, Europe, China, Southeast Asia, India, and other parts of the world [2,3,4,5,6,7]. Furthermore, a body of knowledge is accumulating about the cellular and molecular interactions between these hosts and pathogens that are helping to elucidate the basis of susceptibility and resistance, e.g., References [8,9,10,11,12,13].
Wheat and rice are major sources of nutrition for 40% and over 50% of the world’s population, respectively [14,15]. Wheat, rice, and barley provide about 56% of the caloric content and 50% of required protein for the human diet [16]. In 2017, 16.4 million tons of barley were produced in 107 countries, including Russia, the Middle East, Europe, Australia, UK, North, Central and South America, and Asia [17]. Annual yield losses due to diseases have been estimated at 10%–15% [16,18]. Plant-parasitic nematodes are thought to cause $100–800 billion in losses per year worldwide [19,20,21], with $157 billion in losses due to root-knot nematodes of the genus Meloidogyne [10]. Over $100 billion is attributed to cereal cyst nematodes (Heterodera spp.), with losses ranging from 20% to 90%, depending upon location and year [20,22,23]. Meloidogyne graminicola of rice has resulted in yield losses of up to 80% in Southeast Asia [11,24]. This nematode is also problematic in Latin America, Asia, Southeast Asia, and India where flooding is restricted in the interest of water conservation [4,6]. In the PNW, the root lesion nematodes, Pratylenchus neglectus and P. thornei, contribute up to 35% and 60% yield reductions in wheat, with yearly chronic losses of 5% ($51 million) [25,26]. The cereal cyst nematodes Heterodera avenae and H. filipjevi (Madzhidov, 1981) Stelter, 1984 also are problems on PNW wheat and barley [5].
Solutions for nematode resistance likely will depend on the host–pathogen interaction, particularly on plant genotype and pathogen-specific aspects of the infection processes. Root-knot nematodes enter the root at the zone of elongation, and migrate intercellularly to the vascular cylinder, where they induce feeding sites composed of giant cells [9,13]. Female nematodes deposit eggs onto the root surface in a gelatinous matrix. The nematodes also stimulate plant cell division resulting in an external organ, called the knot or gall. While little cellular damage occurs during the establishment of the feeding site, the nematode draws nutrients from the host, alters host root morphology, and generates progeny. In contrast, cereal cyst nematodes intracellularly migrate to the vascular cylinder, and induce the fusion of multiple host cells to form a multinucleate feeding organ called the syncytium [10,13,27]. The root lesion nematodes also intracellularly migrate through the root, causing significant host cell breakage. However, these nematodes feed from root cortical cells without disrupting the lignin and suberin layers of the vascular cylinder [13,28].
Wheat and barley are generally susceptible to the soilborne fungal pathogens Fusarium culmorum, one of several Fusarium spp. associated with Fusarium crown, stem, and root rot, and R. solani AG-8, the causal agent of Rhizoctonia root rot and bare patch. Fusarium culmorum is a yield-reducing pathogen of durum wheat and bread wheat in warm dry climates, and has been reported in Europe, Australia, Canada, North America, Africa, and Western Asia [8,29,30]. Yield losses due to Fusarium-derived root and stem diseases were reported to be about 25% in Australia and 31%–65% in North America [25,31]. In the dryland wheat-based production systems of the PNW, chronic infection by soilborne pathogens causes 10%–15% annual yield losses [32], or about $101 million yearly for wheat in Washington State [33]. Fusarium culmorum and R. solani anastomosis group 8 (AG-8) were detected in 36% and 50% of surveyed fields in Washington and Oregon, respectively [25,34]. Both of these pathogens are harbored in stubble resulting from direct seeding [32,35].
Management options for soilborne diseases of the small-grain cereals include rotation, application of appropriate amounts of fertilizers, fungicide seed treatments, tillage, and use of partially-resistant crop genotypes, when available. There are excellent articles and extension bulletins describing widely-used management practices for the above mentioned soilborne diseases that are not included in this review. Instead, we consider how recent biotechnological advances can help to identify new genes for disease resistance, and how the genes can be deployed as traits in the field. In this review, we focus on new genetic loci for resistance or tolerance from association mapping and genome-wide association studies (GWAS); the roles of genomics and transcriptomics in identifying specific candidate resistance and parasitism genes; advances in understanding the mechanisms by which pathogen proteins act in a susceptible host to cause the disease condition; and the difficulties of using candidate resistance genes from non-cereals to find and test the corresponding genes in small-grain cereals. Finally, we recognize the utility of a molecular toolbox of wheat, barley, and rice genes and regulatory sequences for generating cis-genic germplasm for pre-breeding. Engineered disease resistance was popular in the 1980s and 1990s, and should be given consideration again in the light of CRISPR/Cas9 biotechnology that will eventually be refined for producing non-GM germplasm for the cereals [36,37,38,39,40].
Accordingly, Section 7 of the review includes new data on wheat root genes induced during infection by Pratylenchus neglectus and P. thornei, the aim of which is to identify regulatory DNA segments with potential to turn on the expression of selected resistance genes when Pratylenchus is present. Candidate defense genes for disease resistance in wheat, barley, or rice are sometimes found in non-cereals, necessitating a search for the cereal gene equivalent (orthologue). In this section, we also discuss attempts to identify the wheat orthologue of the DORN1 gene (also known as P2K1) from Arabidopsis. The DORN1 (Does not Respond to Nucleotides1) protein is a plasma membrane-bound receptor that senses extracellular ATP released during cellular damage [41]. The gene was originally isolated using a genetic screen for an ATP-insensitive mutant [42]. When overexpressed in Arabidopsis (Arabidopsis thaliana (L.) Heynh), tobacco (Nicotiana benthamiana Domin), and potato (Solanum tuberosum L.), the Arabidopsis DORN1 gene conferred host resistance against the biotroph Phytophthora brassicae De Cock and Man in’t Veld 2002, the hemibiotrophs Pseudomonas syringae van Hall and Phytophthora infestans (Mont.) de Bary 1876, and the necrotroph Botrytis cinerea (Pers.:Fr) [43,44,45,46]. Pathogen resistance resulted when the receptor-ATP complex triggered the release of reactive oxygen species (ROS) and activated additional defense pathways in the host [47,48]. Root defense strategies and root interactions with the microbiome are reviewed in De Coninck et al. [49].
2. New Sources of Genetic Resistance to Nematodes of Small-Grain Cereals
Genetic resistance remains a desirable means of controlling soilborne pathogens. New mapping approaches are enabling the screening of populations with a genetically wider range of individuals. For instance, populations for association mapping can include accessions from global collections (diversity panel) or accessions enriched for the trait of interest based on geographic origin (focused identification of germplasm strategy, or FIGS) [1,50]. Mapping approaches now use single-nucleotide polymorphisms, or SNPs (90K iSelect SNP genotyping assay), or hybridization of host DNA to short DNA sequences on an array that represent a nonredundant set of genes located on all 21 wheat chromosomes (diversity array technology, or DArT, platform) [51,52]. A DArT array for barley has also been developed [53]. Genome sequences of individuals from the mapping population (genotyping-by-sequencing) have been used to obtain SNPs at a high density for better mapping resolution [54]. In some cases, the genome sequence of the host can be used to select candidate resistance genes based on physical proximity to a mapped locus or marker. The new mapping approaches, suitable for elite lines and adapted cultivars, enable the screening of a wider variety of genetic resources and avoid the problems associated with inter-specific crosses. An excellent review of genetic resistance to cereal cyst nematodes in wheat and barley is available [20].
Resistance to Meloidogyne graminicola in rice is important because all cultivars generally are susceptible to this nematode. Genetic resistance from Oryza glaberrima, an African cultivated rice, and O. longistaminata, a perennial wild rice relative, have been introgressed into adapted cultivars, although expression of resistance is attenuated in progeny of the latter [55]. Nevertheless, O. glaberrima confers nematode arrest at different stages in the infection process, providing different mechanisms of resistance for deployment. Recently, 332 accessions from a rice diversity panel, comprised of elite lines and landraces from 82 countries and representing five rice subpopulations [56], was screened for resistance to M. graminicola under controlled conditions [3]. Strong resistance was manifest in two accessions as absence of gall formation and very few females after M. graminicola infection. A GWAS approach revealed 11 QTL associated with gall number. Five of the QTL, located on Chr 3, 4, and 11, co-segregated with eight SNP markers. The authors concluded that two loci on Chr 3 and Chr 11 were close to previously reported loci. Another source of resistance to M. graminicola was identified in O. sativa cultivar Zhonghua 11, from the Chinese Academy of Agricultural Sciences rice breeding program [55]. This cultivar displayed reduced reproduction, fewer feeding sites and underdeveloped giant cells, compared to susceptible cultivar IR64. Resistance functioned in both flooded and dryland (aerobic) field conditions, and appeared to be dominant in F1 progeny of a cross between Zhonghua 11 and IT64. Zhonghua 11 shows promise as a source of an effective and readily transmissible trait, although, as pointed out by the authors, further testing is required.
Native sources of resistance to cereal cyst nematodes include the Cre genes from wild relatives of wheat or adapted cultivars. These have been deployed and combined for resistance against Heterodera avenae [14]. The wheat DArT array was used to discover markers for new nematode resistance loci in 107 spring wheat genotypes from the CIMMYT semi-arid wheat screening nursery [14]. Genotypes grown in this nursery are scored for yield under low or high rainfall (depending upon the year) in soil containing nematodes, root rot pathogens, and other biotic challenges. The study revealed three DArT markers for four novel H. avenae resistance QTL. In a separate study, resistance to H. filipjevi was revealed in winter wheat accessions from the International Wheat Improvement Program using the 90K SNP platform [57]. Resistance was based on counts of cysts and females in root and soil samples, and was associated with 11 QTL. The PNW adapted cultivar Madsen harbored resistance against H. avenae and H. filipjevi under both controlled environments and in naturally infested fields in China [5]. Reduced root penetration by juveniles and reduced white females on roots was observed for both species. While the action of these new sources of resistance might be conditioned by environment and genotype of the nematode, transfer of resistance to adapted germplasm will not require extensive pre-breeding.
Resistance to P. neglectus and P. thornei was reported in wheat and barley cultivars in Australia and the Pacific Northwest [7,14,26,28]. Wheat generally sustained more damage than barley, and P. thornei was more frequent, and hence more destructive, than P. neglectus [25,26]. Wheat and barley cultivars were rated for grain yield in the field, with and without application of the nematicide aldicarb; those for which yield was not substantially increased by aldicarb were considered to have native resistance [26]. This resistance was better described as tolerance, because enhanced grain yield was not due to reduced nematode reproduction. Tolerance was observed in 38 of 45 cultivars of spring wheat and barley and 22 of 121 winter wheat cultivars. Additional resistance to P. neglectus was mapped in doubled haploid populations of winter barley using DArT, restriction fragment length polymorphisms, SSR markers, and SNP markers [58]. Resistance, based on nematode counts from inoculations in a controlled environment, was attributed to six QTL. A major QTL conferred resistance to both P. neglectus and P. penetrans, also a pathogen of small-grain cereals. Spring wheat genotypes from the CIMMYT semi-arid wheat screening nursery screens produced seven and six new QTL and associated DArT markers for P. neglectus and P. thornei resistance, respectively [14]. Resistance to these two species also was reported in 32 Iranian wheat landraces [59]. In growth chamber assays, nematode counts in roots plus soil samples were reduced in resistant landraces compared to control lines. Association mapping using simple sequence repeat (SSR) markers resulted in three clusters of markers, indicating that at least three different resistance loci were present among the 32 landraces. In a subsequent study, landrace IWA8608077, with moderate but consistent resistance against P. neglectus and P. thornei, was used as a parent in a cross with the susceptible PNW cultivar Louise [25]. A resulting recombinant inbred line population was screened for visible root browning and reduction in nematode counts. Six major QTL for resistance were identified, four of which mapped to chromosome (Chr) 5A.
3. An Update on Genetic Resistance to Soilborne Fungal Pathogens of Small-Grain Cereals
Adapted small-grain cereal cultivars lack effective resistance or tolerance against Fusarium culmorum and Rhizoctonia solani, and genetic resistance in wild relatives has been scarce. An interesting locus in wheat was identified from mapping the recombinant inbred line population of the landrace IWA8608077 (resistant) crossed to Louise [25]. A major QTL for Pratylenchus resistance on Chr 5A also was associated with partial resistance to F. culmorum and R. solani AG-8. Partial resistance was evident as reduced discoloration of internode and crown tissue for F. culmorum, and reduced foliar stunting for R. solani AG-8, compared to controls. The P. neglectus and R. solani loci co-mapped to the same genetic interval (cM), whereas recombination was observed between that locus, the P. thornei locus, and F. culmorum locus. These and additional findings suggested that three different genes or gene regions from IWA8608077 were contributing to the resistances on Chr 5A. A QTL on Chr 5A of spring wheat accessions was involved in resistance against F. culmorum and F. graminearum, pathogens of the Fusarium head blight complex [60].
Genetic resistance to R. solani AG-8 of wheat and barley has been reported [61,62], but not integrated into breeding programs due to lack of molecular markers or field performance. Recent screening and mapping of populations derived from synthetic or synthetic-derived CIMMYT wheat accessions resulted in five new sources of resistance or partial resistance; mapping of the trait was carried out for two sources [63,64]. For both sources, reduced disease severity was deemed to be due to tolerance, rather than resistance, although soil population densities of the pathogen were not quantified. Two evaluations for shoot-stunting were carried out in field plots where pathogen buildup in the soil was favored. The field screens were interspersed with a greenhouse root rot assay. In 190 recombinant inbred lines from a cross of synthetic-derived SCPB-3104 and susceptible PNW spring wheat cultivar Louise, three tolerance QTL were identified using genotyping-by-sequencing and composite interval mapping [63]. Markers associated with the QTL mapped to Chr 1BL, 2AS, and 2DL. A similar study of a cross between SYN-172 and Louise yielded three consistent QTL on Chr 1AL, 7DS, and 7DL, with 7DL derived from Louise [64]. Transgressive segregation was observed for both the SPCB-3104 and SYN-172 mapping populations.
4. Genes Associated with Disease Resistance against Soilborne Pathogens
Molecular markers are of value to breeders and pre-breeders, but identification of the genes underlying genetic resistance should be identified in order to determine resistance mechanisms. Such knowledge can be applied to other host genotypes, pathogen isolates and, possibly, to other environments. Proceeding from a trait or locus to a gene of interest is now more feasible with whole genome sequencing, sequencing of coding portions of the genome [65], and global sequencing of expressed genes (transcriptomics). Data for multiple individuals can be obtained and analyzed simultaneously due to lower costs of next-generation sequencing and more sophisticated software for assigning function to candidate genes.
In a screen for M. graminicola resistance in O. sativa, Dimpka et al. [3] focused on 493 candidate resistance genes in the regions of 11 QTL associated with reduced gall numbers. Genes most likely to have defense-related functions were identified using a variety of software. Sixteen of the genes were differentially expressed in rice roots during nematode infection [66], and 51 were differentially expressed in giant cells [67], discussed further in Section 5. Genes that were predicted to encode lectin domain proteins or other lectin-associated proteins were well represented in the candidate gene pool. The candidate pool also contained orthologues of the barley Mla gene for powdery mildew resistance, and genes encoding peroxidase, enzymes for ROS generation. In a study of H. filipjevi resistance in wheat, Pariyar et al. [57] reported 11 resistance QTL and 9 candidate protein-coding genes in the regions of the QTL. The candidate proteins included transcription factors that control the expression of specific genes, and regulators of cellular processes.
Alternatively, molecular genetics and biochemical approaches have been used to discover the roles of specific genes for defense functions in the host. For instance, the Cre3 genes from wheat, Triticum turgidum, and its wild wheat relative, Aegilops tauschii, encode resistance gene analogs, a subset of a large and complex family of nucleotide binding site leucine-rich repeat (NBS LRR) proteins [68,69]. The NBS-LRR proteins have a role in early pathogen recognition leading to resistance. A majority of plant genes required for qualitative (race-specific) resistance contain NBS and LRR motifs. The H. avenae resistance genes Cre3, Cre 5, and Cre 7 in wheat acted through a cytosolic ascorbate peroxidase gene [70]. The ascorbate peroxidase gene was induced earlier and to a greater extent in resistant genotypes compared to susceptible ones, and its expression was high around the nematode feeding site. This finding indicated that ROS were contributing to resistance in the case of these Cre genes.
Clues to defense pathways in the host can be obtained from studies of single genes. The wild grass, Aegilops variabilis, carries CreX- and CreY-mediated resistance to H. avenae, which has been deployed in wheat [71]. A gene encoding tryptophan decarboxylase1 was strongly induced in Ae. variabilis roots during early stages of nematode infection compared to non-infected controls, but when silenced using the barley striped mosaic virus gene silencing system, the plant roots showed enhanced susceptibility to infection [72]. Tryptophan decarboxylase converts tryptophan to tryptamine, a precursor for the synthesis of secondary metabolites and the phytohormone IAA. Subsequent studies with the Ae. variabilis gene were done in tobacco, a more tractable host for gene disruption and overexpression experiments. These studies showed that overexpression of the tryptophan decarboxylase1 gene bolstered resistance, and confirmed that gene disruption or inhibition of its protein reduced resistance [72]. The tryptophan decarboxylase gene manipulations resulted in changes in ferulic acid, a cell wall strengthener in the grasses; caffeic acid, an antioxidant and precursor to ferulic acid; serotonin, thought to affect root development and antagonize IAA [73]; and expression of the gene for tryptamine-5-hydroxylase, the enzyme that converts tryptamine to serotonin [74]. However, IAA and IAA biosynthetic genes were not affected. The findings suggest that the Aegilops Cre-mediated resistance involves the production of secondary metabolites that condition cell wall rigidity and root development.
To examine the role of phytohormone signaling in M. graminicola resistance, expression of genes for salicylic acid, jasmonate, and ethylene signaling was compared in resistant and susceptible rice cultivars [4]. The resistant cultivar showed reduced numbers of galls, females, and eggs relative to the susceptible cultivar. In this cultivar, a gene for ethylene biosynthesis, and genes for jasmonic acid biosynthesis and jasmonate signal transduction were induced throughout early infection, whereas in the susceptible cultivar, expression was attenuated or transient. Two genes for phenylpropanoid biosynthesis (salicylic acid pathway), genes for callose and lignin biosynthesis, and a gene for callose metabolism were also elevated during infection in the resistant cultivar. Mechanisms of resistance appear to involve ethylene and jasmonate signaling and modification of cell wall components.
Barley plants carrying the Rha2 gene for resistance to H. avenae differed from susceptible plants in nematode-related morphological changes in the roots, including faster syncytial development, rapid deterioration of the feeding site, and altered syncytial cell wall composition [22]. In resistant barley, levels of (1,3;1,4)-β-glucan also were higher in the cell walls of the feeding site compared to susceptible barley. The authors examined the expression of genes involved in cell-wall synthesis and found higher expression of a cellulose synthase-like F10 gene in resistant barley. This study indicated that resistance was accompanied by alternations in cell wall metabolism and composition.
For the soilborne fungi, there are fewer recent leads about how specific genes or resistance-related QTL contribute to reduced disease symptoms. In a comparison of wheat cultivars with resistance or susceptibility to F. culmorum, applications of the phytohormone methyl jasmonate alleviated crown and foot rot severity in the susceptible cultivar [75]. Six defense genes were induced in the resistant wheat, but only three were induced in the susceptible cultivar, suggesting that the latter had an attenuated jasmonate signal pathway. A number of studies have linked changes in lignin to both the nematode infection process and to host-nematode interactions [4,6,13,58,76]. Thompson et al. [25] found that the resistant landrace IWA8608077 had higher lignin content than the susceptible Louise [77]. Lignin has roles in plant growth, long-distance water transport, and defense, so enhancing its defense function without perturbing development or secondary metabolism will require a more complete understanding of host transcriptional and metabolic pathways for lignin metabolism [78].
5. Mining ‘Omics Data for Leads to Novel Defense Genes
Whole genome sequencing (genomics), and global profiling of gene expression (transcriptomics) and protein expression (proteomics) are valuable in comparing both host and pathogen genotypes. Genome sequencing provides the ultimate genotyping data, and can aid in predicting whether an organism has the capacity to produce a phenotype based on the presence or absence of known genes. The transcriptome represents a collection of expressed genes in a selected group of cells from a selected genotype, at a given time, and under specific conditions. At best, it gives insight into the abundant genes and major molecular pathways associated with a biological response, such as a host resistance. However, it is not always clear how to select specific genes for further testing in the laboratory or field from global gene expression data. The size and quality of reference databases continue to be a critical aspect of gene identification (annotation) in ‘omics studies.
A comprehensive comparative genomics approach recently was used to discover pathogenicity genes of the root-knot nematode, Meloidogyne incognita [18]. The authors collated 15,952 genes common to plant-parasitic nematodes that were not found in other nematode genomes and identified 993 genes predicted to encode secreted nematode proteins with potential to interact with host proteins and result in disease. Sixteen genes that were present as single-copy in M. incognita were targeted for gene silencing experiments. RNA-based gene silencing is discussed in more detail in Section 6. Of the 16 target genes, 12 resulted in reduction in nematode egg masses and/or gall counts in root infection assays when gene function was disrupted. This approach identified novel pathogenicity proteins and narrowed the candidate gene pool to a feasible number for further testing
A transcriptome study of root responses to M. graminicola infection was carried out using the resistant African rice, Oryza glaberrima, and a susceptible japonica cultivar of O. sativa [6]. In the resistant rice, nematode penetration was reduced, and females were smaller and laid fewer eggs compared to the susceptible rice. Over 3-fold more host genes were induced in root tips or whole roots at early infection in the resistant compared to susceptible rice, and about 5.5-fold fewer genes were repressed in the resistant compared to the susceptible genotype. The expression of genes encoding the antifungal protein thionin was high in root tips of the resistant rice without nematode challenge, whereas genes for the phenylpropanoid pathway enzyme phenylalanine ammonia lyase (PAL), for lignin biosynthesis, and the pathogenesis-related proteins PR-2 (glucanase) and PR-10 (RNase-like) were strongly induced during infection. A gene for callose degradation was also induced upon infection in the resistant rice. These findings indicated that resistance involves the jasmonate and ethylene signal pathways, and changes in cell-wall composition. However, cultivar differences in lignin content were not supported by observations of autofluorescence around the giant cells. Finally, the authors examined 10 genes from their study that corresponded to O. sativa candidate-resistant genes shown to be physically linked to QTL for M. graminicola resistance [3]. These included genes for the phenylpropanoid and lignin biosynthetic pathways, thionin and thaumatin-like antifungal genes, jasmonic acid biosynthesis, and a stripe rust resistance gene orthologue.
Transcriptomic responses to H. avenae were reported for roots of resistant and susceptible wheat genotypes [79]. The resistant wheat roots harbored fewer mature nematodes, which was attributed to slowed development of J3- and J4-stage nematodes. The roots of the resistant genotype also displayed a bi-phasic release of ROS, an early transient, and a later sustained burst, typical of many resistance responses in leaves [80,81]. Resistance was associated with induction of genes for phenylpropanoid biosynthesis, a chitin elicitor receptor kinase, the brassinosteroid insensitive1-associated kinase, peroxidases, and lipoxygenases. The latter two genes were thought to be involved in the ROS bursts. Induction of genes involved in oligo- and monosaccharide transport, RNA methylation, DNA repair, and sulfite metabolism were also observed. In a related study, Qiao et al. [82] examined genes induced by H. avenae in the roots of the susceptible wheat at early- to mid-stage infection. The genes encoded cell-wall degrading enzymes, peptide transporters, auxin biosynthesis and signaling, peroxidases, gluthathione S-transferases, and UDP-glycosyltransferases (for glycosylation of secondary stress metabolites and phytohormones). The antifungal lipid transfer proteins, 1,3-β-glucanases, and chitinases also were induced. Additional experiments could be done to demonstrate a genetic or physiological correlation between susceptibility and candidate gene induction, as some of the genes might be involved in the non-specific root immunity response to biotic stress [83]. Twelve of 16 NBS-LRR genes were repressed during nematode challenge, indicating that susceptibility was associated with damping early pathogen recognition mediated by NBS-LRR resistance proteins. The same susceptible wheat cultivar used in the above two studies was challenged with mobile J2 H. avenae to characterize root interactions with the pre-penetration stage of the nematode [84]. Wheat roots produced 27 induced genes and 66 repressed genes, and the nematodes showed 867 induced genes and 12 repressed genes. The repressed wheat genes were enriched for ROS metabolism, whereas over 40% of the induced nematode genes were orthologues of known candidate parasitism genes; genes for primary metabolism and nematode body wall development were also observed. One nematode-expressed gene of interest encoded a chitinase-like protein that suppressed the host cell death resistance response in the tobacco assay.
There is a void of information about how the roots of small-grain cereals respond to R. solani or F. culmorum on a global gene expression scale. However, the resistance response to R. solani AG-8 was examined in the model brassica A. thaliana using the Affymetrix ATH1 Genome array, a DNA hybridzation-based platform [85]. The study revealed that R. solani AG-8 induced root genes for ROS metabolism, including peroxidases, an alternative oxidase, the respiratory burst homolog subunit RbohD of the NADPH oxidase complex, FAD-binding proteins, and glutathione S-transferases. Genes for heat shock proteins, expansins, and modification of xyloglucan, pectin, polygalacturonic acid, and other cell wall-associated proteins components were also induced. The authors generated mutations in two Rboh subunit genes NADPH oxidase that resulted in an enhanced degree of susceptibility to R. solani AG-8. In general, the data indicated that Arabidopsis roots responded to a fungal necrotroph in ways reported for small-grain cereal roots after nematode challenge. One exception was the lack of evidence for the jasmonate signal pathway. Specifically, though, the study pointed to the importance of a functional NADPH oxidase in disease resistance.
A proteomics approach was used to identify R. solani AG-8 genes induced in the presence of wheat seedlings [12]. This interaction represented the susceptibility response of the host, and the pathogenic response for R. solani. Protein fractions were obtained from cultured mycelium and mycelium growing on nitrocellulose membranes laid over wheat seedlings in a system. Among the secreted proteins found only in mycelium exposed to the host, cell-wall-degrading enzymes and proteins associated with ROS metabolism were well-represented. Culture filtrates constitutively expressed proteins for lignin breakdown. In the process of mapping the peptide fragments to the genome of R. solani AG-8, the authors noted that only 1 of 36 pectate lyase genes encoded by the fungal genome was expressed in wheat-exposed mycelium, whereas at least 9 were expressed during infection of the clover Medicago truncatula. These observations suggest that the pathogen adapts its gene expression to the host species, with implications for the specificity of host resistance strategies.
6. Host-Induced Gene Silencing (HIGS)
Gene silencing is a natural process by which organisms manipulate expression of their own genes or the genes other organisms. The process, which involves inhibition of specific genes by hybridization with complementary inhibitory RNA (RNAi), has been elucidated at the molecular level and has been adapted for gene expression studies in plants [86,87]. Host-induced gene silencing makes use of the natural phenomenon of RNA exchange among plants and microbes, through processes not yet fully understood. In the HIGS approach, the host plant is made to produce an RNAi molecule complementary to a pathogen target gene, such as a pathogenicity or virulence gene, which is transferred to the pathogen during infection. Phytopathogenic nematodes and fungi produce parasitism or disease-inducing proteins, also called effectors, which are introduced into host cells or tissues, and interact with host proteins to produce the susceptible response (disease condition). Genes encoding toxins, cell-wall-degrading enzymes, ROS detoxifying enzymes, or other proteins that undermine host defense processes are attractive candidate HIGS targets [7,88,89].
Single-gene approaches for P. thornei or H. avenae resistance were enabled by the identification of orthologues of known parasitism genes. Silencing of genes for two components of nematode body wall muscle structure and function, troponin C and calponin, reduced reproduction of P. thornei and P. zeae (maize pathogen) in a carrot disk culture system [90]. Nematodes soaked in a solution containing the troponin C and calponin RNAi molecules showed a 77%–81% reduction in numbers of juveniles, adults, and eggs. Genes from either species were effective for both species, but P. thornei was more responsive to silencing than P. zeae, and there was a gene-by-species effect. Two other H. avenae orthologues of parasitism genes from other nematode species were silenced in wheat roots [91,92]. The RNAi molecules were introduced using the barley striped mosaic virus HIGS system. Numbers of H. avenae juveniles and females were reduced by 66%–73% and 47%–73%, respectively, compared to non-silenced controls. One parasitism gene was expressed in pre-parasitic J2 [91], whereas the other was highest in J4-stage nematodes [92]. Additional experiments done in tobacco indicated that both parasitism genes suppressed specific points in the innate plant immunity response.
Alternatively, transcriptomics has been used to survey nematode genes expressed during infection, with the aim of identifying nematode parasitism genes that would be effective targets for HIGS. For instance, the transcriptome of H. avenae during colonization of roots of resistant Ae. variabilis was examined at the pre-penetration, early, and late stages of infection [23]. The study led to the identification of 56 predicted parasitism genes, 32 of which encoded cell-wall-modifying proteins, possibly for host wall penetration or breakdown, and 10 for ROS formation or detoxification. The findings suggested that resistance in Ae. variabilis surmounted or circumvented the action of these parasitism genes. Three nematode targets having no orthologues in humans, insects, and plants were selected for HIGS experiments. The silencing was accomplished by soaking J2 nematodes in solutions of the RNAi molecules. The silencing of the protease calpain and neuropeptide FLP-4 genes promoted nematode mortality.
A similar study of the global transcriptional response of Pratylenchus penetrans (Cobb, 1917) Chitwood and Oteifa, 1952 to soybean root colonization provided molecular leads for HIGS in a relative of P. thornei and P. neglectus [93]. This Pratylenchus species is not a major pathogen of wheat in the PNW, but recently has been a problem in certain locations [94,95,96]. Candidate parasitism genes identified in the study included 12 orthologues from other nematode species and 41 genes encoding ROS-associated genes, and five types of cell wall-modifying enzymes, thought to protect nematode from host defenses.
The transcriptome of M. graminicola was obtained from J2 stage nematodes collected from rice roots [97]. Of 1328 candidate secreted protein genes, the authors focused on four that were expressed in the esophageal gland cells or olfactosensory organs, and were common to Meloidogyne and cyst nematodes, but without sequence orthologues in humans, plants, and insects. Two genes lacked functional annotation (hypothetical protein), the third encoded a prion-like (Q/N-rich) domain-bearing protein [98] associated with conversion of soluble protein to protein aggregates in other organisms, and the fourth encoded a trehalose-6-phosphate phosphatase, thought to reduce accumulation of toxic trehalose-6-phosphate in the nematode gut [99]. After HIGS, nematode viability was 20%–56%, compared to 100% for the water control. Furthermore, the M. graminicola transcriptome [97] was used to obtain a full-length copy of a novel parasitism gene called MgGPP [100]. This gene encoded a protein that had limited sequence identity (37%–41%) to a family of Meloidogyne virulence proteins of unknown function. The MgGPP protein was expressed in the esophageal gland cells of the nematode and accumulated in early parasitism. Overexpression of the gene resulted in enhanced susceptibility of transgenic rice lines to M. graminicola; female nematode numbers increased 23%–67%, depending on the line. Conversely, rice plants transformed with RNAi species showed reduction in MgGPP gene expression and 50%–72% reduction in female nematode numbers. Clues to possible mechanism(s) of MgGPP action were revealed using molecular and histochemical approaches. The protein underwent host-mediated post-translational modifications that resulted in suppression of the normal host cell death resistance response and localization of the modified protein to the nucleus.
Fungal pathogenicity factors, including secreted cell-wall degrading enzymes, phytotoxins, and cell death-inducing proteins, have been described [89,101]. Certain isolates of F. culmorum produce the toxins nivalenol and deoxynivalenol, which contribute to yield losses caused by F. graminearum in Fusarium head blight of cereals [8,35,102,103]. A possible candidate for HIGS in F. culmorum arose from observations of developmental and metabolic defects associated with mutations in the StuA transcription factor gene [30]. The stuA mutants of F. culmorum displayed reduced conidiation, delayed spore germination, decreased mycelial diameter, length and density, decreased catalase and polygalacturonase activities, and loss of pathogenicity to the stem and root [30,104]. These phenotypes were independent of toxin production, rendering StuA a promising target for HIGS of stem and root rot of wheat and barley. Another promising HIGS target for F. culmorum is the CYP51B gene, which encodes a lanosterol C14-α-demethylase essential for plasma membrane permeability and ascospore germination. Simultaneous silencing of the CYP51A, CYP51A, and CYP51C genes in F. graminearum resulted in abnormal morphology and growth inhibition of the mycelium, and the resistance response in detached leaves of the model plant Arabidopsis and transgenic barley expressing the RNAi molecules [105].
7. Building a Molecular Toolbox for Engineering of Targeted Disease Resistance
7.1. Materials and Methods for Transcriptomics of Pratylenchus-Challenged Wheat Roots
Pratylenchus neglectus and P. thornei, obtained from soil samples, were kind gifts from Drs. Yan and Smiley, Oregon State University, Pendleton, Oregon, USA. Nematodes were cultured on sterilized carrot disks according to Yan et al. [106]. Seeds of susceptible spring wheat cultivar Scarlet [107] were sown singly in 6-inch plastic cones (Stuewe and Sons, Corvallis, OR, USA) containing autoclaved sand. Seedlings were grown at 25 °C, 16h lt/20 °C, 8 h dk. Nematodes were introduced to roots of 14-day-old plants as soil drenches of 2000 mixed-stage (J2-J4) juveniles. Each experiment consisted of four treatments, three plants per treatment: (1) Pratylenchus neglectus only (Pn); (2) P. thornei only (Pt); (3) a 1:1 mixture of P. neglectus and P. thornei (PnPt); and (4) no-nematode control (Cont). After a 21-day treatment period, roots were rinsed to remove sand, blotted gently, and pooled by treatment into tubes containing liquid nitrogen. Harvested roots were stored at −80 °C prior to RNA isolation. The experiment was repeated twice for a total of three biological replications and 12 RNA samples.
Total RNA was obtained from treated roots using a modified TRIzol method [83]. RNA samples were processed and sequenced by MRDNA, Shallowater, TX, USA. RNA concentration was determined using the Qubit® RNA Assay Kit (Life Technologies, Carlsbad, CA, USA), and information for RNA quality and average library size was obtained using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA quality was based on sizes of all RNA fragments obtained using the Bioanalyzer and the Agilent 2100 software [108]. Libraries of each of the 12 RNA samples were prepared from 1.0 μg of total RNA using the TruSeq™ RNA LT Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were indexed and prepared for polymerase chain reaction amplification using barcodes and adaptors. A total of 2 nM of each library were pooled; 5 pM of the pooled mix was subjected to Illumina HiSeq 2500 paired-end 2 × 150 sequencing.
Raw sequence reads were filtered for quality, and adaptors, index barcodes, and ambiguous bases were removed using the CLC Genomics Workbench Trim Sequences tools (Qiagen Bioinformatics, Redwood City, CA, USA). De novo assembly of filtered reads was conducted using the De Bruijn graphing algorithm [109]. Paired-end reads exhibiting overlapping sequences were merged and reads greater than 85 bp (20–22 bp overlap) were assembled into contiguous sequences (contigs) representing the expressed transcripts. Expressed transcript groups were mapped to the Triticum aestivum available at the International Wheat Genome Sequencing Consortium (IWGSC) RefSeq v1.0 coding sequence database [110]. The IWGSC database represented putative coding sequences on the long and short arms of each of the seven chromosomes of the A, B, and D genomes of hexaploid wheat. Transcript groups also were annotated using Blast2GO (BioBam Bioinformatics S.L., Valencia, Spain), which provided initial sequence and gene function information. Numbers of transcripts among samples were normalized using reads per kilobase of transcript per million mapped reads (RPKM) [111] so that transcript abundance between treatments could be compared. Transcript groups having an RKPM value >50 for at least one of four treatments (Pn, Pt, Pn and Pt, or control) were examined further to identify those that were induced in the presence of nematodes. Transcript groups that showed a Pn/Cont, Pt/Cont, and/or PnPt/Cont RPKM ratio of >2.0 were considered to be induced during Pratylenchus challenge. The arbitrary cutoff of RPKM >50 was intended to rise above “background” expression and increase the identity of causative loci.
7.2. Results and Discussion of the Pratylenchus-Wheat Root Transcriptome
Properties of the RNA and RNA libraries generated for the wheat root-Pratylenchus transcriptomes are shown in Supplementary Table S1. Total RNA concentrations were sufficient for library preparation. RNA quality (RIN), ranging from 3.9 to 5.4, indicated partial degradation, likely occurring during the root washing step prior to freezing in liquid nitrogen. However, the RNA produced libraries of sufficient quantity and size for Illumina HiSeq sequencing, and allowed us to analyze RPKM data for candidate pathogen-inducible promoters.
A total of 262,908 wheat transcript groups were annotated using Blast2GO (BioBam Bioinformatics S.L.) and the IWGSC coding sequence database for hexaploid wheat; 4,17 groups harbored at least one treatment having an RPKM value >50.0. The 24 non-paired read files are available as accession numbers SAMN10686471-10686492 under NCBI BioProject ID PRJNA512537.
We selected 44 transcript groups in which induction of >2.0 was observed for one or more of the Pratylenchus treatments relative to the non-challenged control (Table 1). One important caveat of our RNA-seq data was that initial RNA sample quality was lower than ideal. Therefore, induction of an annotated gene was deemed more reliable if they were represented by multiple transcript groups, all of which were up-regulated by nematode treatment. Nine annotated transcripts, such as those encoding three late embryogenesis abundant proteins, were comprised at least two members and accounted for 36 of the 44 transcript groups. Induction was often specific to one nematode species, indicating that the two species of Pratylenchus were interacting differently with the roots of cv. Scarlet. For instance, ascorbate peroxidase and catalase peroxidase genes showed induction by P. neglectus, and the glutathione S-transferases and late embryogenesis abundant proteins were induced by P. thornei at the time of root harvest. A similar degree of induction was observed in the corresponding PnPt treatments, indicating that the presence of the second species did not have a strongly positive or negative effect on transcription. The CAP superfamily, actin-11, and three transcript groups of the major pollen allergen appeared to be induced by both Pn and Pt. Induction of the CAP superfamily also seemed to be additive. Authentication of induction will require additional experiments, such as quantitative reverse-transcription PCR. Gene expression will also be quantified using the resistant Scarlet mutant [112]. A similar approach could be used to find regulatory DNA segments that respond to other pathogens.

Table 1.
Wheat root transcripts showing induction after challenge with root lesion nematodes Pratylenchus neglectus and/or P. thornei.
7.3. Materials and Methods for the Wheat Orthologue of Arabidopsis DORN1
The action of extracellular ATP in Arabidopsis, acting through its receptor DORN1, has led us to consider characterizing the wheat orthologue of DORN1 in interactions with nematodes and fungal necrotrophic root pathogens. A collection of wheat and barley orthologues predicted to contain lectin-binding, kinase, and transmembrane domains were obtained from published literature [113,114]. Candidate wheat or barley orthologues were identified using HMMER 3.0 [115] and TMHMM [116] and databases IWGSC URGI and International Barley Sequencing Consortium genome coding sequences [110], for wheat and barley, respectively. Another wheat orthologue search based on amino acid sequence identity was conducted in 2019 using Blast and the IWGSC RefSeq v1.0 coding sequence database. For this search, GenBank accessions of DORN1 nucleotide sequences from the brassicas Arabidopsis thaliana, A. lyrata, and Camelina sativa were obtained. The coding regions of the brassica sequences were used to find IWGSC (wheat) accessions with the highest sequence identity. Deduced amino acid sequences of the cereal DORN1 candidates were compared to those from the brassica species using Clustal Omega [117], and regions of identity were visualized using GeneDoc [118].
7.4. Results and Discussion of Candidate Wheat Orthologues of DORN1
Because DORN1 is highly expressed in roots [47], DORN1-mediated resistance is dependent upon the jasmonate signal pathway [45,46,119], and wheat roots appear to possess an active jasmonate signal pathway [120], we hypothesize that expression of the wheat orthologue of the DORN1 gene will confer resistance to the necrotroph R. solani AG-8, hemibiotroph F. culmorum, and possibly to nematodes that cause cellular damage while migrating through host root tissue.
Searches of the wheat and barley databases using HMM and TMHMM resulted in 23 candidate barley DORN1 orthologues containing both lectin-binding and transmembrane domains, but not the kinase domain. In contrast, 205 candidate wheat orthologues were retrieved that contained lectin-binding and kinase domains, regardless of whether transmembrane domains were present. The numbers of candidates and the absence of at least one key domain discouraged us from pursuing cereal DORN1 genes at the time. More recently, three accessions (Traes_2BS_80806CA5F.1, Traes_2AS_DE7DCB294.1, and Traes_1BS_A999A190E.3) from the IWGSC RefSeq v1.0 database were identified based on highest amino acid sequence identity to DORN1 from A. thaliana, A. lyrata, and Camelina sativa, respectively (Supplementary Table S2). Identification of the wheat orthologue was not clear-cut, as certain features of the DORN1 protein vary among species [42]. The brassica amino acid sequences shared 90.4%–96.4% identity with each other, whereas the candidate wheat orthologues were 58.4%–59.3% identical to each other at the amino acid level. For the sake of clarity, the candidate wheat orthologues were compared only to the A. thaliana-deduced amino acid sequence (Figure 1). Wheat sequences 80806CA5F.1, A999A190E.3, and DE7DCB294.1 showed 45.9%, 45.5%, and 44.0% identity, respectively, to the Arabidopsis DORN1, but there was no single wheat orthologue that stood out for future gene transfer to wheat and subsequent testing in soilborne fungal pathogenicity assays. However, the pool of wheat DORN1 candidate sequences was narrowed from 205 to 3 genes.
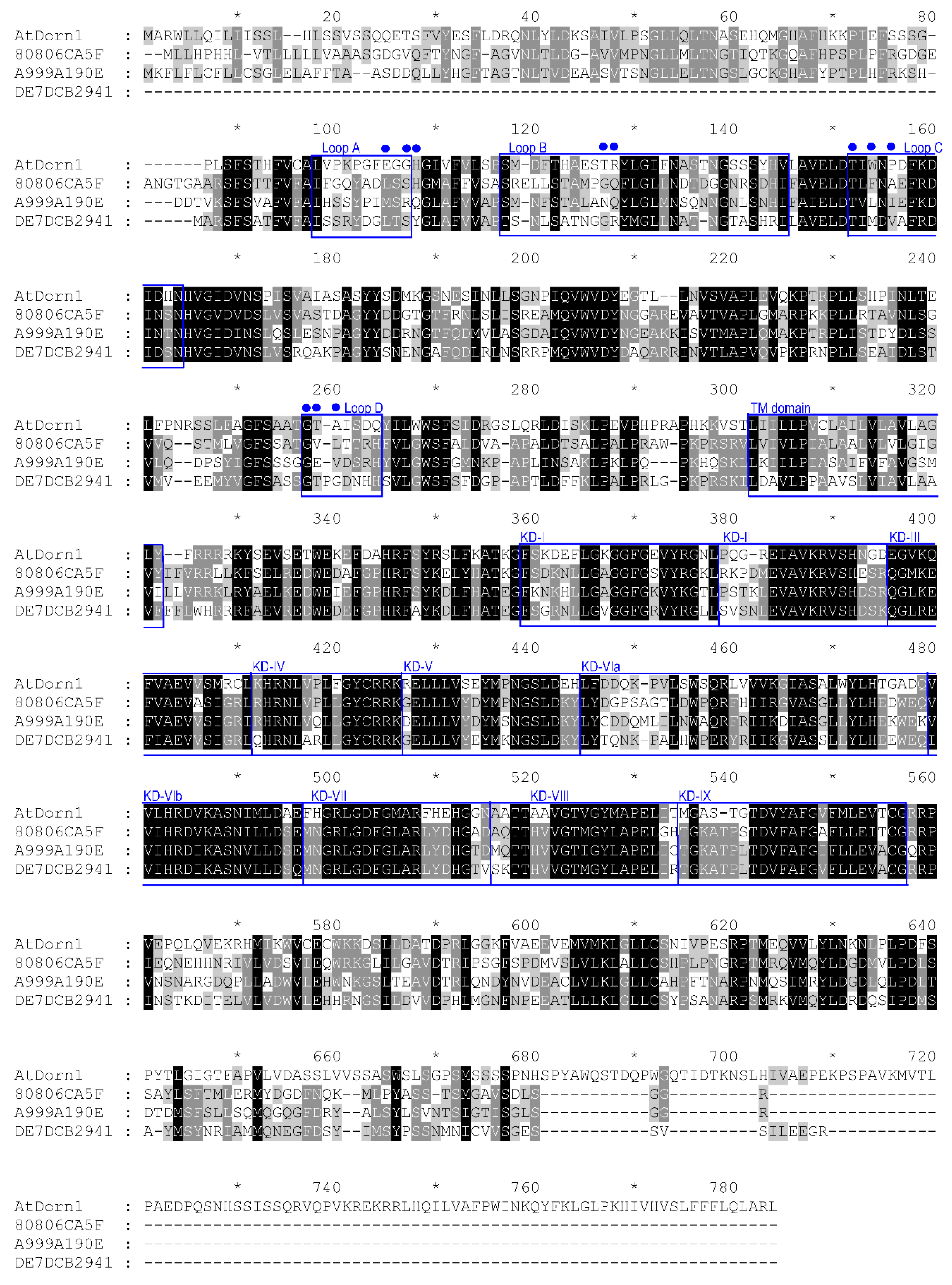
Figure 1.
Alignment of deduced amino acid sequence of DORN1 from A. thaliana (derived from GenBank NM_001085304.1) with three candidate wheat orthologues Traes_2BS_80806CA5F.1, Traes_2AS_DE7DCB294.1, and Traes_1BS_A999A190E.3. Loops are extracellular and contain ATP binding residues (blue dots). Boxes delineate a conserved transmembrane domain (TM) and kinase domains (KD). Domain KD-X is poorly conserved and is not indicated [113,114].
Identifying gene orthologues can be complicated by evolutionary differences among different species. For instance, recognition of extracellular ATP by a transmembrane receptor might depend on conservation of structural features rather than on conserved amino acid sequences. The wheat DORN1 gene, once identified, might function in barley and vice versa, due to the molecular relatedness of these species, whereas the Arabidopsis gene might or might not be operational in the small-grain cereals. The parasitism genes orthologues from H. avenae and M. gramincola often did not share high sequence identity with known parasitism genes. Yang and coauthors [92] noted significant diversity among G16B09 parasitism genes from H. avenae, H. glycines, H. schachtii, and two Globodera spp. Likewise, the annexin orthologue from H. avenae shared 76% amino acid sequence identity to the G. pallida-deduced protein, and differed substantially from proteins of other Heterodera spp. and Globodera spp. It is likely that the H. avenae genes were the correct orthologues, based on their ability to suppress host defenses. In a comparison of fungal CYP51 proteins, Koch and Kogel [121] found 80.9% and 64.8% identity between F. graminearum and F. solani or Magnaporthe oryzae, respectively. The application of CYP51B HIGS for suppression of R. solani AG-8 is being explored. In this case, there is substantial gene sequence diversity relative to Fusarium CYP51B to merit design of a new RNAi molecule for R. solani.
The relatively low efficiency of wheat transformation limits the number genes that can be reasonably tested in planta. However, rice is much more amenable to Agrobacterium transformation [122,123], and a rapid and efficient system for barley root transformation has been developed [124].
8. Conclusions
In conclusion, deploying effective genetic resistance to soilborne nematode and necrotrophic fungal pathogens of the small-grain cereals remains a challenge. Genetic resistance is dependent on the genotypes of the host and pathogen, and can be location-dependent, as was noted for nematode resistance [7]. Nevertheless, marker technology and linkage analysis software are screens of adapted or semi-adapted cereal genotypes. Lignin accumulation and ROS were recurring defense pathways in cereal root-pathogen interactions. A second pathway for the biosynthesis of the lignin precursor 4-coumaric acid has been described in maize and Brachypodium distachyon. Many other grasses, including rice, wheat, and barley, harbor one to six copies of the requisite gene in their genomes [78,125]. It is very likely that enhancement of lignin content to the right degree and at the right time in the root could protect against a range of nematode and fungal pathogens. The use of ‘omics approaches for resistance gene discovery has yielded orthologous and novel parasitism genes from plant-parasitic nematodes, and remains to be fully realized for identifying parasitism genes from the fungal pathogens. Sequencing of coding regions of the genome (exome) can be applied to move from phenotype to genotype at the single polymorphism level with development of species-specific probes [65,126,127,128]. With the increasing number of genome sequences available each year, the pan-genome mining approach [18] will become feasible for more host and pathogen species. One caveat of certain roots is the potential for ectopic expression of genes normally found in other organs, such as chlorophyll- and photosynthesis-related genes [67,76,129]. It is not clear whether the resistance interaction provides better leads for novel gene discovery than the susceptible interaction; both are informative. Many ‘omics studies produced sequences lacking functional annotation (e.g., “hypothetical protein”). These might be considered for HIGS approaches if species- and gene-specificity are desired. Pathogen targets that are important for growth and life cycle in the host should be better HIGS candidates than those involved in the “arms race,” which are continually under selection. Off-target effects of the RNAi molecule will be minimized by selecting for single copies of the RNAi molecule in the host and will need to be monitored in the recipient pathogen. In most cases, the effect(s) of each candidate gene on pathogen suppression and disease severity ultimately needs testing in the field. There is no single approach guaranteed to produce a gene, genetic locus, or trait that is easy to deploy and is effective in the field. However, a variety of approaches, each with advantages and drawbacks, are available.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/9/4/188/s1, Table S1: Properties of the RNA and RNA sample libraries used in wheat root-Pratylenchus transcriptomics, Table S2: Candidate IWGSC wheat DORN1 orthologues of brassica GenBank accessions.
Author Contributions
Conceptualization, P.A.O. and R.M.S.; methodology, P.A.O., A.B.P. and R.M.S.; software, R.M.S.; formal analysis, P.A.O. and R.M.S.; investigation, P.A.O. and A.B.P.; resources, P.A.O., A.B.P. and R.M.S.; data curation, R.M.S.; writing—original draft preparation, P.A.O.; writing—review and editing, P.A.O., A.B.P. and R.M.S.; visualization, P.A.O.
Funding
This research was funded by USDA CRIS Project No. 2090-22000-017-00D (P.A.O.).
Acknowledgments
The authors thank Inga Zasada, USDA-ARS, Corvallis, Oregon, USA, for resources and facilities to generate RNA from Pratylenchus-challenged roots; Scot Dowd at MRDNA, Shallowater, Texas, USA for generating the RNA-seq data; Kiwamu Tanaka, Washington State University, Pullman, for input about the DORN1 sequences, and critical reading of the manuscript; Amit Dhingra, Washington State University, Pullman for providing bioinformatics facilities. Mention of a trademark, proprietary product or vendor does not imply its approval to the exclusion of other products or vendors that may also be suitable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| HIGS | host-induced gene silencing |
| GWAS | genome wide association study |
| PNW | Pacific Northwest, USA |
| QTL | quantitative trait locus/loci |
| RNAi | inhibitory RNA |
| ROS | reactive oxygen species |
| RPKM | reads per kilobase million |
| SNP | single nucleotide polymorphism |
| SSR | simple sequence repeat |
References
- Mondal, S.; Rutkoski, J.E.; Velu, G.; Singh, P.K.; Crespo-Herrera, L.A.; Guzmán, C.; Bhavani, S.; Lan, C.; He, X.; Singh, R.P. Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front. Plant Sci. 2016, 7, 991. [Google Scholar] [CrossRef]
- O’Leary, G.J.; Aggarwal, P.K.; Calderini, D.F.; Connor, D.J.; Craufurd, P.; Eigenbrode, S.D.; Han, X.; Hatfield, J.L. Challenges and responses to ongoing and projected climate change for dryland cereal production systems throughout the world. Agronomy 2018, 8, 34. [Google Scholar] [CrossRef]
- Dimkpa, S.O.N.; Lahari, Z.; Shrestha, R.; Douglas, A.; Gheysen, G.; Price, A.H. A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. J. Exp. Bot. 2016, 67, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Kumari, C.; Dutta, T.K.; Banakar, P.; Rao, U. Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci. Rep. 2016, 6, 22846. [Google Scholar] [CrossRef]
- Wu, L.; Li, H.; Sun, L.; Gao, X.; Qui, D.; Sun, Y.; Wang, X.; Murray, T.D.; Li, H. Characterization of resistance to the cereal cyst nematode in the soft white wheat ‘Madsen’. Plant Dis. 2016, 100, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Petitot, A.-S.; Kyndt, T.; Haidar, R.; Dereeper, A.; Collin, M.; Engler, J.A.; Gheysen, G.; Fernandez, D. Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 2017, 119, 885–899. [Google Scholar] [CrossRef]
- Smiley, R.D.; Dababat, A.A.; Iqbal, S.; Jones, M.G.K.; Maafi, Z.T.; Peng, D.; Subbotin, S.A.; Waeyenberge, L. Cereal cyst nematodes: A complex and destructive group of Heterodera species. Plant Dis. 2017, 101, 1692–1720. [Google Scholar] [CrossRef] [PubMed]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Goto, D.B.; Miyazama, H.; Mar, J.C.; Sato, M. Not to be suppressed? Rethinking the host response at a root-parasite interface. Plant Sci. 2013, 213, 9–17. [Google Scholar] [CrossRef]
- Bohlman, H.; Sobczak, M. The plant cell wall in the feeding sites of cyst nematodes. Front. Plant Sci. 2014, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, T.; Fernandez, D.; Gheysen, G. Plant-parasitic nematode infections in rice: Molecular and cellular insights. Annu. Rev. Phytopathol. 2014, 52, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Hane, J.K.; Stoll, T.; Nicholas Pain, N.; Hastie, M.L.; Kau, P.; Hoogland, C.; Gorman, J.J.; Singh, K.B. Proteomic analysis of Rhizoctonia solani identifies infection-specific, redox associated proteins and insight into adaptation to different plant hosts. Mol. Cell. Proteomics 2016, 15, 1188–1203. [Google Scholar] [CrossRef]
- Holbein, J.; Grundler, F.M.W.; Siddique, S. Plant basal resistance to nematodes: An update. J. Exp. Bot. 2016, 67, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Dababat, A.A.; Ferney, G.-B.H.; Erginbas-Orakci, G.; Dreisigacker, S.; Imren, M.; Toktay, H.; Elekcioglu, H.I.; Mekete, T.; Nicol, J.M.; Ansari, O.; et al. Association analysis of resistance to cereal cyst nematodes (Heterodera avenae) and root lesion nematodes (Pratylenchus neglectus and P. thornei) in CIMMYT advanced spring wheat lines for semi-arid conditions. Breed. Sci. 2016, 66, 692–702. [Google Scholar] [CrossRef]
- IRIN News. 2010. Available online: http://www.irinnews.org/report/91012/asia-key-facts-about-rice (accessed on 11 February 2019).
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-induced gene silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Crops, Barley. 2016. Available online: http://www.fao.org/faostat/ (accessed on 11 February 2019).
- Danchin, E.G.D.; Arguel, M.-J.; Campan-Fournier, A.; Perfus-Barbeoch, L.; Marc Magliano, M.; Rosso, M.-N.; Da Rocha, M.; Da Silva, C.; Nottet, N.; Labadie, K.; Guy, J.; et al. Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathog. 2013, 9, e1003745. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, P.K.; Mitchum, M.G. War of the worms: How plants fight underground attacks. Curr. Opin. Plant Biol. 2013, 16, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Shahzadi, M.; Zahoor, A.; Dababat, A.A.; Toktay, H.; Bakhsh, A.; Nawaz, M.A.; Li, H. Resistance to cereal cyst nematodes in wheat and barley: An emphasis on classical and modern approaches. Int. J. Mol. Sci. 2019, 20, 432. [Google Scholar] [CrossRef]
- Siddique, S.; Grundler, F.M.W. Parasitic nematodes manipulate plant development to establish feeding sites. Curr. Opin. Microbiol. 2018, 46, 102–108. [Google Scholar] [CrossRef]
- Aditya, J.; Lewis, J.; Shirley, N.J.; Tan, H.-T.; Henderson, M.; Fincher, G.B.; Burton, R.A.; Mather, D.E.; Tucker, M.R. The dynamics of cereal cyst nematode infection differ between susceptible and resistant barley cultivars and lead to changes in (1,3;1,4)-b-glucan levels and HvCslF gene transcript abundance. New Phytol. 2015, 207, 135–147. [Google Scholar] [CrossRef]
- Zheng, M.; Long, H.; Zhao, Y.; Li, L.; Xu, D.; Zhang, H.; Liu, F.; Deng, G.; Pan, Z.; Yu, M. RNA-seq based identification of candidate parasitism genes of cereal cyst nematode (Heterodera avenae) during incompatible infection to Aegilops variabilis. PLoS ONE 2015, 10, e0141095. [Google Scholar] [CrossRef]
- Zhan, L.-P.; Peng, D.-L.; Wang, X.-L.; Kong, L.-A.; Peng, H.; Liu, S.-M.; Liu, Y.; Huang, W.-K. Priming effect of root-applied silicon on the enhancement of induced resistance to the root-knot nematode Meloidogyne graminicola in rice. BMC Plant Biol. 2018, 18, 50. [Google Scholar] [CrossRef]
- Thompson, A.L.; Mahoney, A.K.; Smiley, R.W.; Paulitz, T.C.; Hulbert, S.; Garland-Campbell, K. Resistance to multiple soil-borne pathogens of the Pacific Northwest, USA is colocated in a wheat recombinant inbred line population. G3 Genes Genomes Genet. 2017, 7, 1109–1116. [Google Scholar] [CrossRef]
- Smiley, R.D. Root-lesion nematodes reduce yield of intolerant wheat and barley. Agron. J. 2009, 101, 1322–1335. [Google Scholar] [CrossRef]
- Zhang, L.; Lilley, C.J.; Imren, M.; Knox, J.P.; Urwin, P.E. The complex cell wall composition of syncytia induced by plant parasitic cyst nematodes reflects both function and host plant. Front. Plant Sci. 2017, 8, 1087. [Google Scholar] [CrossRef]
- Fosu-Nyarko, J.; Jones, M.G.K. Advances in understanding the molecular mechanisms of root lesion nematode host interactions. Annu. Rev. Phytopathol. 2016, 54, 253–278. [Google Scholar] [CrossRef]
- Kim, D.-W.; Kim, G.-Y.; Kim, H.-K.; Kim, J.; Jeon, S.J.; Lee, C.W.; Lee, H.B.; Yun, S.-H. Characterization of nivalenol-producing Fusarium culmorum isolates obtained from the air at a rice paddy field in Korea. Plant Pathol. J. 2016, 32, 182–189. [Google Scholar] [CrossRef]
- Pasquali, M.; Spanu, F.; Scherm, B.; Balmas, V.; Hoffmann, L.; Hammond-Kosack, K.E.; Beyer, M.; Migheli, Q. FcStuA from Fusarium culmorum controls wheat foot and root rot in a toxin dispensable manner. PLoS ONE 2013, 8, e57429. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Qian, L.; Gabur, I.; Obermeier, C.; Hickey, L.T.; Werner, C.R.; Kontowski, S.; Frisch, M.; Friedt, W.; Snowdon, R.J.; et al. Genetic insights into underground responses to Fusarium graminearum infection in wheat. Sci. Rep. 2018, 8, 13153. [Google Scholar] [CrossRef]
- Paulitz, T.C. Low input no-till cereal production in the Pacific Northwest of the US: The challenges of root diseases. Eur. J. Plant Pathol. 2006, 115, 271–281. [Google Scholar] [CrossRef]
- NASS (National Agricultural Statistics Service). Data and Statistics for Washington State Field Crops. Available online: https://www.nass.usda.gov/Statistics_by_State/Washington/index.php (accessed on 13 February 2019).
- Jaaffar, A.K.M.; Paulitz, T.C.; Schroeder, K.L.; Thomashow, L.S.; Weller, D.M. Molecular characterization, morphological characteristics, virulence, and geographic distribution of Rhizoctonia spp. in Washington State. Phytopathology 2016, 106, 459–473. [Google Scholar] [CrossRef]
- Beccari, G.; Covarelli, L.; Nicholson, P. Infection processes and soft wheat response to root rot and crown rot caused by Fusarium culmorum. Plant Pathol. 2011, 60, 671–684. [Google Scholar] [CrossRef]
- Gasparis, S.; Kała, M.; Przyborowski, M.; Łyżnik, L.A.; Orczyk, W.; Nadolska-Orczyk, A. A simple and efficient CRISPR/Cas9 platform for induction of single and multiple, heritable mutations in barley (Hordeum vulgare L.). Plant Methods 2018, 14, 111. [Google Scholar] [CrossRef]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef]
- Lawrenson, T.; Harwood, W.A. Creating targeted gene knockouts in barley using CRISPR/Cas9. Methods Mol. Biol. 2019, 1900, 217–232. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Wang, K. Generation of marker-free transgenic rice using CRISPR/Cas9 system controlled by floral specific promoters. J. Genet. Genomics 2019, 46, 61–64. [Google Scholar] [CrossRef]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Tanaka, K.; Choi, J.; Cao, Y.; Stacey, G. Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 2014, 5, 446. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I.9 is a novel phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef]
- Bouwmeester, K.; Han, M.; Blanco-Portales, R.; Song, W.; Weide, R.; Guo, L.-Y.; van der Vossen, E.A.G.; Govers, F. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol. J. 2014, 12, 10–16. [Google Scholar] [CrossRef]
- Balagué, C.; Gouge, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signaling. Mol. Plant Biol. 2017, 18, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Zhang, T.; Koo, A.J.; Stacey, G.; Tanaka, K. Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 2018, 176, 511–523. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Liang, Y.; Cao, Y.; Lee, S.Y.; Stacey, G. Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis. Biochem. J. 2014, 463, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Cao, Y.; Li, H.; Kim, D.; Ahsan, N.; Thelen, J.; Stacey, G. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.A.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef]
- Khazaei, H.; Street, K.; Bari, A.; Mackay, M.; Stoddard, F.L. The FIGS (Focused Identification of Germplasm Strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 2013, 8, e63107. [Google Scholar] [CrossRef]
- Marone, D.; Panio, G.; Ficco, D.B.M.; Russo, M.A.; De Vita, P.; Papa, R.; Rubiales, D.; Cattivelli, L.; Mastrangelo, A.M. Characterization of wheat DArT markers: Genetic and functional features. Mol. Genet. Genomics 2012, 287, 741–753. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Eilam, T.; Sela, H.; Alekperov, C.; Peleg, Z.; Dvorak, J.; Korol, A.; Distelfeld, A. Ultra-dense genetic map of durum wheat x wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol. Breed. 2014, 34, 1549–1562. [Google Scholar] [CrossRef]
- Wenzl, P.; Carling, J.; Kudrna, D.; Jaccoud, D.; Huttner, E.; Kleinhofs, A.; Kilian, A. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 2004, 101, 9915–9920. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M.; et al. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- Phan, N.T.; De Waele, D.; Lorieux, M.; Xiong, L.; Bellafiore, S. A hypersensitivity-like response to Meloidogyne graminicola in rice (Oryza sativa). Phytopathology 2018, 108, 521–528. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Rafiqul Islam, M.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Pariyar, S.R.; Dababat, A.A.; Sannemann, W.; Erginbas-Orakci, G.; Elashry, A.; Siddique, S.; Morgounov, A.; Leon, J.; Grundler, F.M.W. Genome-wide association study in wheat identifies resistance to the cereal cyst nematode Heterodera filipjevi. Phytopathology 2016, 106, 1128–1138. [Google Scholar] [CrossRef]
- Galal, A.; Sharma, S.; Abou-Elwafa, S.F.; Sharma, S.; Kopisch-Obuch, F.; Laubach, E.; Perovic, D.; Ordon, F.; Jung, C. Comparative QTL analysis of root lesion nematode resistance in barley. Theor. Appl. Genet. 2014, 127, 1399–1407. [Google Scholar] [CrossRef]
- Thompson, A.L.; Smiley, R.W.; Paulitz, T.C.; Garland-Campbell, K. Identification of resistance to Pratylenchus neglectus and Pratylenchus thornei in Iranian landrace accessions of wheat. Crop Sci. 2016, 56, 654–672. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Steiner, B.; Hartl, L.; Griesser, M.; Angerer, N.; Lengauer, D.; Miedaner, T.; Schneider, B.; Lemmens, M. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 2003, 107, 503–508. [Google Scholar] [CrossRef]
- Okubara, P.A.; Jones, S.S. Seedling resistance to Rhizoctonia and Pythium in wheat chromosome group 4 addition lines from Thinopyrum spp. Can. J. Plant Pathol. 2011, 33, 415–422. [Google Scholar] [CrossRef]
- Okubara, P.A.; Dickman, M.B.; Blechl, A.E. Molecular and genetic aspects of controlling the soilborne necrotrophic pathogens Rhizoctonia and Pythium. Plant Sci. 2014, 128, 61–70. [Google Scholar] [CrossRef]
- Mahoney, A.; Babiker, E.; Paulitz, T.; See, D.; Okubara, P.; Hulbert, S. Characterizing and mapping resistance in synthetic-derived wheat to Rhizoctonia root rot in a green bridge environment. Phytopathology 2016, 106, 1170–1176. [Google Scholar] [CrossRef]
- Mahoney, A.K.; Babiker, E.M.; See, D.R.; Paulitz, T.C.; Okubara, P.A.; Hulbert, S.H. Analysis and mapping of Rhizoctonia root rot resistance traits from the synthetic wheat (Triticum aestivum L.) line SYN-172. Mol. Breed. 2017, 37, 130. [Google Scholar] [CrossRef]
- Warr, A.; Robert, C.; Hume, D.; Archibald, A.; Deeb, N.; Watson, M. Exome sequencing: Current and future perspectives. G3 Genes Genomes Genet. 2015, 5, 1543–1550. [Google Scholar] [CrossRef]
- Kyndt, T.; Nahar, K.; Haegeman, A.; De Vleesschauwer, D.; Hofte, M.; Gheysen, G. Comparing systemic defence-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biol. 2012, 14 (Suppl. 1), 73–82. [Google Scholar] [CrossRef]
- Ji, H.; Gheysen, G.; Denil, S.; Lindsey, K.; Topping, J.F.; Nahar, K.; Haegeman, A.; De Vos, W.H.; Trooskens, G.; Van Criekinge, W.; et al. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 2013, 64, 3885–3898. [Google Scholar] [CrossRef]
- Lagudah, E.S.; Moullet, O.; Appels, R. Map-based cloning of a gene sequence encoding a nucleotide-binding domain and a leucine-rich region at the Cre3 nematode resistance locus of wheat. Genome 1997, 40, 659–665. [Google Scholar] [CrossRef]
- Cortese, M.R.; Fanelli, E.; De Giorgi, C. Characterization of nematode resistance gene analogs in tetraploid wheat. Plant Sci. 2003, 164, 71–75. [Google Scholar] [CrossRef]
- Simonetti, E.; Alba, E.; Montes, M.J.; Delibes, A.; López-Braña, I. Analysis of ascorbate peroxidase genes expressed in resistant and susceptible wheat lines infected by the cereal cyst nematode, Heterodera avenae. Plant Cell Rep. 2010, 29, 1169–1178. [Google Scholar] [CrossRef]
- Barloy, D.; Lemoine, J.; Abelard, P.; Tanguy, A.M.; Rivoal, R.; Jahier, J. Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol. Breed. 2007, 20, 31–40. [Google Scholar] [CrossRef]
- Huang, Q.; Li, L.; Zheng, M.; Chen, F.; Long, H.; Deng, G.; Pan, Z.; Liang, J.; Li, Q.; Yu, M.; et al. The tryptophan decarboxylase 1 gene from Aegilops variabilis no.1 regulate the resistance against cereal cyst nematode by altering the downstream secondary metabolite contents rather than auxin synthesis. Front. Plant Sci. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Pelagio-Flores, R.; Ortíz-Castro, R.; Méndez-Bravo, A.; Macías-Rodríguez, L.; López-Bucio, J. Serotonin, a tryptophan-derived signal conserved in plants and animals, regulates root system architecture probably acting as a natural auxin inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 490–508. [Google Scholar] [CrossRef]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell Rep. 2007, 26, 2009–2015. [Google Scholar] [CrossRef]
- Motallebi, P.; Tonti, S.; Niknam, V.; Ebrahimzadeh, H.; Pisi, A.; Nipoti, P.; Hashemi, M.; Prodi, A. Induction of basal resistance by methyl jasmonate against Fusarium culmorum in bread wheat. Cereal Res. Commun. 2017, 45, 248–259. [Google Scholar] [CrossRef]
- Kyndt, T.; Denil, S.; Haegeman, A.; Trooskens, G.; De Meyer, T.; Van Criekinge, W.; Gheysen, G. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytol. 2012, 196, 887–900. [Google Scholar] [CrossRef]
- Thompson, A.L.; Smiley, R.W.; Garland-Campbell, K. Registration of the LouAu (Louise/IWA8608077) wheat recombinant inbred line mapping population. J. Plant Regist. 2015, 9, 424–429. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.-G.; Muchero, W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef]
- Kong, L.-A.; Wu, D.-Q.; Huang, W.-K.; Peng, H.; Wang, G.-F.; Cui, J.-K.; Liu, S.-M.; Li, Z.-G.; Yang, J.; Peng, D.-L. Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera Biolavenae using comparative transcriptomic analysis. BMC Genomics 2015, 16, 801. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signaling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Qiao, F.; Kong, L.-A.; Peng, H.; Huang, W.-K.; Wu, D.-Q.; Liu, S.-M.; Clarke, J.L.; Qiu, D.-W.; Peng, D.-L. Transcriptional profiling of wheat (Triticum aestivum L.) during a compatible interaction with the cereal cyst nematode Heterodera avenae. Sci. Rep. 2019, 9, 2184. [Google Scholar] [CrossRef]
- Maketon, C.; Fortuna, A.-M.; Okubara, P. Cultivar-dependent transcript accumulation in wheat roots colonized by Pseudomonas fluorescens Q8r1-96 wild type and mutant strains. Biol. Control 2012, 60, 216–224. [Google Scholar] [CrossRef]
- Chen, C.; Cui, L.; Chen, Y.; Zhang, H.; Liu, P.; Wu, P.; Qiu, D.; Zou, J.; Yang, D.; Yang, L.; et al. Transcriptional responses of wheat and the cereal cyst nematode Heterodera avenae during their early contact stage. Sci. Rep. 2017, 7, 14471. [Google Scholar] [CrossRef]
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and Genomic Analysis of Rhizoctonia solani interactions with Arabidopsis; evidence of resistance mediated through NADPH oxidases. PLoS ONE 2013, 8, e56814. [Google Scholar] [CrossRef]
- Guo, G.; Liu, Q.; Smith, N.A.; Liang, G.; Wang, M.-B. RNA silencing in plants: Mechanisms, technologies and applications in horticultural crops. Curr. Genomics 2016, 17, 476–489. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.-H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi—Natures blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Haegeman, A.; Mantelin, S.; Jones, J.T.; Gheysen, G. Functional roles of effectors of plant-parasitic nematodes. Gene 2012, 492, 19–31. [Google Scholar] [CrossRef]
- Presti, L.L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Tan, J.C.H.; Jones, M.G.K.; Fosu-Nyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 2013, 133, 166–178. [Google Scholar] [CrossRef]
- Chen, C.; Liu, S.; Liu, Q.; Niu, J.; Liu, P.; Zhao, J.; Jian, H. An ANNEXIN like protein from the cereal cyst nematode Heterodera avenae suppresses plant defense. PLoS ONE 2015, 10, e0122256. [Google Scholar] [CrossRef]
- Yang, S.; Dai, Y.; Chen, Y.; Yang, J.; Yang, D.; Liu, Q.; Jian, H. Heterodera avenae suppresses plant defenses and promotes parasitism. Front. Plant Sci. 2019, 10, 66. [Google Scholar] [CrossRef]
- Vieira, P.; Eves-van den Akker, S.; Verma, R.; Wantoch, S.; Eisenback, J.D.; Kamo, K. The Pratylenchus penetrans transcriptome as a source for the development of alternative control strategies: Mining for putative genes involved in parasitism and evaluation of in planta RNAi. PLoS ONE 2015, 10, e0144674. [Google Scholar] [CrossRef]
- Smiley, R.; Paulitz, T.; Marshall, J. Controlling Root and Crown Diseases of Small Grain Cereals. PNW 639. Pacific Northwest Extension Publication, 2012; pp. 1–9. Available online: http://smallgrains.wsu.edu/wp-content/uploads/2013/10/Smiley_12_PNW639_RootDis.pdf (accessed on 2 February 2019).
- Thomson, J.; Bertram, L. Crop Damaging Nematodes Found in New Area. Government of Western Australia, Department of Primary Industries and Regional Development, 2014. Available online: https://www.agric.wa.gov.au/news/media-releases/crop-damaging-nematodes-found-new-area (accessed on 2 February 2019).
- Mokrini, F.; Viaene, N.; Waeyenberge, L.; Dababat, A.A.; Moens, M. Investigation of resistance to Pratylenchus penetrans and P. thornei in international wheat lines and its durability when inoculated together with the cereal cyst nematode Heterodera avenae, using qPCR for nematode quantification. Eur. J. Plant Pathol. 2018, 151, 875–889. [Google Scholar] [CrossRef]
- Haegeman, A.; Bauters, L.; Kyndt, T.; Rahman, M.M.; Gheysen, G. Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2013, 14, 379–390. [Google Scholar] [CrossRef]
- Pallarès, I.; de Groot, N.S.; Iglesias, V.; Sant’Anna, R.; Biosca, A.; Fernàndez-Busquets, X.; Ventura, S. Discovering putative prion-like proteins in Plasmodium falciparum: A computational and experimental analysis. Front. Microbiol. 2018, 9, 1737. [Google Scholar] [CrossRef]
- Kormish, J.D.; McGhee, J.D. The C. elegans lethal gut-obstructed gob-1 gene is trehalose-6-phosphate phosphatase. Dev. Biol. 2005, 287, 35–47. [Google Scholar] [CrossRef]
- Chen, J.; Lin, B.; Huang, Q.; Hu, L.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 2017, 13, e1006301. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, N.; Liu, J.; Liu, W.; Wang, G.-L. The role of effectors and host immunity in plant–necrotrophic fungal interactions. Virulence 2014, 5, 722–732. [Google Scholar] [CrossRef]
- Castiblanco, V.; Elena Castillo, H.E.; Miedaner, T. Candidate genes for aggressiveness in a natural Fusarium culmorum population greatly differ between Wheat and Rye Head Blight. J. Fungi 2018, 4, 14. [Google Scholar] [CrossRef]
- Castiblanco, V.; Marulanda, J.J.; Würschum, T.; Miedaner, T. Candidate gene based association mapping in Fusarium culmorum for field quantitative pathogenicity and mycotoxin production in wheat. BMC Genet. 2017, 18, 49. [Google Scholar] [CrossRef]
- Lysøe, E.; Pasquali, M.; Breakspear, A.; Kistler, H.C. The transcription factor FgStuAp influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum. Mol. Plant-Microbe Interact. 2011, 24, 54–56. [Google Scholar] [CrossRef]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef]
- Yan, G.; Smiley, R.W.; Okubara, P.A.; Skantar, A.; Easley, S.A.; Sheedy, J.G.; Thompson, A.L. Detection and discrimination of Pratylenchus neglectus and P. thornei in DNA extracts from soil. Plant Dis. 2008, 92, 1480–1487. [Google Scholar] [CrossRef]
- Kidwell, K.K.; Shelton, G.B.; Morris, C.F.; Line, R.F.; Miller, B.C.; Davis, M.A.; Konzak, C.F. Registration of ‘Scarlet’ wheat. Crop Sci. 1999, 39, 1255. [Google Scholar] [CrossRef]
- Mueller, O.; Lightfoot, S.; Schroeder, A. RNA integrity number (RIN)—Standardization of RNA quality control. Agilent Appl. Note 2004, 1–8, Publication Number 5989-1165EN. Available online: https://www.agilent.com/cs/library/applications/5989-1165EN.pdf (accessed on 5 January 2019).
- Compeau, P.E.C.; Pevzner, P.A.; Tesler, G. How to apply de Bruijn graphs to genome Assembly. Nat. Biotechnol. 2011, 29, 987–991. [Google Scholar] [CrossRef]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Okubara, P.A.; Steber, C.M.; DeMacon, V.L.; Walter, N.L.; Paulitz, T.C.; Kidwell, K.K. EMS-treated hexaploid wheat genotype Scarlet has enhanced tolerance to the soilborne necrotrophic pathogens Rhizoctonia solani AG-8 and R. oryzae. Theor. Appl. Genet. 2009, 119, 293–303. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Tanaka, K.; Cao, Y.; Cho, S.H.; Xu, D.; Stacey, G. Computational analysis of the ligand binding site of the extracellular ATP receptor, DORN1. PLoS ONE 2016, 11, e0161894. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a Hidden Markov Model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B., Jr. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments; Pittsburgh Supercomputing Center’s National Resource for Biomedical Supercomputing: Pittsburgh, PA, USA, 1997. [Google Scholar]
- Jewell, J.B.; Sowders, J.M.; He, R.; Willis, M.A.; Gang, D.R.; Tanaka, K. Extracellular ATP shapes a defense-related transcriptome both independently and along with other defense signaling pathways. Plant Physiol. 2019, 179. [Google Scholar] [CrossRef]
- Okubara, P.A.; Call, D.R.; Kwak, Y.; Skinner, D.Z. Induction of defense gene homologues in wheat roots during interactions with Pseudomonas fluorescens. Biol. Control 2010, 55, 118–125. [Google Scholar] [CrossRef]
- Koch, A.; Kogel, K.-H. New wind in the sails: Improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 2014, 12, 821–831. [Google Scholar] [CrossRef]
- Sahoo, K.K.; Tripathi, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 2011, 7, 49. [Google Scholar] [CrossRef]
- Ozawa, K. A High-efficiency Agrobacterium -mediated transformation system of rice (Oryza sativa L.). Methods Mol. Biol. 2012, 847, 51–57. [Google Scholar] [CrossRef]
- Imani, J.; Li, L.; Schäfer, P.; Kogel, K.-H. STARTS—A stable root transformation system for rapid functional analyses of proteins of the monocot model plant barley. Plant J. 2011, 67, 726–735. [Google Scholar] [CrossRef]
- Barros, J.; Serrani-Yarce1, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Mascher, M.; Richmond, T.A.; Gerhardt, D.J.; Himmelbach, A.; Clissold, L.; Sampath, D.; Ayling, S.; Steuernagel, B.; Pfeifer, M.; D’Ascenzo, M.; et al. Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. 2013, 76, 494–505. [Google Scholar] [CrossRef]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H.; et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 2015, 26, 1382–1397. [Google Scholar] [CrossRef]
- King, R.; Bird, N.; Ramirez-Gonzalez, R.; Coghill, J.A.; Patil, A.; Hassani-Pak, K.; Uauy, C.; Phillips, A.L. Mutation scanning in wheat by exon capture and next-generation sequencing. PLoS ONE 2015, 10, e0137549. [Google Scholar] [CrossRef]
- Okubara, P.A.; Pawlowski, K.; Murphy, T.M.; Berry, A.M. Symbiotic roots nodules of the actinorhizal plant Datisca glomerata express rubisco activase mRNA. Plant Physiol. 1999, 120, 411–420. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).