Screening of Diverse Ethiopian Durum Wheat Accessions for Aluminum Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Germplasm
2.2. Hydroponic Culture
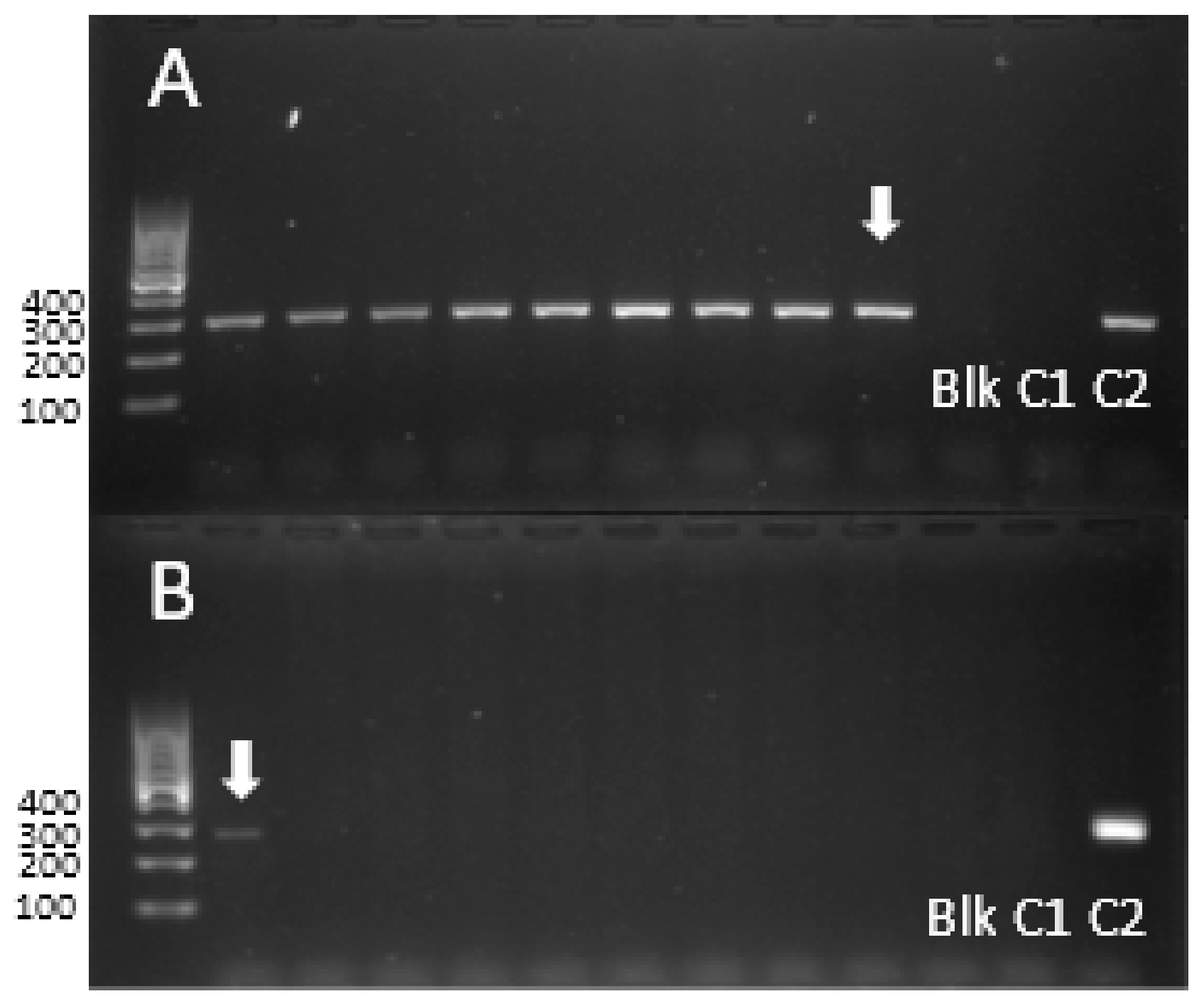
2.3. Determining the Identity of the Al3+-Tolerant Accessions
2.4. Field Experiment
2.5. Statistical Analysis
3. Results
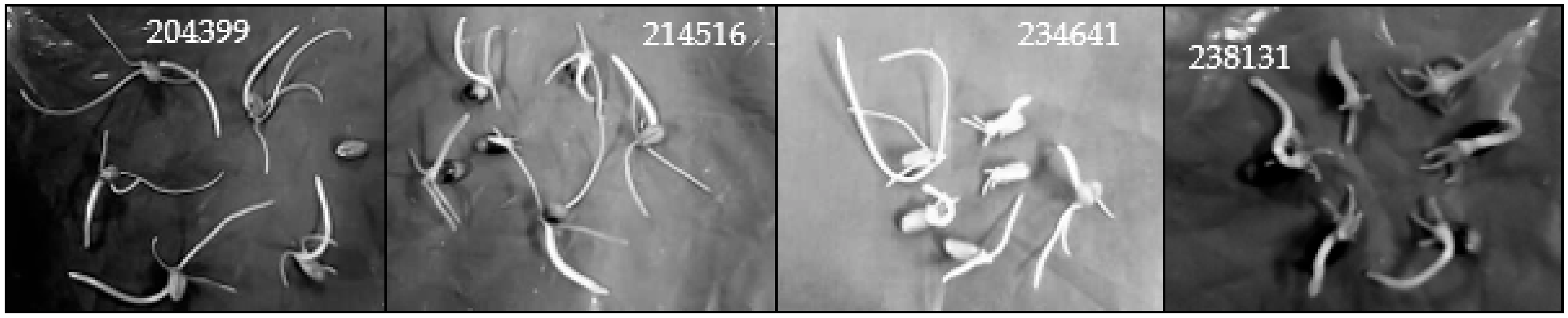
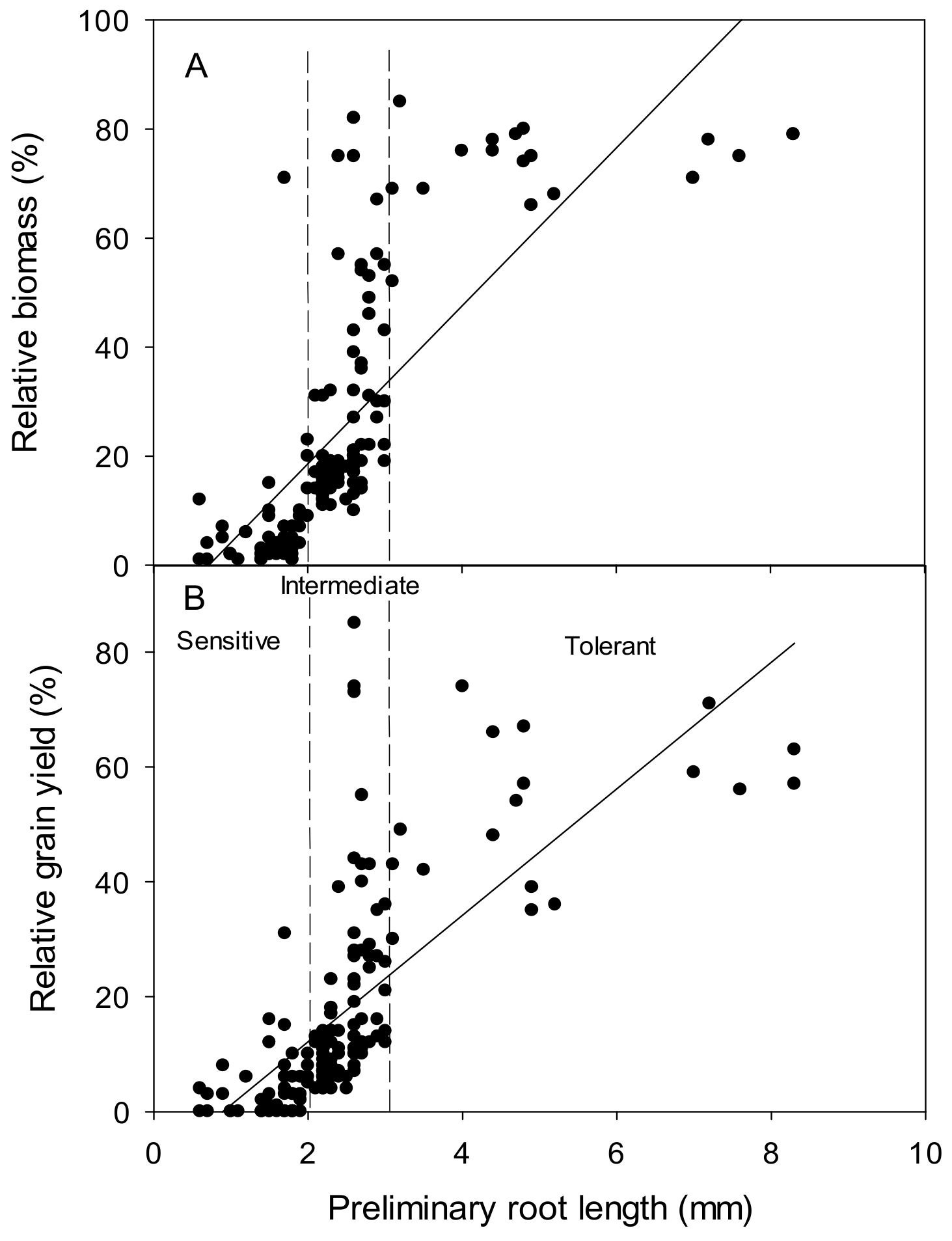
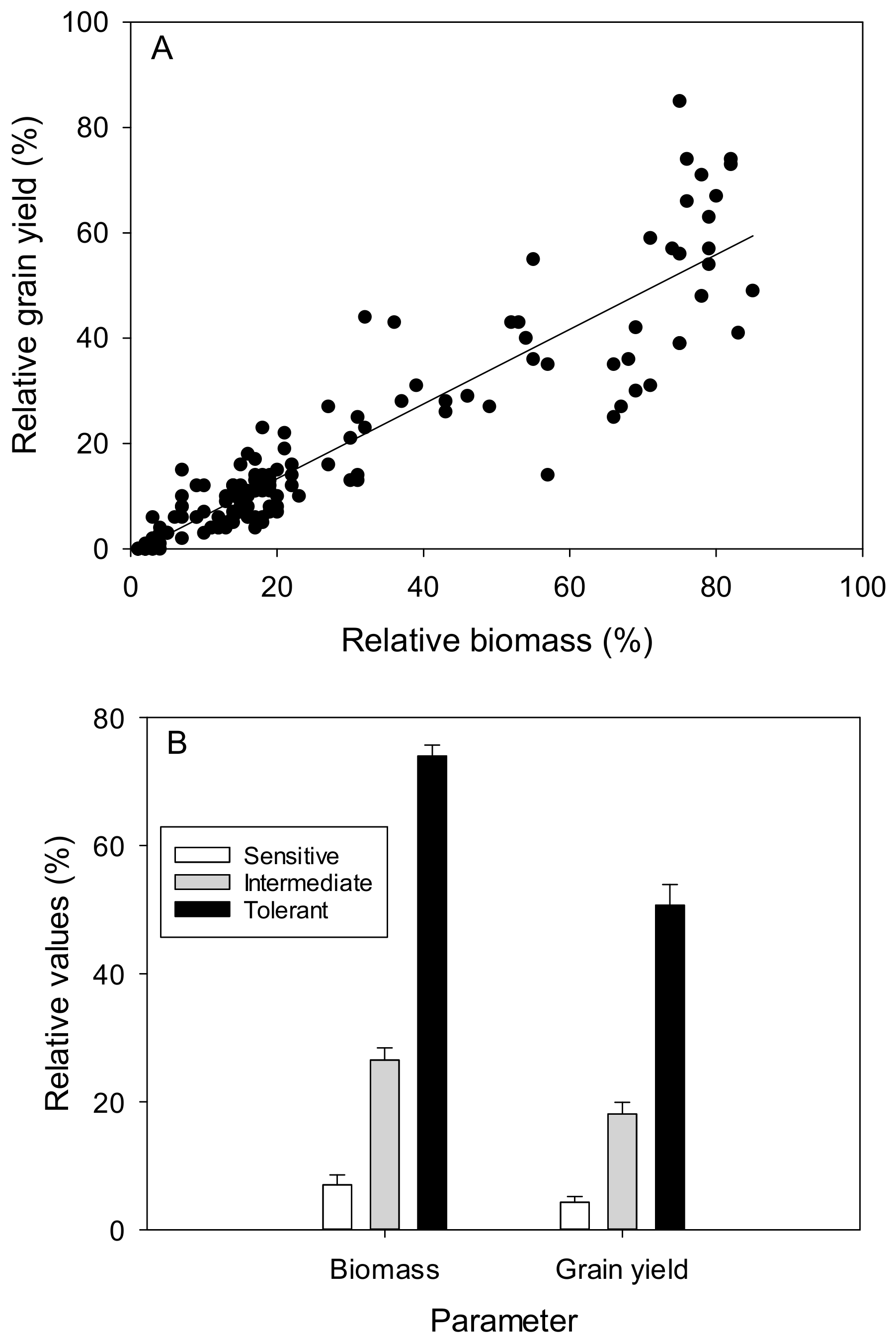
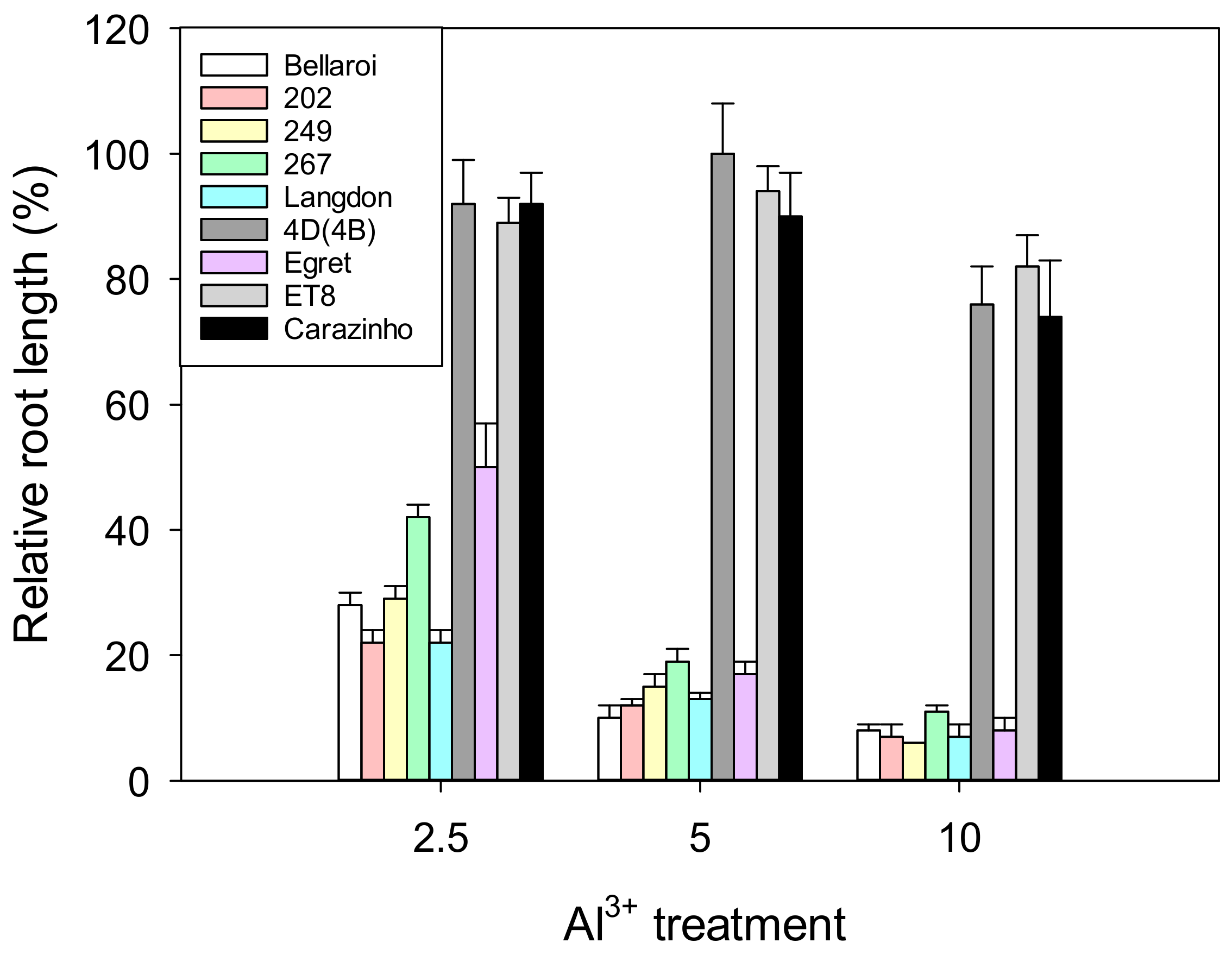
3.1. Hydroponic Screen
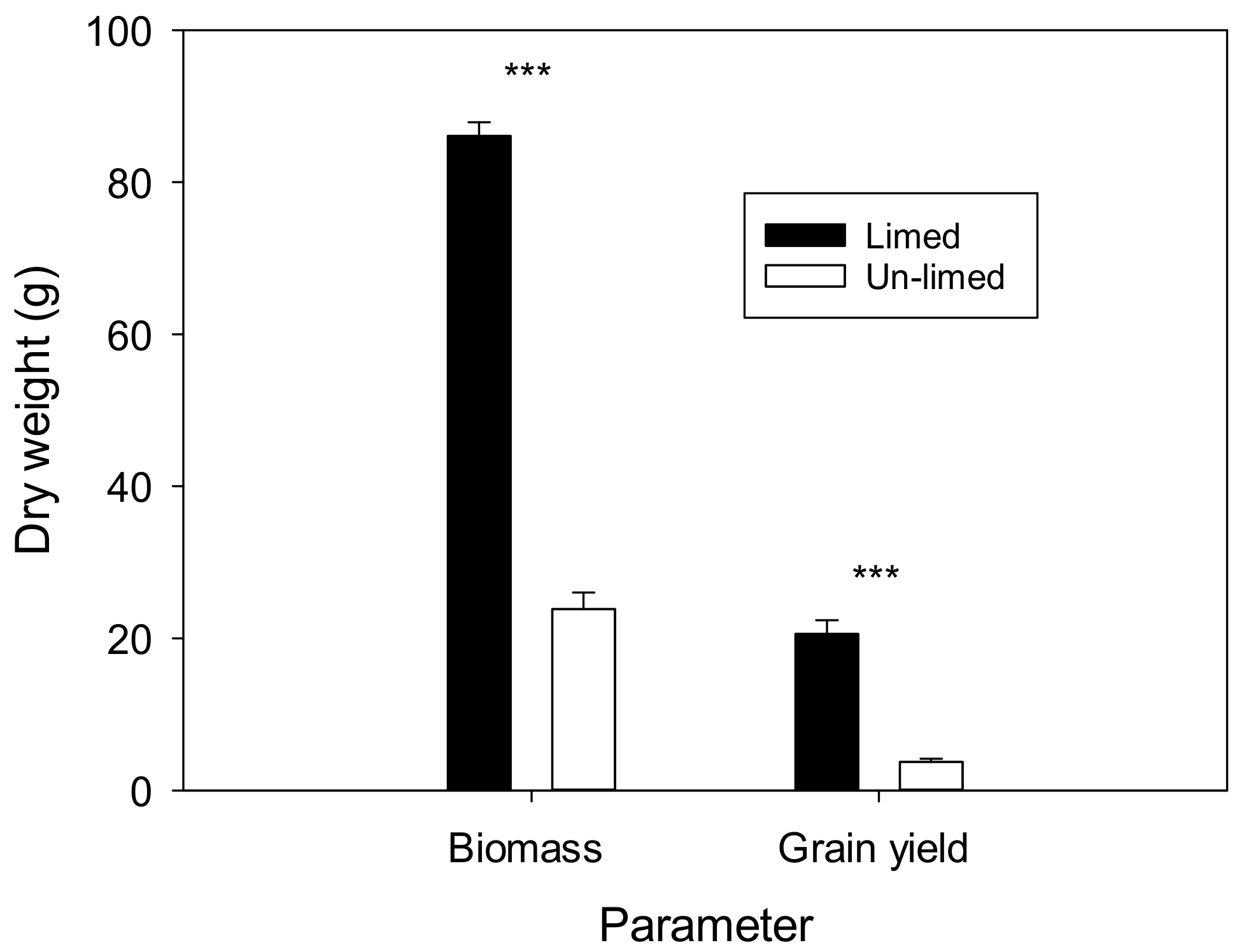
3.2. Field Experiment
3.3. Other Potential Sources of Al3+-Tolerant Durum Wheat
4. Discussion
4.1. A Rapid Screen Proves Robust and Correlates with Grain Yield in Field Trials when Ranking Wheat Germplasm for Al3+ Tolerance
4.2. The Evolution of Al3+ Tolerance in Bread Wheat Occurred Subsequent to the Hybridization of the D-Genome
4.3. Strategies to Enhance the Al3+ Tolerance of Ethiopian Durum Wheat
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Minot, N.; Warner, J.; Lemma, S.; Kasa, L.; Gashaw, A.; Rashid, S. The Wheat Supply Chain in Ethiopia: Patterns, Trends, and Policy Options; International Food Policy Research Institute (IFPRI): Washington, DC, USA, 2015. [Google Scholar]
- Sall, A.T.; Chiari, T.; Legesse, W.; Seid-Ahmed, K.; Ortiz, R.; van Ginkel, M.; Bassi, M.F. Durum wheat (Triticum durum Desf.): Origin, cultivation and potential expansion in sub-Saharan Africa. Agronomy 2019, 9, 263. [Google Scholar] [CrossRef]
- Bona, L.; Wright, R.J.; Baligar, V.C. A rapid method for screening cereals for acid soil tolerance. Cereal Res. Commun. 1991, 19, 465–468. [Google Scholar]
- Raman, H.; Ryan, P.R.; Raman, R.; Stodart, B.J.; Zhang, K.; Martin, P.; Wood, R.; Sasaki, T.; Yamamoto, Y.; Mackay, M.; et al. Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2008, 116, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, J.G.; Minozzo, J.A.D.; Barichello, D.; Fogaca, C.M.; da Silva, J.P.; Consoli, L.; Pereira, J.F. Alleles of organic acid transporter genes are highly correlated with wheat resistance to acidic soil in field conditions. Theor. Appl. Genet. 2016, 129, 1317–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raman, H.; Stodart, B.; Ryan, P.R.; Delhaize, E.; Emebiri, L.; Raman, R.; Coombes, N.; Milgate, A. Genome-wide association analyses of common wheat (Triticum aestivum L.) germplasm identifies multiple loci for aluminium resistance. Genome 2010, 53, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Von Uexküll, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Wayima, E.F. Classification of Ethiopian soils with pH. J. Soil Sci. Environ. Manag. 2019, in press. [Google Scholar]
- Kidanemariam, A.; Gebrekidan, H.; Mamo, T.; Kibret, K. Impact of altitude and land use type on some physical and chemical properties of acidic soils in Tsegede highlands, northern Ethiopia. Open J. Soil Sci. 2012, 2, 223–233. [Google Scholar] [CrossRef]
- Kidanemariam, A.; Gebrekidan, H.; Mamo, T.; Tesfaye, K. Wheat crop response to liming materials and N and P fertilizers in acidic soils of Tsegede highlands, northern Ethiopia. Agric. For. Fish. 2013, 2, 126–135. [Google Scholar] [CrossRef]
- Mosissa, F. Progress of soil acidity management research in Ethiopia. Greener J. Soil Sci. Plant Nutr. 2018, 5, 9–22. [Google Scholar] [CrossRef]
- Kochian, L.V.; Pineros, M.A.; Liu, J.P.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Ann. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.V.; Garvin, D.F.; Wang, Y.H.; Sorrells, M.E.; Klein, P.E.; Schaffert, R.E.; Li, L.; Kochian, L.V. Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 2004, 167, 1905–1914. [Google Scholar] [CrossRef] [PubMed]
- Bian, M.; Waters, I.; Broughton, S.; Zhang, X.Q.; Zhou, M.X.; Lance, R.; Sun, D.F.; Li, C.D. Development of gene-specific markers for acid soil/aluminium tolerance in barley (Hordeum vulgare L.). Mol. Breed. 2013, 32, 155–164. [Google Scholar] [CrossRef]
- Ma, J.F.; Nagao, S.; Sato, K.; Ito, H.; Furukawa, J.; Takeda, K. Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley. J. Exp. Bot. 2004, 55, 1335–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, P.R.; Raman, H.; Gupta, S.; Sasaki, T.; Yamamoto, Y.; Delhaize, E. The multiple origins of aluminium resistance in hexaploid wheat include Aegilops tauschii and more recent cis mutations to TaALMT1. Plant J. 2010, 64, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Foy, C.D. Tolerance of durum wheat lines to an acid, aluminum-toxic subsoil. J. Plant Nutr. 1996, 19, 1381–1394. [Google Scholar] [CrossRef]
- Han, C.; Ryan, P.R.; Yan, Z.; Delhaize, E. Introgression of a 4D chromosomal fragment into durum wheat confers aluminium tolerance. Ann. Bot. 2014, 114, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Delhaize, E.; Ma, J.F.; Ryan, P.R. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012, 17, 341–348. [Google Scholar] [CrossRef]
- Han, C.; Zhang, P.; Ryan, P.R.; Rathjen, T.M.; Yan, Z.H.; Delhaize, E. Introgression of genes from bread wheat enhances the aluminium tolerance of durum wheat. Theor. Appl. Genet. 2016, 129, 729–739. [Google Scholar] [CrossRef]
- Garcia-Oliveira, A.L.; Martins-Lopes, P.; Tolra, R.; Poschenrieder, C.; Guedes-Pinto, H.; Benito, C. Differential physiological responses of Portuguese bread wheat (Triticum aestivum L.) genotypes under aluminium stress. Diversity 2016, 8, 26. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kidane, Y.G.; Fadda, C.; Pe, M.E. Genetic diversity in Ethiopian durum wheat (Triticum turgidum var durum) inferred from phenotypic variations. Plant Genet. Res. Charact. Util. 2018, 16, 39–49. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kiros, A.Y.; Pe, M.E. Phenotypic diversity in Ethiopian durum wheat (Triticum turgidum var. durum) landraces. Crop J. 2015, 3, 190–199. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pe, M.E.; Dell’Acqua, M. High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveals high diversity and potential for wheat breeding. Plant Biotechnol. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front. Plant Sci. 2017, 8, 1277. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Hare, R.; Graham, K.; Coombes, N.; Raman, R. Characterisation of durum germplasm for aluminium resistance using nutrient solution culture. In International Wheat Genetics Symposium; Appels, R., Lagudah, R.E.E., Langridge, P., Mackay, M., McIntyre, L., Sharp, P., Eds.; Sydney University Press: Brisbane, Australia, 2008; pp. 1–3. [Google Scholar]
- Joppa, L.R.; Williams, N.D. Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 1988, 30, 222–228. [Google Scholar] [CrossRef]
- Delhaize, E.; Ryan, P.R.; Hebb, D.M.; Yamamoto, Y.; Sasaki, T.; Matsumoto, H. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc. Natl. Acad. Sci. USA 2004, 101, 15249–15254. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.F.; Delhaize, E.; Zhou, M.X.; Ryan, P.R. The barley MATE gene, HvAACT1, increases citrate efflux and Al+3 tolerance when expressed in wheat and barley. Ann. Bot. 2013, 112, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.J.; Dixon, A.; Gale, M.D.; Wiseman, G. A PCR-based method for the detection of hexaploid bread wheat adulteration of durum wheat and pasta. J. Cereal Sci. 1998, 28, 135–145. [Google Scholar] [CrossRef]
- McNeil, D.; Lagudah, E.S.; Hohmann, U.; Appels, R. Amplification of DNA-sequences in wheat and its relatives—the Dgas44 and R350 families of repetitive sequences. Genome 1994, 37, 320–327. [Google Scholar] [CrossRef]
- Zeven, A.C.; Waninge, J. The presence of 3 groups of Scalavatis and other hexaploid bread wheat plants contaminating durum-wheat fields in Cyprus. Euphytica 1989, 43, 117–124. [Google Scholar] [CrossRef]
- Tovkach, A.; Ryan, P.R.; Richardson, A.E.; Lewis, D.C.; Rathjen, T.M.; Ramesh, S.; Tyerman, S.D.; Delhaize, E. Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol. 2013, 161, 880–892. [Google Scholar] [CrossRef]
- Pereira, J.F.; Ryan, P.R. The role of transposable elements in the evolution of aluminium resistance in plants. J. Exp. Bot. 2019, 70, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Kidane, Y.G.; Mancini, C.; Mengistu, D.K.; Frascaroli, E.; Fadda, C.; Pe, M.E.; Dell’Acqua, M. Genome wide association study to identify the genetic base of smallholder farmer preferences of durum wheat traits. Front. Plant Sci. 2017, 8, 1230. [Google Scholar] [CrossRef] [PubMed]
- Tsegaye, B.; Berg, T. Genetic erosion of Ethiopian tetraploid wheat landraces in Eastern Shewa, Central Ethiopia. Genet. Resour. Crop Evol. 2007, 54, 715–726. [Google Scholar] [CrossRef]
- Pooniya, V.; Palta, J.A.; Chen, Y.; Delhaize, E.; Siddique, K.H. Impact of the TaMATE1B gene on above and below-ground growth of durum wheat grown on an acid and Al3+-toxic soil. Plant Soil 2019, 1–12. [Google Scholar] [CrossRef]

| Soil Parameter | Value | |
|---|---|---|
| pH (water extract; 1:1.25) | 4.92 | |
| Buffer pH (water extract 1:2) | 5.57 | |
| Electrical conductivity (dS/m) | 0.07 | |
| Organic matter (%) | 4.01 | |
| Total N (%) | 0.78 | |
| Available P (mg/kg) | 8.87 | |
| Exchangeable acidity (meq/100 g) | 2.23 | |
| CEC and exchangeable bases (cmol (+)/kg) | CEC | 27.19 |
| Ca | 5.27 | |
| Mg | 0.66 | |
| K | 0.91 | |
| Texture | Clay (%) | 59 |
| Silt (%) | 28 | |
| Sand (%) | 13 | |
| Soil class | Clay | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wayima, E.F.; Ligaba-Osena, A.; Dagne, K.; Tesfaye, K.; Machuka, E.M.; Mutiga, S.K.; Delhaize, E. Screening of Diverse Ethiopian Durum Wheat Accessions for Aluminum Tolerance. Agronomy 2019, 9, 440. https://doi.org/10.3390/agronomy9080440
Wayima EF, Ligaba-Osena A, Dagne K, Tesfaye K, Machuka EM, Mutiga SK, Delhaize E. Screening of Diverse Ethiopian Durum Wheat Accessions for Aluminum Tolerance. Agronomy. 2019; 9(8):440. https://doi.org/10.3390/agronomy9080440
Chicago/Turabian StyleWayima, Edossa Fikiru, Ayalew Ligaba-Osena, Kifle Dagne, Kassahun Tesfaye, Eunice Magoma Machuka, Samuel Kilonzo Mutiga, and Emmanuel Delhaize. 2019. "Screening of Diverse Ethiopian Durum Wheat Accessions for Aluminum Tolerance" Agronomy 9, no. 8: 440. https://doi.org/10.3390/agronomy9080440
APA StyleWayima, E. F., Ligaba-Osena, A., Dagne, K., Tesfaye, K., Machuka, E. M., Mutiga, S. K., & Delhaize, E. (2019). Screening of Diverse Ethiopian Durum Wheat Accessions for Aluminum Tolerance. Agronomy, 9(8), 440. https://doi.org/10.3390/agronomy9080440





