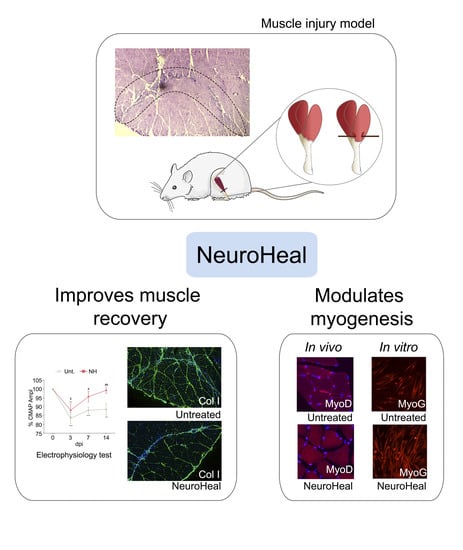
NeuroHeal Improves Muscle Regeneration after Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Muscle Injury Model
2.2. Drug Treatment
2.3. Electrophysiology
2.4. Measurement of Muscle Force
2.5. Histology
2.6. Myoblast Differentiation and Analysis
2.7. Statistical Analysis
3. Results
3.1. Text
3.1.1. NeuroHeal Accelerates Muscle Fiber Function Recovery
3.1.2. NeuroHeal Modulates Fiber Regeneration and Reduces Collagen Deposition
3.1.3. NeuroHeal Influences Muscle Satellite Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Special Statement
Acknowledgments
Conflicts of Interest
References
- Chan, O.; Del Buono, A.; Best, T.M.; Maffulli, N. Acute muscle strain injuries: A proposed new classification system. Knee Surg. Sport. Traumatol. Arthrosc. 2012, 20, 2356–2362. [Google Scholar] [CrossRef] [PubMed]
- Huard, J.; Li, Y.; Fu, F.H. Muscle injuries and repair: Current trends in research. J. Bone Jt. Surg.-Am. Vol. 2002, 84, 822–832. [Google Scholar] [CrossRef]
- Canata, G.L.; D’Hooghe, P.; Hunt, K.J. Muscle and Tendon Injuries; Canata, G.L., D’Hooghe, P., Hunt, K.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 978-3-662-54183-8. [Google Scholar]
- Starkey, J.D.; Yamamoto, M.; Yamamoto, S.; Goldhamer, D.J. Skeletal Muscle Satellite Cells Are Committed to Myogenesis and Do Not Spontaneously Adopt Nonmyogenic Fates. J. Histochem. Cytochem. 2011, 59, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Pallafacchina, G.; Blaauw, B.; Schiaffino, S. Role of satellite cells in muscle growth and maintenance of muscle mass. Nutr. Metab. Cardiovasc. Dis. 2013, 23, S12–S18. [Google Scholar] [CrossRef]
- Rocheteau, P.; Vinet, M.; Chretien, F. Dormancy and Quiescence of Skeletal Muscle Stem Cells. In Results and Problems in Cell Differentiation; Springer Verlag: New York, NY, USA, 2015; Volume 56, pp. 215–235. [Google Scholar]
- Lepper, C.; Partridge, T.A.; Fan, C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011, 138, 3639–3646. [Google Scholar] [CrossRef] [Green Version]
- Sambasivan, R.; Yao, R.; Kissenpfennig, A.; Van Wittenberghe, L.; Paldi, A.; Gayraud-Morel, B.; Guenou, H.; Malissen, B.; Tajbakhsh, S.; Galy, A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 2011, 138, 3647–3656. [Google Scholar] [CrossRef] [Green Version]
- Grassi, A.; Napoli, F.; Romandini, I.; Samuelsson, K.; Zaffagnini, S.; Candrian, C.; Filardo, G. Is Platelet-Rich Plasma (PRP) Effective in the Treatment of Acute Muscle Injuries? A Systematic Review and Meta-Analysis. Sport. Med. 2018, 48, 971–989. [Google Scholar] [CrossRef]
- Contreras-Muñoz, P.; Torrella, J.R.; Serres, X.; Rizo-Roca, D.; De La Varga, M.; Viscor, G.; Martínez-Ibáñez, V.; Peiró, J.L.; Järvinen, T.A.H.H.; Rodas, G.; et al. Postinjury Exercise and Platelet-Rich Plasma Therapies Improve Skeletal Muscle Healing in Rats but Are Not Synergistic When Combined. Am. J. Sports Med. 2017, 45, 2131–2141. [Google Scholar] [CrossRef]
- Qazi, T.H.; Duda, G.N.; Ort, M.J.; Perka, C.; Geissler, S.; Winkler, T. Cell therapy to improve regeneration of skeletal muscle injuries. J. Cachexia. Sarcopenia Muscle 2019, 10, 501–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo-Guitart, D.; Forés, J.; Navarro, X.; Casas, C. Boosted Regeneration and Reduced Denervated Muscle Atrophy by NeuroHeal in a Pre-clinical Model of Lumbar Root Avulsion with Delayed Reimplantation. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo-Guitart, D.; Casas, C. Network-centric medicine for peripheral nerve injury: Treating the whole to boost endogenous mechanisms of neuroprotection and regeneration. Neural Regen. Res. 2019, 14, 1122. [Google Scholar] [CrossRef] [PubMed]
- Romeo-Guitart, D.; Marcos-DeJuana, C.; Marmolejo-Martínez-Artesero, S.; Navarro, X.; Casas, C. Novel neuroprotective therapy with NeuroHeal by autophagy induction for damaged neonatal motoneurons. Theranostics 2020, 10, 5154–5168. [Google Scholar] [CrossRef] [PubMed]
- Romeo-Guitart, D.; Leiva-Rodriguez, T.; Forés, J.; Casas, C. Improved Motor Nerve Regeneration by SIRT1/Hif1a-Mediated Autophagy. Cells 2019, 8, 1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo-Guitart, D.; Casas, C. NeuroHeal Treatment Alleviates Neuropathic Pain and Enhances Sensory Axon Regeneration. Cells 2020, 9, 808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmolejo-Martínez-Artesero, S.; Romeo-Guitart, D.; Mañas-García, L.; Barreiro, E.; Casas, C. NeuroHeal Reduces Muscle Atrophy and Modulates Associated Autophagy. Cells 2020, 9, 1575. [Google Scholar] [CrossRef]
- Romeo-Guitart, D.; Forés, J.; Herrando-Grabulosa, M.; Valls, R.; Leiva-Rodríguez, T.; Galea, E.; González-Pérez, F.; Navarro, X.; Petegnief, V.; Bosch, A.; et al. Neuroprotective Drug for Nerve Trauma Revealed Using Artificial Intelligence. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harriss, D.; Atkinson, G. Ethical Standards in Sport and Exercise Science Research: 2014 Update. Int. J. Sports Med. 2013, 34, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Muñoz, P.; Fernández-Martín, A.; Torrella, R.; Serres, X.; De La Varga, M.; Viscor, G.; Järvinen, T.A.H.; Martínez-Ibáñez, V.; Peiró, J.L.; Rodas, G.; et al. A New Surgical Model of Skeletal Muscle Injuries in Rats Reproduces Human Sports Lesions. Int. J. Sports Med. 2016, 37, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Gayraud-Morel, B.; Chrétien, F.; Tajbakhsh, S. Skeletal muscle as a paradigm for regenerative biology and medicine. Regen. Med. 2009, 4, 293–319. [Google Scholar] [CrossRef]
- Dugdale, H.F.; Hughes, D.C.; Allan, R.; Deane, C.S.; Coxon, C.R.; Morton, J.P.; Stewart, C.E.; Sharples, A.P. The role of resveratrol on skeletal muscle cell differentiation and myotube hypertrophy during glucose restriction. Mol. Cell. Biochem. 2018, 444, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Amat, R.; Planavila, A.; Chen, S.L.; Iglesias, R.; Giralt, M.; Villarroya, F. SIRT1 Controls the Transcription of the Peroxisome Proliferator-activated Receptor-γ Co-activator-1α (PGC-1α) Gene in Skeletal Muscle through the PGC-1α Autoregulatory Loop and Interaction with MyoD. J. Biol. Chem. 2009, 284, 21872–21880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducreux, S.; Gregory, P.; Schwaller, B. Inverse Regulation of the Cytosolic Ca2+ Buffer Parvalbumin and Mitochondrial Volume in Muscle Cells via SIRT1/PGC-1α Axis. PLoS ONE 2012, 7, e44837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo-Guitart, D.; Leiva-Rodríguez, T.; Espinosa-Alcantud, M.; Sima, N.; Vaquero, A.; Domínguez- Martín, H.; Ruano, D.; Casas, C.; Domínguez-Martín, H.; Ruano, D.; et al. SIRT1 activation with neuroheal is neuroprotective but SIRT2 inhibition with AK7 is detrimental for disconnected motoneurons. Cell Death Dis. 2018, 9, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Luo, J.; Yu, B.; Zheng, P.; Huang, Z.; Mao, X.; He, J.; Yu, J.; Chen, J.; Chen, D. Dietary resveratrol supplementation improves meat quality of finishing pigs through changing muscle fiber characteristics and antioxidative status. Meat Sci. 2015, 102, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lagouge, M.; Argmann, C.; Gerhart-Hines, Z.; Meziane, H.; Lerin, C.; Daussin, F.; Messadeq, N.; Milne, J.; Lambert, P.; Elliott, P.; et al. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1α. Cell 2006, 127, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Ljubicic, V.; Burt, M.; Lunde, J.A.; Jasmin, B.J. Resveratrol induces expression of the slow, oxidative phenotype in mdx mouse muscle together with enhanced activity of the SIRT1-PGC-1α axis. Am. J. Physiol. Physiol. 2014, 307, C66–C82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Wang, B.; Du, F.; Li, H.; Wang, S.; Hu, C.; Zhu, C.; Yu, X. The Involvement of PI3K-Mediated and L-VGCC-Gated Transient Ca2+ Influx in 17β-Estradiol-Mediated Protection of Retinal Cells from H2O2-Induced Apoptosis with Ca2+ Overload. PLoS ONE 2013, 8, e77218. [Google Scholar] [CrossRef] [Green Version]
- Brack, A.S.; Rando, T.A. Tissue-Specific Stem Cells: Lessons from the Skeletal Muscle Satellite Cell. Cell Stem Cell 2012, 10, 504–514. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [Green Version]
- Ng, F.; Tang, B.L. Sirtuins’ modulation of autophagy. J. Cell. Physiol. 2013, 228, 2262–2270. [Google Scholar] [CrossRef]
- Tang, A.H.; Rando, T.A. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 2014, 33, 2782–2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryall, J.G.; Orso, S.D.; Derfoul, A.; Juan, A.; Zare, H.; Feng, X.; Clermont, D.; Koulnis, M.; Gutierrez-cruz, G.; Sartorelli, V. The NAD+ -Dependent SIRT1 Deacetylase Translates a Metabolic Switch into Regulatory Epigenetics in Skeletal Muscle Stem Cells. Cell Stem Cell 2016, 16, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms Controlling Mitochondrial Biogenesis and Respiration through the Thermogenic Coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Tonkin, J.; Villarroya, F.; Puri, P.L.; Vinciguerra, M. SIRT1 signaling as potential modulator of skeletal muscle diseases. Curr. Opin. Pharmacol. 2012, 12, 372–376. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marmolejo-Martínez-Artesero, S.; Romeo-Guitart, D.; Venegas, V.; Marotta, M.; Casas, C. NeuroHeal Improves Muscle Regeneration after Injury. Cells 2021, 10, 22. https://doi.org/10.3390/cells10010022
Marmolejo-Martínez-Artesero S, Romeo-Guitart D, Venegas V, Marotta M, Casas C. NeuroHeal Improves Muscle Regeneration after Injury. Cells. 2021; 10(1):22. https://doi.org/10.3390/cells10010022
Chicago/Turabian StyleMarmolejo-Martínez-Artesero, Sara, David Romeo-Guitart, Vanesa Venegas, Mario Marotta, and Caty Casas. 2021. "NeuroHeal Improves Muscle Regeneration after Injury" Cells 10, no. 1: 22. https://doi.org/10.3390/cells10010022