Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications
Abstract
:1. Exosomes Mediate Crosstalk between Cancer and Non-Cancer Cells within the Tumor Microenvironment
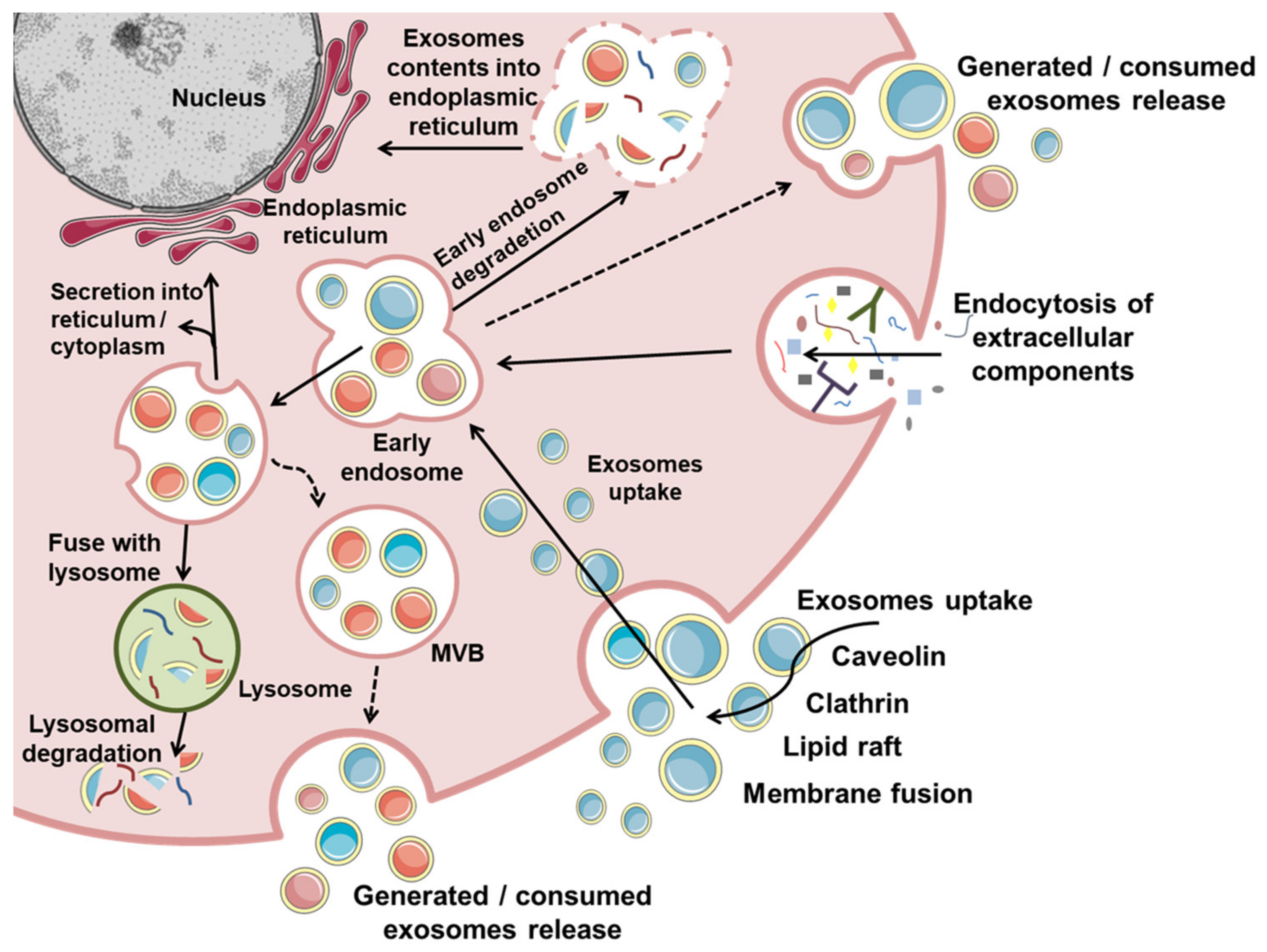
2. Exosome Biogenesis
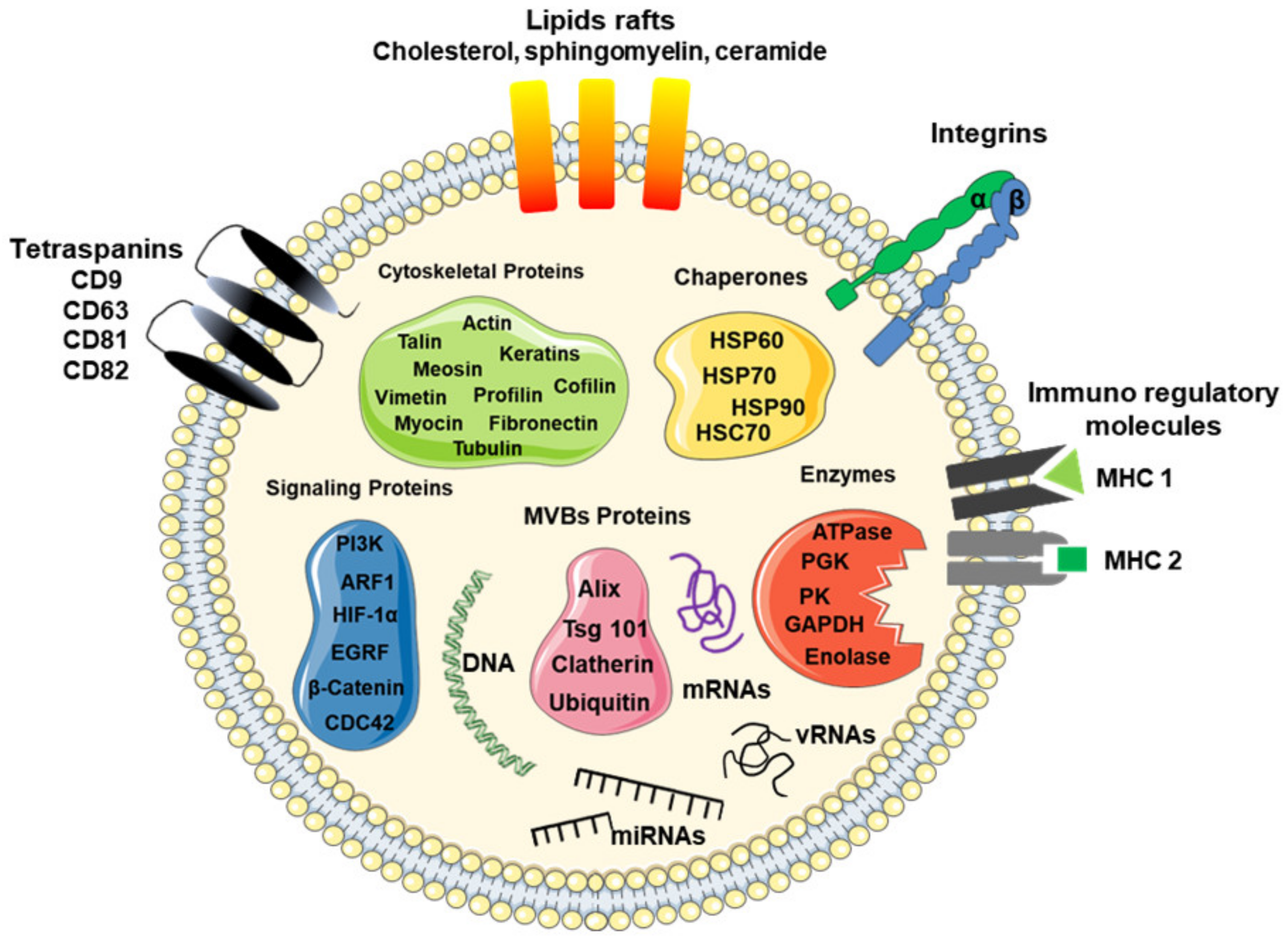
3. Molecular Cargo
4. Cancer-Derived Exosomes in Carcinogenesis
4.1. Cancer-Derived Exosomes Mediate Crosstalk between Inflammation and Cancer Initiation
4.2. Cancer-Derived Exosomes Regulate Tumor Promotion and Progression
4.3. Cancer-Derived Exosomes Regulate Several Steps of the Metastatic Process
4.3.1. Cancer-Derived Exosomes as a Key Regulator of the Epithelial–Mesenchymal Transition (EMT)
4.3.2. Exosomes in Angiogenesis
4.3.3. Cancer-Derived Exosomes Contribute to Pre-Metastatic Niche (PMN) Formation
4.3.4. Exosomes in Cancer Stem Cell (CSC) Formation
5. Mesenchymal Stem Cell (MSC) Recruitment to the Tumor Microenvironment (TME)
6. Mesenchymal Stem Cells as a Source of Exosomes for Cancer Treatment
7. Clinical Applications of MSC-Derived Exosomes for Cancer Treatment
8. Future Prospects of Cell-Free Therapy for Cancer Treatment and Challenges to Be Overcome
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Omran, A.R. The epidemiologic transition: A theory of the epidemiology of population change. Milbank Q. 2005, 83, 731–757. [Google Scholar] [CrossRef] [Green Version]
- IARC. Estimate Number of Deaths from 2020 to 2040. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/bars?mode=population&types=1 (accessed on 27 August 2021).
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Bu, L.; Baba, H.; Yoshida, N.; Miyake, K.; Yasuda, T.; Uchihara, T.; Tan, P.; Ishimoto, T. Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene 2019, 38, 4887–4901. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-based cell-cell communication in the tumor microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Lv, D.; Zhu, X.; Tang, H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019, 12, 53. [Google Scholar] [CrossRef]
- Golchin, A.; Hosseinzadeh, S.; Ardeshirylajimi, A. The exosomes released from different cell types and their effects in wound healing. J. Cell. Biochem. 2018, 119, 5043–5052. [Google Scholar] [CrossRef]
- Baixauli, F.; LÃ3pez-Otà n, C.; Mittelbrunn, M. Exosomes and autophagy: Coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 2014, 5, 403. [Google Scholar] [CrossRef] [Green Version]
- Baek, G.; Choi, H.; Kim, Y.; Lee, H.C.; Choi, C. Mesenchymal stem cell-derived extracellular vesicles as therapeutics and as a drug delivery platform. Stem Cells Transl. Med. 2019, 8, 880–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, H.; He, D.; He, X.; Wang, K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. ChemBioChem 2019, 20, 451–461. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [Green Version]
- Antounians, L.; Tzanetakis, A.; Pellerito, O.; Catania, V.D.; Sulistyo, A.; Montalva, L.; McVey, M.J.; Zani, A. The regenerative potential of amniotic fluid stem cell extracellular vesicles: Lessons learned by comparing different isolation techniques. Sci. Rep. 2019, 9, 1837. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, K.; Wu, S.; Cui, M.; Xu, T. Focus on mesenchymal stem cell-derived exosomes: Opportunities and challenges in cell-free therapy. Stem Cells Int. 2017, 2017, 6305295. [Google Scholar] [CrossRef]
- Exosomes, E.; Vivo, I.; Verweij, F.J.; Revenu, C.; Arras, G.; Del Bene, F.; Van Niel, G.; Verweij, F.J.; Revenu, C.; Arras, G.; et al. Live Tracking of Inter-organ Communication by Resource Live Tracking of Inter-organ Communication by Endogenous Exosomes In Vivo. Dev. Cell 2019, 48, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Araldi, R.P.; D’Amelio, F.; Vigerelli, H.; de Melo, T.C.; Kerkis, I. Stem cell-derived exosomes as therapeutic approach for neurodegenerative Ddsorders: From biology to biotechnology. Cells 2020, 9, 2663. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M.; Adam, M.; Hammonds, J.R.; Turbide, C. Vesicle formation during reticulocyte maturation. J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Harding, C.; Stahl, P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem. Biophys. Res. Commun. 1983, 113, 650–658. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Pan, B.T. The fate of the transferrin receptor during maturation of sheep reticulocytes in vitro. Can. J. Biochem. Cell Biol. 1984, 62, 1246–1254. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Al-Sowayan, B.S.; Al-Shareeda, A.T.; Alrfaei, B.M. Cancer stem cell-exosomes, unexposed player in tumorigenicity. Front. Pharmacol. 2020, 11, 384. [Google Scholar] [CrossRef] [Green Version]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Haider, K.H.; Aramini, B. Mircrining the injured heart with stem cell-derived exosomes: An emerging strategy of cell-free therapy. Stem Cell Res. Ther. 2020, 11, 23. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.-L.; Sun, P.; Li, Y.; Liu, S.-S.; Lu, Y. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl. Cancer Res. 2019, 8, 298–311. [Google Scholar] [CrossRef]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Fader, C.M.; Colombo, M.I. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2009, 16, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140203. [Google Scholar] [CrossRef]
- Rezaie, J.; Ajezi, S.; Avci, Ç.B.; Karimipour, M.; Geranmayeh, M.H.; Nourazarian, A.; Sokullu, E.; Rezabakhsh, A.; Rahbarghazi, R. Exosomes and their application in biomedical field: Difficulties and advantages. Mol. Neurobiol. 2018, 55, 3372–3393. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Airola, M.V.; Hannun, Y.A. Sphingolipid metabolism and neutral sphingomyelinases. Handb. Exp. Pharmacol. 2013, 215, 57–76. [Google Scholar]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, more than markers of extracellular vesicles in reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Ghossoub, R.; Chéry, M.; Audebert, S.; Leblanc, R.; Egea-Jimenez, A.L.; Lembo, F.; Mammar, S.; Le Dez, F.; Camoin, L.; Borg, J.-P.; et al. Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. USA 2020, 117, 5913–5922. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Jaiswal, R.; Sedger, L.M. Intercellular vesicular transfer by exosomes, microparticles and oncosomes-implications for cancer biology and treatments. Front. Oncol. 2019, 9, 125. [Google Scholar] [CrossRef] [Green Version]
- Meehan, B.; Rak, J.; Di Vizio, D. Oncosomes—large and small: What are they, where they came from? J. Extracell. Vesicles 2016, 5, 33109. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; You, S.; Spinelli, C.; Morley, S.; Zandian, M.; Aspuria, P.-J.; Cavallini, L.; Ciardiello, C.; Sobreiro, M.R.; Morello, M.; et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 2015, 6, 11327–11341. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015, 25, 24–38. [Google Scholar] [CrossRef] [Green Version]
- Sumrin, A.; Moazzam, S.; Khan, A.A.; Ramzan, I.; Batool, Z.; Kaleem, S.; Ali, M.; Bashir, H.; Bilal, M. Exosomes as biomarker of cancer. Braz. Arch. Biol. Technol. 2018, 61, 1–14. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Liao, K.; Zhou, W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells Int. 2018, 2018, 4837370. [Google Scholar] [CrossRef]
- Othman, N.; Jamal, R.; Abu, N. Cancer-derived exosomes as effectors of key Inflammation-related players. Front. Immunol. 2019, 10, 2103. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Stefanius, K.; Servage, K.; de Souza Santos, M.; Gray, H.F.; Toombs, J.E.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife 2019, 8, e40226. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.D.; Wong, W.; Lee, M.M.; Cho, W.C.; Yee, B.K.; Kwan, Y.W.; Tai, W.C. Exosomes in inflammation and inflammatory disease. Proteomics 2019, 19, 1800149. [Google Scholar] [CrossRef]
- Weber, D. Inflammation and cancer: Tumor initiation, progression and metastasis, and Chinese botanical medicines. J. Chin. Integr. Med. 2010, 8, 1006–1013. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Singel, K.L.; Segal, B.H. Neutrophils in the tumor microenvironment: Trying to heal the wound that cannot heal. Immunol. Rev. 2016, 273, 329–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanius, K.; Servage, K.; Orth, K. Exosomes in cancer development. Curr. Opin. Genet. Dev. 2021, 66, 83–92. [Google Scholar] [CrossRef]
- Bretz, N.P.; Ridinger, J.; Rupp, A.-K.; Rimbach, K.; Keller, S.; Rupp, C.; Marmé, F.; Umansky, L.; Umansky, V.; Eigenbrod, T.; et al. Body Fluid Exosomes Promote Secretion of Inflammatory Cytokines in Monocytic Cells via Toll-like Receptor Signaling. J. Biol. Chem. 2013, 288, 36691–36702. [Google Scholar] [CrossRef] [Green Version]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z.; et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci. Rep. 2015, 4, 5750. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhang, X.; Zhang, B.; Shi, H.; Yuan, X.; Sun, Y.; Pan, Z.; Qian, H.; Xu, W. Exosomes derived from gastric cancer cells activate NF-κB pathway in macrophages to promote cancer progression. Tumor Biol. 2016, 37, 12169–12180. [Google Scholar] [CrossRef]
- Butin-Israeli, V.; Bui, T.M.; Wiesolek, H.L.; Mascarenhas, L.; Lee, J.J.; Mehl, L.C.; Knutson, K.R.; Adam, S.A.; Goldman, R.D.; Beyder, A.; et al. Neutrophil-induced genomic instability impedes resolution of inflammation and wound healing. J. Clin. Investig. 2019, 129, 712–726. [Google Scholar] [CrossRef] [PubMed]
- De-Sá-Júnio, P.; Câmara, D.; Porcacchia, A.; Fonseca, P.; Jorge, S.; Araldi, R.; Ferreira, A. The roles of ROS in cancer heterogeneity and therapy. Oxid. Med. Cell. Longev. 2017, 2017, 2467940. [Google Scholar]
- Araldi, R.; Mazzuchelli-de-souza, J.; Modolo, D.; Souza, E.; Melo, T.; Spadacci-Morena, D.; Magnelli, R.; Rocha, M.; De-Sá-Júnior, P.; Carvalho, R.; et al. Mutagenic potential of Bos taurus papillomavirus type 1 E6 recombinant protein: First description. Biomed. Res. Int. 2015, 2015, 806361. [Google Scholar] [CrossRef] [Green Version]
- Shang, A.; Gu, C.; Liu, W.; Wang, X.; Zeng, B.; Chen, C.; Chang, W.; Ping, Y.; Sun, J.; Ji, P.; et al. Exosomal KRAS Mutation Promotes Promotion of Colorectal Cancer by the Formation of Tumor-Associated Neutrophil Extracellular Traps. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Leal, A.C.; Mizurini, D.M.; Gomes, T.; Rochael, N.C.; Saraiva, E.M.; Dias, M.S.; Werneck, C.C.; Sielski, M.S.; Vicente, C.P.; Monteiro, R.Q. Tumor-Derived Exosomes Induce the Formation of Neutrophil Extracellular Traps: Implications for the Establishment of Cancer-Associated Thrombosis. Sci. Rep. 2017, 7, 6438. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, C.; Harikumar, K.B. The origin and functions of exosomes in cancer. Front. Oncol. 2018, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Abd Elmageed, Z.Y.; Yang, Y.; Thomas, R.; Ranjan, M.; Mondal, D.; Moroz, K.; Fang, Z.; Rezk, B.M.; Moparty, K.; Sikka, S.C.; et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014, 32, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Sugimoto, H.; Connell, J.T.O.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Adjiri, A. DNA mutations may not be the cause of cancer. Oncol. Ther. 2017, 5, 85–101. [Google Scholar] [CrossRef]
- Zhang, D.; Huo, D.; Xie, H.; Wu, L.; Zhang, J.; Liu, L.; Jin, Q.; Chen, X. CHG: A systematically integrated database of cancer hallmark. Front Genet. 2020, 11, 29. [Google Scholar] [CrossRef]
- Koga, K.; Matsumoto, K.; Akiyoshi, T.; Kubo, M.; Yamanaka, N.; Tasaki, A.; Nakashima, H.; Nakamura, M.; Kuroki, S.; Tanaka, M.; et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005, 25, 3703–3707. [Google Scholar]
- Yang, L.; Wu, X.-H.; Wang, D.; Luo, C.-L.; Chen, L.-X. Bladder cancer cell-derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Mol. Med. Rep. 2013, 8, 1272–1278. [Google Scholar] [CrossRef] [Green Version]
- Osaki, M.; Okada, F. Exosomes and their role in cancer progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Cao, H.; Shen, B.; Feng, J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget 2015, 6, 37151–37168. [Google Scholar] [CrossRef] [Green Version]
- Qu, J.-L.; Qu, X.-J.; Zhao, M.-F.; Teng, Y.-E.; Zhang, Y.; Hou, K.-Z.; Jiang, Y.-H.; Yang, X.-H.; Liu, Y.-P. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009, 41, 875–880. [Google Scholar] [CrossRef]
- Kogure, T.; Lin, W.-L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef] [Green Version]
- Ke, X.; Yan, R.; Sun, Z.; Cheng, Y.; Meltzer, A.; Lu, N.; Shu, X.; Wang, Z.; Huang, B.; Liu, X.; et al. Esophageal adenocarcinoma–derived extracellular vesicle microRNAs induce a neoplastic phenotype in gastric organoids. Neoplasia 2017, 19, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.-D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Huang, L.; Li, Y.; Zhang, X.; Gu, J.; Yan, Y.; Xu, X.; Wang, M.; Qian, H.; Xu, W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012, 315, 28–37. [Google Scholar] [CrossRef]
- Xiao, H.; Lässer, C.; Shelke, G.V.; Wang, J.; Rådinger, M.; Lunavat, T.R.; Malmhäll, C.; Lin, L.H.; Li, J.; Li, L.; et al. Mast cell exosomes promote lung adenocarcinoma cell proliferation—Role of KIT-stem cell factor signaling. Cell Commun. Signal. 2014, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wu, F.; Zhou, H. Macrophage-derived exosomes in cancers: Biogenesis, functions and therapeutic applications. Immunol. Lett. 2020, 227, 102–108. [Google Scholar] [CrossRef]
- Compton, C. Cancer initiation, promotion, and progression and the acquisition of key behavioral traits. In Cancer: The Enemy from Within; Springer International Publishing: Cham, Switzerland, 2020; pp. 25–48. [Google Scholar]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Tsai, J.H.; Yang, J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013, 27, 2192–2206. [Google Scholar] [CrossRef] [Green Version]
- Araldi, R.P.; de Melo, T.C.; Levy, D.; de Souza, D.M.; Maurício, B.; Colozza-Gama, G.A.; Bydlowski, S.P.; Peng, H.; Rauscher, F.J.; Cerutti, J.M. LIMD2 regulates key steps of metastasis cascade in papillary thyroid cancer cells via MAPK crosstalk. Cells 2020, 9, 2522. [Google Scholar] [CrossRef]
- Araldi, R.; Módolo, D.; De-Sá-Júnior, P.; Consonni, S.; Carvalho, R.; Roperto, F.; Beçak, W.; Stocco, R. Genetics and metabolic deregulation following cancer initiation: A world to explore. Biomed. Pharmacother. 2016, 82, 449–458. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Cichon, M.; Nelson, C.; Radisky, D. Regulation of epithelial-mesenchymal transition in breast cancer cells by cell contact and adhesion. Cancer Inform. 2015, 14, CIN–S18965. [Google Scholar] [CrossRef]
- Lee, K.; Nelson, C. New Insights into the Regulation of Epithelial-Mesenchymal Transition and Tissue Fibrosis, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2012; Volume 294, ISBN 9780123943057. [Google Scholar]
- Tian, Y.; Xie, Q.; He, J.; Luo, X.; Zhou, T.; Liu, Y.; Huang, Z.; Tian, Y.; Sun, D.; Yao, K. Radioactive 125 I seeds inhibit cell growth and epithelial-mesenchymal transition in human glioblastoma multiforme via a ROS-mediated signaling pathway. BMC Cancer 2015, 15, 1. [Google Scholar] [CrossRef]
- Yan, C.; Grimm, W.A.; Garner, W.L.; Qin, L.; Travis, T.; Tan, N.; Han, Y.-P. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor mecrosis factor-α through bone morphogenic protein-2. Am. J. Pathol. 2010, 176, 2247–2258. [Google Scholar] [CrossRef] [Green Version]
- Radisky, D.; LaBarge, M. Epithelial-mesenchymal transition and the stem cell phenotype. Cell Stem Cell 2008, 2, 511–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haensel, D.; Dai, X. Epithelial-to-mesenchymal transition in cutaneous wound healing: Where we are and where we are heading. Dev. Dyn. 2018, 247, 473–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, R.C.; Pastar, I.; Ojeh, N.; Chen, V.; Liu, S.; Garzon, K.I.; Tomic-Canic, M. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016, 365, 495–506. [Google Scholar] [CrossRef]
- Wever, O.; Pauwels, P.; De Craene, B.; Sabbah, M.; Emami, S.; Redeuilh, G.; Gespach, C.; Bracke, M.; Berx, G. Molecular and pathological signatures of epithelial-mesenchymal transitions at the cancer invasion front. Histochem. Cell Biol. 2008, 130, 481–494. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Gillies, R.; Gatenby, R. Adaptation to hypoxia and acidosis in carcinogenesis and tumor progression. Semin. Cancer Biol. 2008, 18, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, Y.; Feng, Y.; Gao, S. SNAIL gene inhibited by hypoxia-inducible factor 1α (HIF-1α) in epithelial ovarian cancer. Int. J. Immunopathol. Pharmacol. 2016, 29, 364–375. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.; Yuan, J.; Huang, T.; Zhang, C.; Zhu, Z.; Wang, L.; Jiang, G.; Zeng, F. Stabilization of Snail by HIF-1α and TNF-α is required for hypoxia-induced invasion in prostate cancer PC3 cells. Mol. Biol. Rep. 2014, 41, 4573–4582. [Google Scholar] [CrossRef]
- Sun, S.; Ning, X.; Zhang, Y.; Lu, Y.; Nie, Y.; Han, S.; Liu, L.; Du, R.; Xia, L.; He, L.; et al. Hypoxia-inducible factor-1α induces Twist expression in tubular epithelial cells subjected to hypoxia, leading to epithelial-to-mesenchymal transition. Kidney Int. 2009, 75, 1278–1287. [Google Scholar] [CrossRef] [Green Version]
- Araldi, R.; Lima, T.; Módolo, D.; Mazzuchelli-de-Souza, J.; Magnelli, R.; Maurício, B.; Spadacci-Morena, D.; De-Sá-Júnior, P.; Carvalho, R.; Beçak, W.; et al. Analysis of stem-cell and migratory phenotype in primary cultures derived from BPV-infected benign and malignant neoplasms. J. Cancer Res. Ther. Oncol. 2017, 5, 1–13. [Google Scholar]
- Yang, S.; Zhang, Z.; Hao, Y.; Zhao, Y.; Qian, F.; Shi, Y.; Li, P.; Liu, C.; Yu, P. HIF-1α induces the epithelial-mesenchymal transition in gastric cancer stem cells through the Snail pathway. Oncotarget 2017, 8, 9535–9545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hapke, R.Y.; Haake, S.M. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020, 487, 10–20. [Google Scholar] [CrossRef]
- Payen, V.L.; Mina, E.; Van Hée, V.F.; Porporato, P.E.; Sonveaux, P. Monocarboxylate transporters in cancer. Mol. Metab. 2020, 33, 48–66. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, I.; Bang, H.; Kim, H.C.; Lee, W.Y.; Yun, S.H.; Lee, J.; Lee, S.J.; Park, Y.S.; Kim, K.-M.; et al. MCT4 expression is a potential therapeutic target in colorectal cancer with peritoneal carcinomatosis. Mol. Cancer Ther. 2018, 17, 838–848. [Google Scholar] [CrossRef] [Green Version]
- De la Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9, 1143. [Google Scholar] [CrossRef] [Green Version]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar] [CrossRef] [Green Version]
- Mortezaee, K. Immune scape: A critical hallmark in solid tumors. Life iSci. 2020, 258, 118110. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; García-Hernández, A.A.; Ramos, C. Matrix Metalloproteinases’ Role in Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 97–131. [Google Scholar] [PubMed]
- Panigrahi, G.K.; Praharaj, P.P.; Peak, T.C.; Long, J.; Singh, R.; Rhim, J.S.; Abd Elmageed, Z.Y.; Deep, G. Hypoxia-induced exosome secretion promotes survival of African-American and Caucasian prostate cancer cells. Sci. Rep. 2018, 8, 3853. [Google Scholar] [CrossRef]
- Meng, W.; Hao, Y.; He, C.; Li, L.; Zhu, G. Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 2019, 18, 57. [Google Scholar] [CrossRef] [Green Version]
- Kore, R.A.; Edmondson, J.L.; Jenkins, S.V.; Jamshidi-Parsian, A.; Dings, R.P.M.; Reyna, N.S.; Griffin, R.J. Hypoxia-derived exosomes induce putative altered pathways in biosynthesis and ion regulatory channels in glioblastoma cells. Biochem. Biophys. Rep. 2018, 14, 104–113. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhou, X.; Yao, Q.; Liu, Y.; Zhang, H.; Dong, Z. HIF-1-mediated production of exosomes during hypoxia is protective in renal tubular cells. Am. J. Physiol.-Ren. Physiol. 2017, 313, F906–F913. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, L.; Rong, Y.; Qian, D.; Chen, J.; Zhou, Z.; Luo, Y.; Jiang, D.; Cheng, L.; Zhao, S.; et al. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020, 103, 196–212. [Google Scholar] [CrossRef]
- Cheng, H.; Chang, S.; Xu, R.; Chen, L.; Song, X.; Wu, J.; Qian, J.; Zou, Y.; Ma, J. Hypoxia-challenged MSC-derived exosomes deliver miR-210 to attenuate post-infarction cardiac apoptosis. Stem Cell Res. Ther. 2020, 11, 224. [Google Scholar] [CrossRef]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2016, 38, 119–133. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Boussadia, Z.; Zanetti, C.; Parolini, I. Role of microenvironmental acidity and tumor exosomes in cancer immunomodulation. Transl. Cancer Res. 2020, 9, 5775–5786. [Google Scholar] [CrossRef]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [Green Version]
- Svensson, K.J.; Kucharzewska, P.; Christianson, H.C.; Sköld, S.; Löfstedt, T.; Johansson, M.C.; Mörgelin, M.; Bengzon, J.; Ruf, W.; Belting, M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13147–13152. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yang, C.; Yang, S.; Cheng, F.; Rao, J.; Wang, X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018, 9, 954. [Google Scholar] [CrossRef] [Green Version]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Lee, S.; Shin, E.; Seong, K.M.; Jin, Y.W.; Youn, H.; Youn, B. The emerging roles of exosomes as EMT regulators in cancer. Cells 2020, 9, 861. [Google Scholar] [CrossRef]
- Boopathy, G.T.K.; Hong, W. Role of Hippo pathway-YAP/TAZ signaling in angiogenesis. Front. Cell Dev. Biol. 2019, 7, 49. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/β-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Nam, R.K.; Benatar, T.; Wallis, C.J.D.; Amemiya, Y.; Yang, W.; Garbens, A.; Naeim, M.; Sherman, C.; Sugar, L.; Seth, A. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate 2016, 76, 869–884. [Google Scholar] [CrossRef]
- Hu, X.; Mu, Y.; Liu, J.; Mu, X.; Gao, F.; Chen, L.; Wu, H.; Wu, H.; Liu, W.; Zhao, Y. Exosomes derived from hypoxic colorectal cancer cells transfer miR-410-3p to regulate tumor progression. J. Cancer 2020, 11, 4724–4735. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Rana, S.; Zöller, M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia 2013, 15, 875–887. [Google Scholar] [CrossRef] [Green Version]
- Ge, R.; Tan, E.; Sharghi-Namini, S.; Asada, H.H. Exosomes in cancer microenvironment and beyond: Have we overlooked these extracellular messengers? Cancer Microenviron. 2012, 5, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kling, M.J.; Chaturvedi, N.K.; Kesherwani, V.; Coulter, D.W.; McGuire, T.R.; Sharp, J.G.; Joshi, S.S. Exosomes secreted under hypoxia enhance stemness in Ewing’s sarcoma through miR-210 delivery. Oncotarget 2020, 11, 3633–3645. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, Y.; Dong, L.; Zhang, Y.; Zhang, Y.; Wang, C.; Zhang, Q.; Yang, S.; Cao, L.; Zhang, X.; et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J. Exp. Clin. Cancer Res. 2019, 38, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, N.D.; Elias, M.; Guiro, K.; Bhatia, R.; Greco, S.J.; Bryan, M.; Gergues, M.; Sandiford, O.A.; Ponzio, N.M.; Leibovich, S.J.; et al. Exosomes from differentially activated macrophages influence dormancy or resurgence of breast cancer cells within bone marrow stroma. Cell Death Dis. 2019, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, A.; Tousif, S.; Wang, Y.; Hough, K.; Khan, S.; Strenkowski, J.; Chacko, B.K.; Darley-Usmar, V.M.; Deshane, J.S. Lung tumor cell-derived exosomes promote M2 macrophage polarization. Cells 2020, 9, 1303. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Wu, T.-M.; Ling, C.-C.; Yu, F.; Zhang, J.; Cao, P.-S.; Gu, L.-P.; Wang, H.-M.; Xu, H.; Li, L.; et al. M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B. Mol. Ther. Oncolytics 2021, 20, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Luo, Q.; Wang, W.; Li, J.; Wang, T.; Wang, P.; Chen, L.; Zhang, P.; Chen, H.; Liu, Y.; et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018, 9, 434. [Google Scholar] [CrossRef]
- Shi, Y.-H.; Fang, W.-G. Hypoxia-inducible factor-1 in tumour angiogenesis. World J. Gastroenterol. 2004, 10, 1082. [Google Scholar] [CrossRef]
- Hashimoto, T.; Shibasaki, F. Hypoxia-inducible factor as an angiogenic master switch. Front. Pediatr. 2015, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, H.; Kizaka-Kondoh, S.; Li, G.; Itasaka, S.; Shibuya, K.; Inoue, M.; Hiraoka, M. Significance of HIF-1-active cells in angiogenesis and radioresistance. Oncogene 2007, 26, 7508–7516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; McGough, R.; Aswad, B.; Block, J.A.; Terek, R. Hypoxia induces HIF-1Œ± and VEGF expression in chondrosarcoma cells and chondrocytes. J. Orthop. Res. 2004, 22, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Cébe-Suarez, S.; Zehnder-Fjällman, A.; Ballmer-Hofer, K. The role of VEGF receptors in angiogenesis; complex partnerships. Cell. Mol. Life Sci. 2006, 63, 601–615. [Google Scholar] [CrossRef] [Green Version]
- Aslan, C.; Maralbashi, S.; Salari, F.; Kahroba, H.; Sigaroodi, F.; Kazemi, T.; Kharaziha, P. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J. Cell. Physiol. 2019, 234, 16885–16903. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Ko, S.Y.; Lee, W.; Kenny, H.A.; Dang, L.H.; Ellis, L.M.; Jonasch, E.; Lengyel, E.; Naora, H. Cancer-derived small extracellular vesicles promote angiogenesis by heparin-bound, bevacizumab-insensitive VEGF, independent of vesicle uptake. Commun. Biol. 2019, 2, 386. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Wang, Y.; Zhang, X.; Feng, M.; Ma, J.; Li, J.; Yang, X.; Fang, F.; Xia, Q.; Zhang, Z.; et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol. Cancer 2019, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Sheng, Y.; Li, B.; Wang, Q.; Liu, X.; Han, J. Ovarian cancer derived PKR1 positive exosomes promote angiogenesis by promoting migration and tube formation in vitro. Cell Biochem. Funct. 2021, 39, 308–316. [Google Scholar] [CrossRef]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- Ribatti, D.; Mangialardi, G.; Vacca, A. Stephen Paget and the ‘seed and soil’ theory of metastatic dissemination. Clin. Exp. Med. 2006, 6, 145–149. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, H.; Jiang, X.; Qian, C.; Liu, Z.; Luo, D. Factors involved in cancer metastasis: A better understanding to “seed and soil” hypothesis. Mol. Cancer 2017, 16, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psaila, B.; Lyden, D. The metastatic niche: Adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.P. The lymph node pre-metastatic niche. J. Mol. Med. 2015, 93, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.-N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical significance of PD-L1 + exosomes in plasma of head and neck cancer patients. Clin. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.-S.; Sharma, P.; Yerneni, S.S.; Simms, P.; Jackson, E.K.; Whiteside, T.L.; Boyiadzis, M. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myeloid leukemia. Sci. Rep. 2017, 7, 14684. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, S.; Floros, T.; Theodoraki, M.-N.; Hong, C.-S.; Jackson, E.K.; Lang, S.; Whiteside, T.L. Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin. Cancer Res. 2017, 23, 4843–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czernek, L.; Düchler, M. Functions of cancer-derived extracellular vesicles in immunosuppression. Arch. Immunol. Ther. Exp. 2017, 65, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringner, M.; Morgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [Green Version]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Clarke, M.; Fuller, M. Stem cells and cancer: Two faces of eve. Cell 2006, 124, 1111–1115. [Google Scholar] [CrossRef] [Green Version]
- Islam, F.; Qiao, B.; Smith, R.A.; Gopalan, V.; Lam, A.K.Y. Cancer stem cell: Fundamental experimental pathological concepts and updates. Exp. Mol. Pathol. 2015, 98, 184–191. [Google Scholar] [CrossRef]
- Begicevic, R.-R.; Falasca, M. ABC transporters in cancer stem cells: Beyond chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Kim, J.; Kim, M.; Kim, J.; Lee, K.; Kim, B.; Oh, H.; Kim, D.; Kang, S.; Kim, H.; et al. Prognostic implication of ABC transporters and cancer stem cell markers in patients with stage III colon cancer receiving adjuvant FOLFOX-4 chemotherapy. Oncol. Lett. 2019, 17, 5572–5580. [Google Scholar] [CrossRef] [Green Version]
- Abbasifarid, E.; Sajjadi-Jazi, S.M.; Beheshtian, M.; Samimi, H.; Larijani, B.; Haghpanah, V. The role of ATPbBinding cassette transporters in the chemoresistance of anaplastic thyroid cancer: A systematic review. Endocrinology 2019, 160, 2015–2023. [Google Scholar] [CrossRef]
- Olivares-Urbano, M.A.; Griñán-Lisón, C.; Marchal, J.A.; Núñez, M.I. CSC radioresistance: A therapeutic challenge to improve radiotherapy effectiveness in cancer. Cells 2020, 9, 1651. [Google Scholar] [CrossRef]
- Arnold, C.R.; Mangesius, J.; Skvortsova, I.-I.; Ganswindt, U. The role of cancer stem cells in radiation resistance. Front. Oncol. 2020, 10, 164. [Google Scholar] [CrossRef] [Green Version]
- Nimmakayala, R.K.; Batra, S.K.; Ponnusamy, M.P. Unraveling the journey of cancer stem cells from origin to metastasis. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 50–63. [Google Scholar] [CrossRef]
- Eun, K.; Ham, S.W.; Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC targeting. BMB Rep. 2017, 50, 117–125. [Google Scholar] [CrossRef]
- Lagasse, E. Cancer stem cells with genetic instability: The best vehicle with the best engine for cancer. Gene Ther. 2008, 15, 136–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, J.; Oliveira, T.; Greten, F. The architect who never sleeps: Tumor-induced plasticity. FEBS Lett. 2014, 588, 2422–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shipitsin, M.; Polyak, K. The cancer stem cell hypothesis: In search of definitions, markers, and relevance. Lab. Investig. 2008, 88, 459–463. [Google Scholar] [CrossRef]
- Li, L.; Borodyansky, L.; Yang, Y. Genomic instability en route to and from cancer stem cells. Cell Cycle 2009, 8, 1000–1002. [Google Scholar] [CrossRef]
- Wang, B.; Tan, Z.; Guan, F. Tumor-derived exosomes mediate the instability of cadherins and promote tumor progression. Int. J. Mol. Sci. 2019, 20, 3652. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Liu, Z.; Niu, B.; Zhang, J.; Tan, T.K.; Lee, S.R.; Zhao, Y.; Harris, D.C.H.; Zheng, G. E-cadherin/β -catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, 2011, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.K.; Kwon, Y.; Jang, M.; Park, M.; Kim, J.; Cho, S.; Jang, D.G.; Lee, W.-B.; Jung, S.H.; Choi, H.J.; et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci. Rep. 2019, 9, 18440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavila, N.; Thundimadathil, J. The emerging roles of cancer stem cells and Wnt/Beta-catenin signaling in hepatoblastoma. Cancers 2019, 11, 1406. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Kawaguchi, M.; Shoda, S.; Miyoshi, T.; Iwasaki, R.; Hyodo, F.; Mori, T.; Hara, A.; Tomita, H.; Matsuo, M. Nuclear accumulation of β-catenin in cancer stem Cell radioresistance and stemness in human colon cancer. Anticancer Res. 2019, 39, 6575–6583. [Google Scholar] [CrossRef]
- Pandit, H.; Li, Y.; Li, X.; Zhang, W.; Li, S.; Martin, R.C.G. Enrichment of cancer stem cells via β-catenin contributing to the tumorigenesis of hepatocellular carcinoma. BMC Cancer 2018, 18, 783. [Google Scholar] [CrossRef]
- Bach, D.-H.; Hong, J.-Y.; Park, H.J.; Lee, S.K. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 2017, 141, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef] [Green Version]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2017, 36, 1770–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, K. Overcoming multidrug resistance in cancer stem cells. Biomed. Res. Int. 2015, 2015, 635745. [Google Scholar] [CrossRef] [Green Version]
- Dean, M. ABC Transporters, Drug Resistance, and Cancer Stem Cells. J. Mammary Gland Biol. Neoplasia 2009, 14, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.-I. Therapy resistance mediated by exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Bai, X.; Ni, J.; Zhang, H.; Duan, W.; Graham, P.; Li, Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020, 11, 987. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhu, X.; Chen, W.; Zhong, S.; Hu, Q.; Ma, T.; Zhang, J.; Chen, L.; Tang, J.; Zhao, J. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumor Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-Resistance in Prostate Cancer: Evaluating Associated Phenotypic Changes and Potential for Resistance Transfer via Exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, MicroRNA and Protein Profiles of Melanoma Exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [Green Version]
- Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Baldini, N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int. J. Oncol. 2016, 49, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Xu, J.; Zhong, S.; Liu, Y.; Xiao, H.; Geng, L.; Liu, H. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol. Rep. 2019, 41, 1893–1900. [Google Scholar] [CrossRef]
- Mutschelknaus, L.; Peters, C.; Winkler, K.; Yentrapalli, R.; Heider, T.; Atkinson, M.J.; Moertl, S. Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. PLoS ONE 2016, 11, e01522131. [Google Scholar] [CrossRef] [PubMed]
- Aravindhan, S.; Ejam, S.S.; Lafta, M.H.; Markov, A.; Yumashev, A.V.; Ahmadi, M. Mesenchymal stem cells and cancer therapy: Insights into targeting the tumour vasculature. Cancer Cell Int. 2021, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Nwabo, K.A.H.; Kamga, P.T.; Simo, R.T.; Vecchio, L.; Seke, E.P.F.; Muller, J.M.; Bassi, G.; Lukong, E.; Goel, R.K.; Amvene, J.M.; et al. Mesenchymal stromal cells’ role in tumor microenvironment: Involvement of signaling pathways. Cancer Biol. Med. 2017, 14, 129. [Google Scholar] [CrossRef]
- Rattigan, Y.; Hsu, J.-M.; Mishra, P.J.; Glod, J.; Banerjee, D. Interleukin 6 mediated recruitment of mesenchymal stem cells to the hypoxic tumor milieu. Exp. Cell Res. 2010, 316, 3417–3424. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.R.L.V. The unusual paradox of cancer-associated inflammation: An update. J. Bras. Patol. Med. Lab. 2019, 55, 321–332. [Google Scholar] [CrossRef]
- Lin, W.; Huang, L.; Li, Y.; Fang, B.; Li, G.; Chen, L.; Xu, L. Mesenchymal Stem Cells and Cancer: Clinical Challenges and Opportunities. Biomed. Res. Int. 2019, 2019, 2820853. [Google Scholar] [CrossRef] [Green Version]
- Cheteh, E.H.; Sarne, V.; Ceder, S.; Bianchi, J.; Augsten, M.; Rundqvist, H.; Egevad, L.; Östman, A.; Wiman, K.G. Interleukin-6 derived from cancer-associated fibroblasts attenuates the p53 response to doxorubicin in prostate cancer cells. Cell Death Discov. 2020, 6, 42. [Google Scholar] [CrossRef]
- Maffey, A.; Storini, C.; Diceglie, C.; Martelli, C.; Sironi, L.; Calzarossa, C.; Tonna, N.; Lovchik, R.; Delamarche, E.; Ottobrini, L.; et al. Mesenchymal stem cells from tumor microenvironment favour breast cancer stem cell proliferation, cancerogenic and metastatic potential, via ionotropic purinergic signalling. Sci. Rep. 2017, 7, 13162. [Google Scholar] [CrossRef]
- Hill, B.S.; Pelagalli, A.; Passaro, N.; Zannetti, A. Tumor-educated mesenchymal stem cells promote pro-metastatic phenotype. Oncotarget 2017, 8, 73296–73311. [Google Scholar] [CrossRef] [Green Version]
- Sai, B.; Dai, Y.; Fan, S.; Wang, F.; Wang, L.; Li, Z.; Tang, J.; Wang, L.; Zhang, X.; Zheng, L.; et al. Cancer-educated mesenchymal stem cells promote the survival of cancer cells at primary and distant metastatic sites via the expansion of bone marrow-derived-PMN-MDSCs. Cell Death Dis. 2019, 10, 941. [Google Scholar] [CrossRef]
- Chen, J.; Ji, T.; Wu, D.; Jiang, S.; Zhao, J.; Lin, H.; Cai, X. Human mesenchymal stem cells promote tumor growth via MAPK pathway and metastasis by epithelial mesenchymal transition and integrin α5 in hepatocellular carcinoma. Cell Death Dis. 2019, 10, 425. [Google Scholar] [CrossRef] [Green Version]
- Ridge, S.M.; Sullivan, F.J.; Glynn, S.A. Mesenchymal stem cells: Key players in cancer progression. Mol. Cancer 2017, 16, 31. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.; Liang, S.; Ghosh, S.; Hornsby, P.J.; Li, R. Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 2009, 28, 2745–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Hong, I. Double-edged sword of mesenchymal stem cells: Cancer-promoting versus therapeutic potential. Cancer Sci. 2017, 108, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, T.; Wu, W.; Ding, G. Interplay between mesenchymal stem cell and tumor and potential application. Hum. Cell 2020, 33, 444–458. [Google Scholar] [CrossRef]
- Nakamura, K.; Ito, Y.; Kawano, Y.; Kurozumi, K.; Kobune, M.; Tsuda, H.; Bizen, A.; Honmou, O.; Niitsu, Y.; Hamada, H. Antitumor effect of genetically engineered mesenchymal stem cells in a rat glioma model. Gene Ther. 2004, 11, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Xu, Z.; Zhao, T.; Zhao, Z.; Shi, M.; Zhao, R.C.; Ye, L.; Zhang, X. Suppression of tumorigenesis by human mesenchymal stem cells in a hepatoma model. Cell Res. 2008, 18, 500–507. [Google Scholar] [CrossRef] [Green Version]
- Atiya, H.; Frisbie, L.; Pressimone, C.; Coffman, L. Mesenchymal Stem Cells in the Tumor Microenvironment. Tumor Microenviron. 2020, 1234, 31–42. [Google Scholar]
- Hmadcha, A.; Martin-Montalvo, A.; Gauthier, B.R.; Soria, B.; Capilla-Gonzalez, V. Therapeutic Potential of Mesenchymal Stem Cells for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 43. [Google Scholar] [CrossRef]
- Fujita, Y.; Kadota, T.; Araya, J.; Ochiya, T.; Kuwano, K. Clinical application of mesenchymal stem cell-derived extracellular vesicle-based therapeutics for inflammatory lung diseases. J. Clin. Med. 2018, 7, 355. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kandoi, S.; Misra, R.; Vijayalakshmi, S.; Rajagopal, K.; Verma, R.S. The mesenchymal stem cell secretome: A new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 2019, 46, 1–9. [Google Scholar] [CrossRef]
- Klymiuk, M.C.; Balz, N.; Elashry, M.I.; Heimann, M.; Wenisch, S.; Arnhold, S. Exosomes isolation and identification from equine mesenchymal stem cells 06 Biological Sciences 0601 Biochemistry and Cell Biology. BMC Vet. Res. 2019, 15, 42. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhyan, R.K.; Gerasimov, U.V. Bone marrow osteogenic stem cells: In vitro cultivation and transplantation in diffusion chambers. Cell Prolif. 1987, 20, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, A.; Bramanti, P.; Mazzon, E. Mesenchymal stem cells: A potential therapeutic approach for amyotrophic lateral sclerosis? Stem Cells Int. 2019, 2019, 3675627. [Google Scholar] [CrossRef]
- Joyce, N.; Annett, G.; Wirthlin, L.; Olson, S.; Bauer, G.; Nolta, J.A. Mesenchymal stem cells for the treatment of neurodegenerative disease. Regen. Med. 2010, 5, 933–946. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, O.J.; Lee, K.O.; Jung, H.; Oh, S.-H.; Kim, N.K. Enhancing the therapeutic potential of CCL2-overexpressing mesenchymal stem cells in acute stroke. Int. J. Mol. Sci. 2020, 21, 7795. [Google Scholar] [CrossRef]
- Gomzikova, M.O.; Rizvanov, A.A. Current trends in regenerative medicine: From cell to cell-free therapy. Bionanoscience 2017, 7, 240–245. [Google Scholar] [CrossRef]
- Duffield, J.S.; Park, K.M.; Hsiao, L.-L.; Kelley, V.R.; Scadden, D.T.; Ichimura, T.; Bonventre, J.V. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J. Clin. Investig. 2005, 115, 1743–1755. [Google Scholar] [CrossRef]
- Biancone, L.; Bruno, S.; Deregibus, M.C.; Tetta, C.; Camussi, G. Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol. Dial. Transplant. 2012, 27, 3037–3042. [Google Scholar] [CrossRef]
- Eggenhofer, E.; Benseler, V.; Kroemer, A.; Popp, F.C.; Geissler, E.K.; Schlitt, H.J.; Baan, C.C.; Dahlke, M.H.; Hoogduijn, M.J. Mesenchymal stem cells are short-lived and do not migrate beyond the lungs after intravenous infusion. Front. Immunol. 2012, 3, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.S.; Lee, H.J.; Doo, S.H.; Lee, S.J.; Lim, I.; Chang, K.-T.; Kim, S.U. Mesenchymal stem cells overexpressing hepatocyte growth factor (HGF) inhibit collagen deposit and improve bladder function in rat model of bladder outlet obstruction. Cell Transplant. 2012, 21, 1641–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Dennis, J.E.; Muzic, R.F.; Lundberg, M.; Caplan, A.I. The dynamic in vivo distribution of bone marrow-derived mesenchymal stem cells after infusion. Cells Tissues Organs 2001, 169, 12–20. [Google Scholar] [CrossRef]
- Lee, R.H.; Pulin, A.A.; Seo, M.J.; Kota, D.J.; Ylostalo, J.; Larson, B.L.; Semprun-Prieto, L.; Delafontaine, P.; Prockop, D.J. Intravenous hMSCs improve myocardial Infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell 2009, 5, 54–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal stem cell secretome: Toward cell-free therapeutic strategies in regenerative medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Yang, Q.; Wang, Q.; Shi, C.; Wang, D.; Armato, U.; Prà, I.D.; Chiarini, A. Mesenchymal stromal cells-exosomes: A promising cell-free therapeutic tool for wound healing and cutaneous regeneration. Burn. Trauma 2019, 7, s41038-019-0178-8. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Sun, X.; Liu, L.; Jiang, H.; Shen, Y.; Xu, X.; Li, J.; Zhang, G.; Huang, J.; Lin, Z.; et al. Exosomes and their therapeutic potentials of stem cells. Stem Cells Int. 2016, 2016, 7653489. [Google Scholar] [CrossRef] [Green Version]
- Boltze, J. The dark side of the force—Constraints and complications of cell therapies for stroke. Front. Neurol. 2015, 6, 155. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367. [Google Scholar] [CrossRef]
- Agliano, A.; Calvo, A.; Box, C. The challenge of targeting cancer stem cells to halt metastasis. Semin. Cancer Biol. 2017, 44, 25–42. [Google Scholar] [CrossRef]
- Kim, D.S.; Jang, I.K.; Lee, M.W.; Ko, Y.J.; Lee, D.-H.; Lee, J.W.; Sung, K.W.; Koo, H.H.; Yoo, K.H. Enhanced immunosuppressive properties of human mesenchymal stem cells primed by interferon-γ. EBioMedicine 2018, 28, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A system of cytokines encapsulated in extracellular vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. Author correction: A system of cytokines encapsulated in extraCellular vesicles. Sci. Rep. 2020, 10, 18935. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tan, X.; Tan, Y.; Li, Q.; Ma, J.; Wang, G. Mesenchymal Stem Cell Derived Exosomes in Cancer Progression, Metastasis and Drug Delivery: A Comprehensive Review. J. Cancer 2018, 9, 3129–3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberger, L.; Ezquer, M.; Lillo-Vera, F.; Pedraza, P.L.; Ortúzar, M.I.; González, P.L.; Figueroa-Valdés, A.I.; Cuenca, J.; Ezquer, F.; Khoury, M.; et al. Stem cell exosomes inhibit angiogenesis and tumor growth of oral squamous cell carcinoma. Sci. Rep. 2019, 9, 663. [Google Scholar] [CrossRef]
- Yuan, Z.; Kolluri, K.K.; Gowers, K.H.C.; Janes, S.M. TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J. Extracell. Vesicles 2017, 6, 1265291. [Google Scholar] [CrossRef]
- Joo, H.S.; Suh, J.H.; Lee, J.H.; Bang, E.S.; Lee, J.M. Current Knowledge and Future Perspectives on Mesenchymal Stem Cell-Derived Exosomes as a New Therapeutic Agent. Int. J. Mol. Sci. 2020, 21, 727. [Google Scholar] [CrossRef] [Green Version]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Yang, C.; Shi, Y.; Zhao, L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J. Nanobiotechnol. 2018, 16, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Gui, S.; Liu, Y.; Qiu, X.; Zhang, G.; Zhang, X.; Pan, J.; Fan, J.; Qi, S.; Qiu, B. Exosomes derived from microRNA-199a-overexpressing mesenchymal stem cells inhibit glioma progression by down-regulating AGAP2. Aging 2019, 11, 5300–5318. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, L.; Li, F.; Yang, J.; Wan, X.; Ouyang, M. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J. Cell. Physiol. 2019, 234, 21380–21394. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; von der Ohe, J.; Hass, R. Anti-Tumor Effects of Exosomes Derived from Drug-Incubated Permanently Growing Human MSC. Int. J. Mol. Sci. 2020, 21, 7311. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhang, Y.; Zhang, H.; Xu, P.; Liu, J.; Ma, J.; Lv, M.; Li, D.; Katirai, F.; Shen, G.-X.; et al. Delivery of chemotherapeutic drugs in tumour cell-derived microparticles. Nat. Commun. 2012, 3, 1282. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Zhang, Y.; Tang, K.; Zhang, H.; Yin, X.; Li, Y.; Xu, P.; Sun, Y.; Ma, R.; Ji, T.; et al. Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles. Cell Res. 2016, 26, 713–727. [Google Scholar] [CrossRef] [Green Version]
- Saari, H.; Lázaro-Ibáñez, E.; Viitala, T.; Vuorimaa-Laukkanen, E.; Siljander, P.; Yliperttula, M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release 2015, 220, 727–737. [Google Scholar] [CrossRef] [Green Version]
- Ota, Y.; Takahashi, K.; Otake, S.; Tamaki, Y.; Okada, M.; Aso, K.; Makino, Y.; Fujii, S.; Ota, T.; Haneda, M. Extracellular vesicle-encapsulated miR-30e suppresses cholangiocarcinoma cell invasion and migration via inhibiting epithelial-mesenchymal transition. Oncotarget 2018, 9, 16400–16417. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, Z.; Han, X.; Shi, J.; Tu, D.; Song, W.; Zhang, J.; Qiu, X.; Ren, Y.; Zhen, L. MiR-30e inhibits tumor growth and chemoresistance via targeting IRS1 in Breast Cancer. Sci. Rep. 2017, 7, 15929. [Google Scholar] [CrossRef]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.-H.; Bachelier, R.; Allioli, N.; Hong, S.-S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis–Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhao, L.; Zhao, S.; Guo, T.; Li, F.; Li, Z.; Fang, L.; Wu, T.; Gu, C. MicroRNA-30e inhibits proliferation and invasion of non-small cell lung cancer via targeting SOX9. Hum. Cell 2019, 32, 326–333. [Google Scholar] [CrossRef]
- Xu, G.; Cai, J.; Wang, L.; Jiang, L.; Huang, J.; Hu, R.; Ding, F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp. Cell Res. 2018, 362, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xie, F.; Gao, A.; Zhang, R.; Zhang, L.; Xiao, Z.; Hu, Q.; Huang, W.; Huang, Q.; Lin, B.; et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol. Cancer 2017, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Laudato, S.; Patil, N.; Abba, M.L.; Leupold, J.H.; Benner, A.; Gaiser, T.; Marx, A.; Allgayer, H. P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int. J. Cancer 2017, 141, 1879–1890. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xu, J.; He, J.; Zheng, Y.; Li, H.; Lu, Y.; Qian, J.; Lin, P.; Weber, D.M.; Yang, J.; et al. A critical role of autocrine sonic hedgehog signaling in human CD138+ myeloma cell survival and drug resistance. Blood 2014, 124, 2061–2071. [Google Scholar] [CrossRef]
- NguyenThai, Q.-A.; Sharma, N.; Luong, D.H.; Sodhi, S.S.; Kim, J.-H.; Kim, N.; Oh, S.-J.; Jeong, D.K. Targeted inhibition of osteosarcoma tumor growth by bone marrow-derived mesenchymal stem cells expressing cytosine deaminase/5-fluorocytosine in tumor-bearing mice. J. Gene Med. 2015, 17, 87–99. [Google Scholar] [CrossRef]
- Lee, K.S.; Choi, J.S.; Cho, Y.W. Reprogramming of cancer stem cells into non-tumorigenic cells using stem cell exosomes for cancer therapy. Biochem. Biophys. Res. Commun. 2019, 512, 511–516. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Dias Câmara, D.A.; Luiz de Sá Junior, P.; Alexandre de Azevedo, R.; Figueiredo, C.R.; Araldi, R.P.; Levy, D.; Madeiro de Souza, D.; Kerkis, I. Identification of very small cancer stem cells expressing hallmarks of pluripotency in B16F10 melanoma cells and their reoccurrence in B16F10-derived clones. Exp. Cell Res. 2020, 391, 111938. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef]
- Souza, A.G.; Silva, I.B.B.; Campos-Fernandez, E.; Barcelos, L.S.; Souza, J.B.; Marangoni, K.; Goulart, L.R.; Alonso-Goulart, V. Comparative Assay of 2D and 3D Cell Culture Models: Proliferation, Gene Expression and Anticancer Drug Response. Curr. Pharm. Des. 2018, 24, 1689–1694. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros, A.S.; Costa, E.C.; Nunes, A.S.; de Melo-Diogo, D.; Correia, I.J. Comparative study of the therapeutic effect of Doxorubicin and Resveratrol combination on 2D and 3D (spheroids) cell culture models. Int. J. Pharm. 2018, 551, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Thippabhotla, S.; Zhong, C.; He, M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 2019, 9, 13012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kratochvil, M.J.; Seymour, A.J.; Li, T.L.; Paşca, S.P.; Kuo, C.J.; Heilshorn, S.C. Engineered materials for organoid systems. Nat. Rev. Mater. 2019, 4, 606–622. [Google Scholar] [CrossRef]
- Szvicsek, Z.; Oszvald, Á.; Szabó, L.; Sándor, G.O.; Kelemen, A.; Soós, A.Á.; Pálóczi, K.; Harsányi, L.; Tölgyes, T.; Dede, K.; et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell. Mol. Life Sci. 2019, 76, 2463–2476. [Google Scholar] [CrossRef] [Green Version]
- Rocha, S.; Carvalho, J.; Oliveira, P.; Voglstaetter, M.; Schvartz, D.; Thomsen, A.R.; Walter, N.; Khanduri, R.; Sanchez, J.C.; Keller, A.; et al. 3D Cellular Architecture Affects MicroRNA and Protein Cargo of Extracellular Vesicles. Adv. Sci. 2019. [Google Scholar] [CrossRef] [Green Version]
- Villasante, A.; Marturano-Kruik, A.; Ambati, S.R.; Liu, Z.; Godier-Furnemont, A.; Parsa, H.; Lee, B.W.; Moore, M.A.S.; Vunjak-Novakovic, G. Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics 2016, 6, 1119. [Google Scholar] [CrossRef]
- Van De Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; Van Houdt, W.; Van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salahudeen, A.A.; Kuo, C.J. Toward recreating colon cancer in human organoids. Nat. Med. 2015, 21, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.A.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [Green Version]
- Yeon, J.H.; Jeong, H.E.; Seo, H.; Cho, S.; Kim, K.; Na, D.; Chung, S.; Park, J.; Choi, N.; Kang, J.Y. Cancer-derived exosomes trigger endothelial to mesenchymal transition followed by the induction of cancer-associated fibroblasts. Acta Biomater. 2018, 76, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Oszvald, Á.; Szvicsek, Z.; Pápai, M.; Kelemen, A.; Varga, Z.; Tölgyes, T.; Dede, K.; Bursics, A.; Buzás, E.I.; Wiener, Z. Fibroblast-Derived Extracellular Vesicles Induce Colorectal Cancer Progression by Transmitting Amphiregulin. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Huleihel, L.; Hussey, G.S.; Naranjo, J.D.; Zhang, L.; Dziki, J.L.; Turner, N.J.; Stolz, D.B.; Badylak, S.F. Matrix-bound nanovesicles within ECM bioscaffolds. Sci. Adv. 2016, 2, e1600502. [Google Scholar] [CrossRef] [Green Version]
- Belgodere, J.A.; King, C.T.; Bursavich, J.B.; Burow, M.E.; Martin, E.C.; Jung, J.P. Engineering breast cancer microenvironments and 3D bioprinting. Front. Bioeng. Biotechnol. 2018, 6, 1–26. [Google Scholar] [CrossRef]
- Maiullari, F.; Chirivì, M.; Costantini, M.; Ferretti, A.M.; Recchia, S.; Maiullari, S.; Milan, M.; Presutti, D.; Pace, V.; Raspa, M.; et al. In vivo organized neovascularization induced by 3D bioprinted endothelial-derived extracellular vesicles. Biofabrication 2021, 13, 35014. [Google Scholar] [CrossRef] [PubMed]
- Seol, Y.-J.; Kang, H.-W.; Lee, S.J.; Atala, A.; Yoo, J.J. Bioprinting technology and its applications. Eur. J. Cardio-Thorac. Surg. 2014, 46, 342–348. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, V.R.; Araldi, R.P.; Vigerelli, H.; D’Ámelio, F.; Mendes, T.B.; Gonzaga, V.; Policíquio, B.; Colozza-Gama, G.A.; Valverde, C.W.; Kerkis, I. Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications. Cells 2021, 10, 2617. https://doi.org/10.3390/cells10102617
da Costa VR, Araldi RP, Vigerelli H, D’Ámelio F, Mendes TB, Gonzaga V, Policíquio B, Colozza-Gama GA, Valverde CW, Kerkis I. Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications. Cells. 2021; 10(10):2617. https://doi.org/10.3390/cells10102617
Chicago/Turabian Styleda Costa, Vitor Rodrigues, Rodrigo Pinheiro Araldi, Hugo Vigerelli, Fernanda D’Ámelio, Thais Biude Mendes, Vivian Gonzaga, Bruna Policíquio, Gabriel Avelar Colozza-Gama, Cristiane Wenceslau Valverde, and Irina Kerkis. 2021. "Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications" Cells 10, no. 10: 2617. https://doi.org/10.3390/cells10102617
APA Styleda Costa, V. R., Araldi, R. P., Vigerelli, H., D’Ámelio, F., Mendes, T. B., Gonzaga, V., Policíquio, B., Colozza-Gama, G. A., Valverde, C. W., & Kerkis, I. (2021). Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications. Cells, 10(10), 2617. https://doi.org/10.3390/cells10102617






