DNA Methylation and Immune Memory Response
Abstract
:1. Introduction
2. DNA Methylation
3. DNA Methylation and Memory T Cells
3.1. CD4 T Cells
3.2. CD8 T Cells
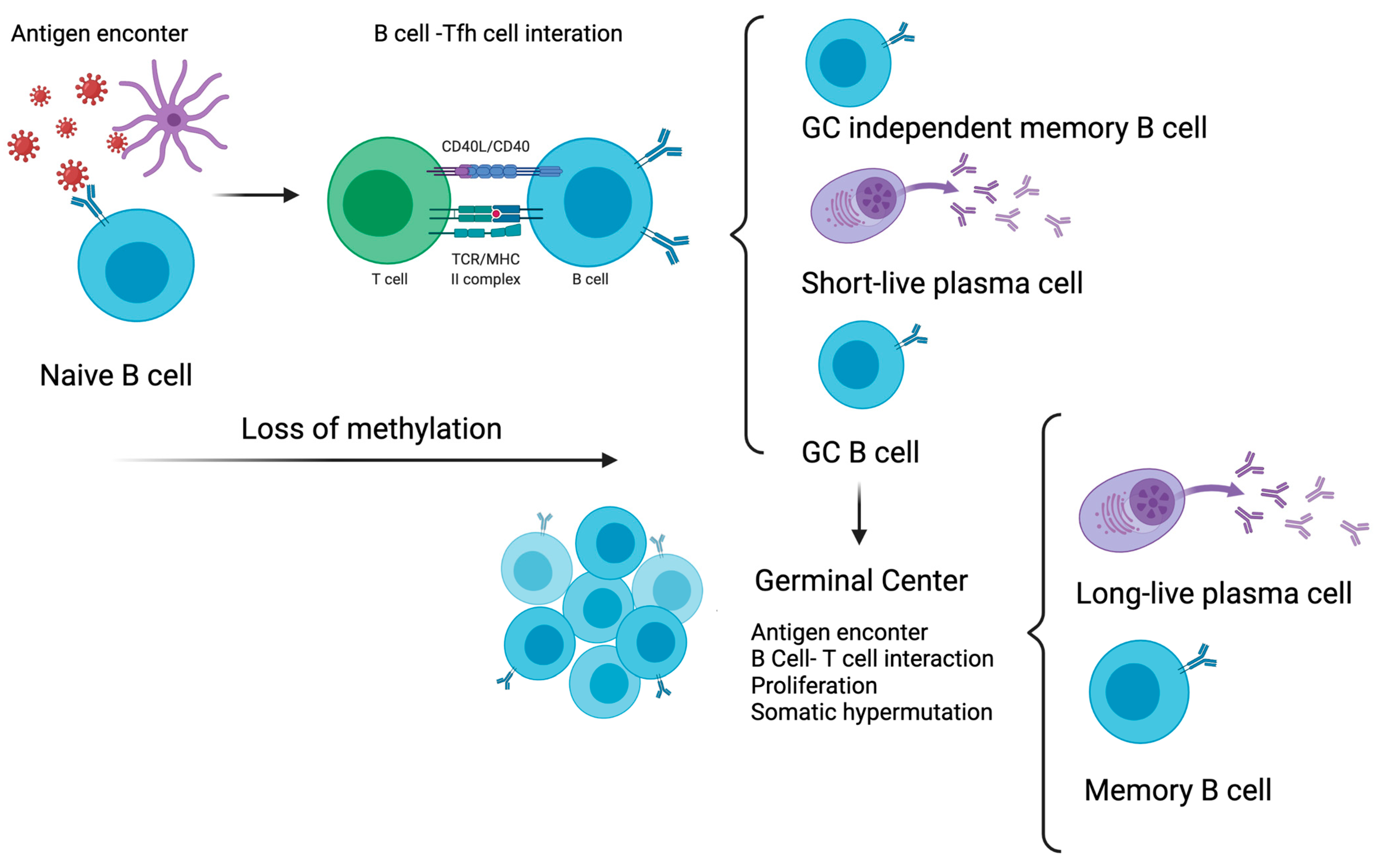
4. DNA Methylation and Memory B Cells
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Mueller, S.N.; Gebhardt, T.; Carbone, F.R.; Heath, W. Memory T Cell Subsets, Migration Patterns, and Tissue Residence. Annu. Rev. Immunol. 2013, 31, 137–161. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, M.; Kwak, K.; Pierce, S.K. B cell memory: Building two walls of protection against pathogens. Nat. Rev. Immunol. 2020, 20, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Zebley, C.C.; Gottschalk, S.; Youngblood, B. Rewriting History: Epigenetic Reprogramming of CD8+ T Cell Differentiation to Enhance Immunotherapy. Trends Immunol. 2020, 41, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Carter, B.; Zhao, K. The epigenetic basis of cellular heterogeneity. Nat. Rev. Genet. 2020, 22, 235–250. [Google Scholar] [CrossRef]
- Noble, D. Conrad Waddington and the origin of epigenetics. J. Exp. Biol. 2015, 218, 816–818. [Google Scholar] [CrossRef] [Green Version]
- Barski, A.; Cuddapah, S.; Kartashov, A.V.; Liu, C.; Imamichi, H.; Yang, W.; Peng, W.; Lane, H.C.; Zhao, K. Rapid Recall Ability of Memory T cells is Encoded in their Epigenome. Sci. Rep. 2017, 7, 39785. [Google Scholar] [CrossRef] [Green Version]
- Henning, A.N.; Roychoudhuri, R.; Restifo, N.P. Epigenetic control of CD8+ T’cell differentiation. Nat. Rev. Immunol. 2018, 18, 340–356. [Google Scholar] [CrossRef]
- Araki, Y.; Fann, M.; Wersto, R.; Weng, N. Histone Acetylation Facilitates Rapid and Robust Memory CD8 T Cell Response through Differential Expression of Effector Molecules (Eomesodermin and Its Targets: Perforin and Granzyme B). J. Immunol. 2008, 180, 8102–8108. [Google Scholar] [CrossRef]
- Russ, B.E.; Olshanksy, M.; Smallwood, H.S.; Li, J.; Denton, A.E.; Prier, J.E.; Stock, A.T.; Croom, H.A.; Cullen, J.G.; Nguyen, M.L.T.; et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 2014, 41, 853–865. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Zhang, K.; Milner, J.J.; Toma, C.; Chen, R.; Scott-Browne, J.P.; Pereira, R.M.; Crotty, S.; Chang, J.T.; Pipkin, M.E.; et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 2017, 18, 573–582. [Google Scholar] [CrossRef] [Green Version]
- Araki, Y.; Wang, Z.; Zang, C.; Wood, W.H.; Schones, D.; Cui, K.; Roh, T.Y.; Lhotsky, B.; Wersto, R.P.; Peng, W.; et al. Genome-wide Analysis of Histone Methylation Reveals Chromatin State-Based Regulation of Gene Transcription and Function of Memory CD8+ T Cells. Immunity 2009, 30, 912–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jima, D.; Moffitt, A.B.; Liu, Q.; Czader, M.; Hsi, E.D.; Fedoriw, Y.; Dunphy, C.H.; Richards, K.L.; Gill, J.I.; et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 2014, 123, 2988–2996. [Google Scholar] [CrossRef]
- Monticelli, S. MicroRNAs in T helper cell differentiation and plasticity. Semin. Immunol. 2013, 25, 291–298. [Google Scholar] [CrossRef]
- Zan, H.; Casali, P. Epigenetics of peripheral B-cell differentiation and the antibody response. Front. Immunol. 2015, 6, 631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, C.; Li, F.; Zhang, B.; Zhang, Y. Regulation of memory CD8+ T cell differentiation by MicroRNAs. Cell. Physiol. Biochem. 2018, 47, 2187–2198. [Google Scholar] [CrossRef]
- Yang, L.; Boldin, M.P.; Yu, Y.; Liu, C.S.; Ea, C.K.; Ramakrishnan, P.; Taganov, K.D.; Zhao, J.L.; Baltimore, D. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 2012, 209, 1655–1670. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Wieland, A.; Araki, K.; Davis, C.W.; Ye, L.; Hale, J.S.; Ahmed, R. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. USA 2012, 109, 9965–9970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.-Y.; Allie, S.R.; Zhang, W.; Usherwood, E.J. MicroRNA miR-155 Affects Antiviral Effector and Effector Memory CD8 T Cell Differentiation. J. Virol. 2013, 87, 2348–2351. [Google Scholar] [CrossRef] [Green Version]
- Baumjohann, D.; Kageyama, R.; Clingan, J.M.; Morar, M.M.; Patel, S.; De Kouchkovsky, D.; Bannard, O.; Bluestone, J.A.; Matloubian, M.; Ansel, K.M.; et al. The microRNA cluster miR-17∼92 promotes T FH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 2013, 14, 840–848. [Google Scholar] [CrossRef]
- Salunkhe, S.; Vaidya, T. CD40-miRNA axis controls prospective cell fate determinants during B cell differentiation. Mol. Immunol. 2020, 126, 46–55. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Jeltsch, A.; Jurkowska, R.Z. New concepts in DNA methylation. Trends Biochem. Sci. 2014, 39, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.L.; Veenstra, G.J.C.; Wade, P.A.; Vermaak, D.; Kass, S.U.; Landsberger, N.; Strouboulis, J.; Wolffe, A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998, 19, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Han, H.; DeCarvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef] [Green Version]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 2018, 9, 1–16. [Google Scholar]
- Han, H.; Cortez, C.C.; Yang, X.; Nichols, P.W.; Jones, P.A.; Liang, G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum. Mol. Genet. 2011, 20, 4299–4310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Probst, A.V.; Dunleavy, E.; Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009, 10, 192–206. [Google Scholar] [CrossRef]
- Kareta, M.S.; Botello, Z.M.; Ennis, J.J.; Chou, C.; Chédin, F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006, 281, 25893–25902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhutani, N.; Brady, J.J.; Damian, M.; Sacco, A.; Corbel, S.Y.; Blau, H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 2010, 463, 1042. [Google Scholar] [CrossRef] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Scicluna, B.P.; van der Poll, T. The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front. Immunol. 2021, 12, 3073. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Hodges, E.; Molaro, A.; Dos Santos, C.O.; Thekkat, P.; Song, Q.; Uren, P.J.; Park, J.; Butler, J.; Rafii, S.; McCombie, W.R.; et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol. Cell 2011, 44, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Bock, C.; Beerman, I.; Lien, W.H.; Smith, Z.D.; Gu, H.; Boyle, P.; Gnirke, A.; Fuchs, E.; Rossi, D.J.; Meissner, A. DNA Methylation Dynamics during In Vivo Differentiation of Blood and Skin Stem Cells. Mol. Cell 2012, 47, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Hogart, A.; Lichtenberg, J.; Ajay, S.S.; Anderson, S.; Margulies, E.H.; Bodine, D.M. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012, 22, 1407–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Good-Jacobson, K.L. Epigenetic regulation of B cell fate and function during an immune response. Immunol. Rev. 2019, 288, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.M.; Adams, N.M.; Geary, C.D.; Weizman, O. El Rapp, M.; Pritykin, Y.; Leslie, C.S.; Sun, J.C. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 2018, 19, 963–972. [Google Scholar] [CrossRef]
- Reiner, S.L. Epigenetic control in the immune response. Hum. Mol. Genet. 2005, 14, R41–6. [Google Scholar] [CrossRef] [Green Version]
- Tough, D.F.; Rioja, I.; Modis, L.K.; Prinjha, R.K. Epigenetic Regulation of T Cell Memory: Recalling Therapeutic Implications. Trends Immunol. 2020, 41, 29–45. [Google Scholar] [CrossRef]
- Caza, T.; Landas, S. Functional and Phenotypic Plasticity of CD4+ T Cell Subsets. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.U.; Agarwal, S.; Rao, A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 2002, 16, 649–660. [Google Scholar] [CrossRef] [Green Version]
- Wilson, C.B.; Rowell, E.; Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009, 9, 91–105. [Google Scholar] [CrossRef]
- Schmidl, C.; Delacher, M.; Huehn, J.; Feuerer, M. Epigenetic mechanisms regulating T-cell responses. J. Allergy Clin. Immunol. 2018, 142, 728–743. [Google Scholar] [CrossRef] [Green Version]
- Soon, M.S.; Engel, J.A.; Lee, H.J.; Haque, A. Development of circulating CD4+ T-cell memory. Immunol. Cell Biol. 2019, 97, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Pepper, M.; Pagán, A.J.; Igyártó, B.Z.; Taylor, J.J.; Jenkins, M.K. Opposing Signals from the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity 2021, 35, 583–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hale, J.S.; Youngblood, B.; Latner, D.R.; Mohammed, A.U.R.; Ye, L.; Akondy, R.S.; Wu, T.; Iyer, S.S.; Ahmed, R. Distinct Memory CD4+ T Cells with Commitment to T Follicular Helper- and T Helper 1-Cell Lineages Are Generated after Acute Viral Infection. Immunity 2013, 38, 805–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubo, N.J.; Fife, B.; Pagan, A.J.; Kotov, D.; Goldberg, M.F.; Jenkins, M.K. Most microbe-specific naive CD4+ T cells produce memory cells during infection. Science 2016, 351, 511–514. [Google Scholar] [CrossRef] [Green Version]
- Snook, J.P.; Kim, C.; Williams, M.A. TCR signal strength controls the differentiation of CD4 + effector and memory T cells. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Gramaglia, I.; Jember, A.; Pippig, S.D.; Weinberg, A.D.; Killeen, N.; Croft, M. The OX40 Costimulatory Receptor Determines the Development of CD4 Memory by Regulating Primary Clonal Expansion. J. Immunol. 2000, 165, 3043–3050. [Google Scholar] [CrossRef]
- Dooms, H.; Abbas, A.K. Control of CD4+ T-cell memory by cytokines and costimulators. Immunol. Rev. 2006, 21, 23–38. [Google Scholar] [CrossRef]
- DiToro, D.; Winstead, C.J.; Pham, D.; Witte, S.; Andargachew, R.; Singer, J.R.; Wilson, C.G.; Zindl, C.L.; Luther, R.J.; Silberger, D.J.; et al. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 2018, 361, eaao2933. [Google Scholar] [CrossRef]
- Polonsky, M.; Rimer, J.; Kern-Perets, A.; Zaretsky, I.; Miller, S.; Bornstein, C.; David, E.; Kopelman, N.M.; Stelzer, G.; Porat, Z.; et al. Induction of CD4 T cell memory by local cellular collectivity. Science 2018, 360, eaaj1853. [Google Scholar] [CrossRef] [Green Version]
- Groom, J.R.; Richmond, J.; Murooka, T.T.; Sorensen, E.W.; Sung, J.H.; Bankert, K. CXCR3 Chemokine Receptor-Ligand Interactions in the Lymph Node Optimize CD4+ T Helper 1 Cell Differentiation. Immunity 2012, 37, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, S.-I.; Ogoshi, K.; Sasaki, A.; Abe, J.; Qu, W.; Nakatani, Y.; Ahsan, B.; Oshima, K.; Shand, F.H.W.; Ametani, A.; et al. Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells. J. Immunol. 2013, 190, 4076–4091. [Google Scholar] [CrossRef] [Green Version]
- Miyao, T.; Floess, S.; Setoguchi, R.; Luche, H.; Fehling, H.J.; Waldmann, H.; Huehn, J.; Hori, S. Plasticity of Foxp3+ T Cells Reflects Promiscuous Foxp3 Expression in Conventional T Cells but Not Reprogramming of Regulatory T Cells. Immunity 2012, 36, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Chang, H.D.; Ivascu, C.; Qian, Y.; Rezai, S.; Okhrimenko, A. Loss of methylation at the IFNG promoter and CNS-1 is associated with the development of functional IFN-γ memory in human CD4+ T lymphocytes. Eur. J. Immunol. 2013, 43, 793–804. [Google Scholar] [CrossRef]
- Hedrich, C.M.; Crispin, J.C.; Rauen, T.; Ioannidis, C.; Apostolidis, S.A.; Lo, M.S. cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. USA 2012, 109, 16606–16611. [Google Scholar] [CrossRef] [Green Version]
- Schmidl, C.; Hansmann, L.; Andreesen, R.; Edinger, M.; Hoffmann, P.; Rehli, M. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naïve Treg. Eur. J. Immunol. 2011, 41, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.C.; Xiao, X.; Sun, R.; Tian, Z.; Wei, H. CD4+CD62L+ Central Memory T Cells Can Be Converted to Foxp3+ T Cells. PLoS ONE 2013, 8, e77322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, H.K.; Hart, T.; LaMere, S.A.; Chew, P.V.; Salomon, D.R. Defining CD4 T Cell Memory by the Epigenetic Landscape of CpG DNA Methylation. J. Immunol. 2015, 194, 1565–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durek, P.; Nordström, K.; Gasparoni, G.; Salhab, A.; Kressler, C.; de Almeida, M. Epigenomic Profiling of Human CD4+ T Cells Supports a Linear Differentiation Model and Highlights Molecular Regulators of Memory Development. Immunity 2016, 45, 1148–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Yao, L.J.; Saidin, S.; Paleja, B.; van Loosdregt, J.; Chua, C.; Arkachaisri, T.; Consolaro, A.; Gattorno, M.; Martini, A.; et al. Molecular mechanisms of autophagic memory in pathogenic T cells in human arthritis. J. Autoimmun. 2018, 94, 90–98. [Google Scholar] [CrossRef]
- Szilagyi, B.A.; Triebus, J.; Kressler, C.; De Almeida, M.; Tierling, S.; Durek, P.; Mardahl, M.; Floess, S.; Huehn, J.; Syrbe, U.; et al. Gut memories do not fade: Epigenetic regulation of lasting gut homing receptor expression in CD4+ memory T cells. Mucosal Immunol. 2017, 10, 1443–1454. [Google Scholar] [CrossRef] [Green Version]
- Nakayama-Hosoya, K.; Ishida, T.; Youngblood, B.; Nakamura, H.; Hosoya, N.; Koga, M.; Koibuchi, T.; Iwamoto, A.; Kawana-Tachikawa, A. Epigenetic Repression of Interleukin 2 Expression in Senescent CD4+T Cells During Chronic HIV Type 1 Infection. J. Infect. Dis. 2014, 211, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Kaech, S.M.; Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 2012, 12, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Kaech, S.M.; Tan, J.T.; Wherry, E.J.; Konieczny, B.T.; Surh, C.D.; Ahmed, R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003, 4, 1191–1198. [Google Scholar] [CrossRef]
- Herndler-Brandstetter, D.; Ishigame, H.; Shinnakasu, R.; Plajer, V.; Stecher, C.; Zhao, J.; Lietzenmayer, M.; Kroehling, L.; Takumi, A.; Kometani, K.; et al. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 2018, 48, 716–729.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurachi, M. CD8+ T cell exhaustion. Semin. Immunopathol. 2019, 41, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Joshi, N.S.; Cui, W.; Chandele, A.; Lee, H.K.; Urso, D.R.; Hagman, J.; Gapin, L.; Kaech, S.M. Inflammation Directs Memory Precursor and Short-Lived Effector CD8+ T Cell Fates via the Graded Expression of T-bet Transcription Factor. Immunity 2007, 27, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Intlekofer, A.; Banerjee, A.; Takemoto, N.; Gordon, S.; DeJong, C.S.; Shin, H.; Hunter, C.A.; Wherry, E.J.; Lindsten, T.; Reiner, S.L. Anomalous Type 17 Response to Viral Infection by CD8+ T Cells Lacking T-bet and Eomesodermin. Science 2008, 321, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Rutishauser, R.L.; Martins, G.A.; Kalachikov, S.; Chandele, A.; Parish, I.A.; Meffre, E.; Jacob, J.; Calame, K.; Kaech, S.M. Transcriptional Repressor Blimp-1 Promotes CD8+ T Cell Terminal Differentiation and Represses the Acquisition of Central Memory T Cell Properties. Immunity 2009, 31, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Masson, F.; Minnich, M.; Olshansky, M.; Bilic, I.; Mount, A.M.; Kallies, A.; Speed, T.P.; Busslinger, M.; Nutt, S.; Belz, G.T. Id2-Mediated Inhibition of E2A Represses Memory CD8+ T Cell Differentiation. J. Immunol. 2013, 190, 4585–4594. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yu, S.; Zhao, D.-M.; Harty, J.; Badovinac, V.; Xue, H.-H. Differentiation and Persistence of Memory CD8+ T Cells Depend on T Cell Factor 1. Immunity 2010, 33, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zander, R.; Khatun, A.; Schauder, D.M.; Cui, W. Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front. Immunol. 2018, 9, 2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akue, A.D.; Lee, J.-Y.; Jameson, S.C. Derivation and Maintenance of Virtual Memory CD8 T Cells. J. Immunol 2012, 188, 2516–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wissink, E.; Watson, N.B.; Smith, N.L.; Grimson, A.; Rudd, B.D. Fetal and adult progenitors give rise to unique populations of CD8+ T cells. Blood 2016, 128, 3073–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.T.; Cross, E.W.; Kedl, R.M. Antigen-inexperienced memory CD8+ T cells: Where they come from and why we need them. Nat. Rev. Immunol. 2017, 17, 391–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, L.; Goudot, C.; Zueva, E.; Gueguen, P.; Burgdorf, N.; Waterfall, J.J.; Quivy, J.-P.; Almouzni, G.; Amigorena, S. The epigenetic control of stemness in CD8+T cell fate commitment. Science 2018, 359, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Youngblood, B.; Hale, J.S.; Kissick, H.T.; Ahn, E.; Xu, X.; Wieland, A.; Araki, K.; West, E.E.; Ghoneim, H.E.; Fan, Y.; et al. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nat. Cell Biol. 2017, 552, 404–409. [Google Scholar] [CrossRef]
- Kersh, E.N.; Fitzpatrick, D.R.; Murali-Krishna, K.; Shires, J.; Speck, S.H.; Boss, J.M.; Ahmed, R. Rapid Demethylation of the IFN-γ Gene Occurs in Memory but Not Naive CD8 T Cells. J. Immunol. 2006, 176, 4083–4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, S.M.; Amezquita, R.A.; Guan, T.; Kleinstein, S.H.; Kaech, S.M. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity 2017, 46, 596–608. [Google Scholar] [CrossRef] [Green Version]
- Shosaku, J. Genome-wide DNA methylation analysis of senescence in repetitively infected memory cytotoxic T lymphocytes. Immunol. 2017, 153, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carty, S.A.; Gohil, M.; Banks, L.B.; Cotton, R.M.; Johnson, M.E.; Stelekati, E.; Wells, A.D.; Wherry, E.J.; Koretzky, G.A.; Jordan, M.S. The Loss of TET2 Promotes CD8+ T Cell Memory Differentiation. J. Immunol. 2018, 200, 82–91. [Google Scholar] [CrossRef] [Green Version]
- Ladle, B.H.; Li, K.-P.; Phillips, M.J.; Pucsek, A.B.; Haile, A.; Powell, J.D.; Jaffee, E.M.; Hildeman, D.A.; Gamper, C.J. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. USA 2016, 113, 10631–10636. [Google Scholar] [CrossRef] [Green Version]
- Youngblood, B.; Oestreich, K.J.; Ha, S.-J.; Duraiswamy, J.; Akondy, R.; West, E.E.; Wei, Z.; Lu, P.; Austin, J.W.; Riley, J.; et al. Chronic Virus Infection Enforces Demethylation of the Locus that Encodes PD-1 in Antigen-Specific CD8+ T Cells. Immunity 2011, 35, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakim, L.M.; Gupta, N.; Mintern, J.; Villadangos, J.A. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 2013, 14, 238–245. [Google Scholar] [CrossRef]
- Yao, Y.; Li, H.; Ding, J.; Xia, Y.; Wang, L. Progesterone impairs antigen-non-specific immune protection by CD8 T memory cells via interferon-γ gene hypermethylation. PLoS Pathog. 2017, 13, e1006736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Araújo-Souza, P.S.; Hanschke, S.C.H.; Nardy, A.F.F.R.; Sécca, C.; Oliveira-Vieira, B.; Silva, K.L. Differential interferon-γ production by naive and memory-like CD8 T cells. J. Leukoc. Biol. 2020, 108, 1329–1337. [Google Scholar] [CrossRef]
- Rodriguez, R.M.; Suarez-Alvarez, B.; Lavín, J.L.; Mosén-Ansorena, D.; Raneros, A.B.; Márquez-Kisinousky, L.; Aransay, A.M.; Lopez-Larrea, C. Epigenetic Networks Regulate the Transcriptional Program in Memory and Terminally Differentiated CD8+ T Cells. J. Immunol. 2016, 198, 937–949. [Google Scholar] [CrossRef] [Green Version]
- Akondy, R.S.; Fitch, M.; Edupuganti, S.; Yang, S.; Kissick, H.T.; Juliana, M.M.; Youngblood, B.; Abdelsamed, H.A.; McGuire, D.; Cohen, K.W.; et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nat. Cell Biol. 2017, 552, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.S.; You, S.; Kang, Y.; Lee, N.; Yoo, S.-A.; Park, K. DNA Methylation Regulates the Differential Expression of CX3CR1 on Human IL-7Rα low and IL-7Rα high Effector Memory CD8+ T Cells with Distinct Migratory Capacities to the Fractalkine. J. Immunol. 2015, 195, 2861–2869. [Google Scholar] [CrossRef] [Green Version]
- Hartana, C.A.; Bergman, E.A.; Broomé, A.; Berglund, S.; Johansson, M.; Alamdari, F.; Jakubczyk, T.; Huge, Y.; Aljabery, F.; Palmqvist, K.; et al. Tissue-resident memory T cells are epigenetically cytotoxic with signs of exhaustion in human urinary bladder cancer. Clin. Exp. Immunol. 2018, 194, 39–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelsamed, H.A.; Zebley, C.C.; Nguyen, H.; Rutishauser, R.L.; Fan, Y.; Ghoneim, H.E.; Crawford, J.C.; Alfei, F.; Alli, S.; Ribeiro, S.P.; et al. Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 2020, 21, 578–587. [Google Scholar] [CrossRef]
- Seifert, M.; Küppers, R. Human memory B cells. Leukemia 2021, 30, 2283–2292. [Google Scholar] [CrossRef] [PubMed]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Dalla-Favera, R. Germinal centres: Role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef]
- Taylor, J.J.; Pape, K.A.; Jenkins, M. A germinal center–independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 2012, 209, 597–606. [Google Scholar] [CrossRef]
- Jacob, J.; Kelsoe, G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J. Exp. Med. 1992, 176, 679–687. [Google Scholar] [CrossRef]
- Defrance, T.; Taillardet, M.; Genestier, L. T cell-independent B cell memory. Curr. Opin. Immunol. 2011, 23, 330–336. [Google Scholar] [CrossRef]
- Bernasconi, N.L.; Traggiai, E.; Lanzavecchia, A. Maintenance of Serological Memory by Polyclonal Activation of Human Memory B Cells. Science 2002, 298, 2199–2202. [Google Scholar] [CrossRef]
- Good, K.L.; Avery, D.T.; Tangye, S.G. Resting Human Memory B Cells Are Intrinsically Programmed for Enhanced Survival and Responsiveness to Diverse Stimuli Compared to Naive B Cells. J. Immunol. 2009, 182, 890–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barwick, B.G.; Scharer, C.D.; Martinez, R.J.; Price, M.J.; Wein, A.N.; Haines, R.R.; Bally, A.P.R.; Kohlmeier, J.E.; Boss, J.M. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 2018, 9, 1900. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-T.; Xiao, Y.; Muench, M.; Xiao, J.; Fomin, M.E.; Wiencke, J.K.; Zheng, S.; Dou, X.; De Smith, A.; Chokkalingam, A.; et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012, 40, 11339–11351. [Google Scholar] [CrossRef]
- Lai, A.Y.; Mav, D.; Shah, R.; Grimm, S.A.; Phadke, D.; Hatzi, K.; Melnick, A.; Geigerman, C.; Sobol, S.E.; Jaye, D.; et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013, 23, 2030–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neri, F.; Rapelli, S.; Krepelova, A.; Incarnato, D.; Parlato, C.; Basile, G.; Maldotti, M.; Anselmi, F.; Oliviero, S. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017, 543, 72–77. [Google Scholar] [CrossRef]
- Chen, M.; Hong, M.J.; Sun, H.; Wang, L.; Shi, X.; Gilbert, B.E.; Corry, D.B.; Kheradmand, F.; Wang, J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 2014, 20, 503–510. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Kodali, S.; Jang, A.; Kuai, L.; Wang, J. Requirement for Autophagy in the Long-Term Persistence but not Initial Formation of Memory B cells. J. Immunol. 2015, 194, 2607–2615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, V.C.R.; Molina, L.D.P.; Rodriguez-Ubreva, J.; Ciudad, L.; Gomez-Cabrero, D.; Company, C.; Urquiza, J.M.; Tegnér, J.; Rodríguez-Gallego, C.; López-Granados, E.; et al. Monozygotic twins discordant for common variable immunodeficiency reveal impaired DNA demethylation during naïve-to-memory B-cell transition. Nat. Commun. 2015, 6, 7335. [Google Scholar] [CrossRef]
- Gatto, S.; Gagliardi, M.; Franzese, M.; Leppert, S.; Papa, M.; Cammisa, M.; Grillo, G.; Velasco, G.; Francastel, C.; Toubiana, S.; et al. ICF-specific DNMT3B dysfunction interferes with intragenic regulation of mRNA transcription and alternative splicing. Nucleic Acids Res. 2017, 45, 5739–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Català-Moll, F.; Ferreté-Bonastre, A.G.; Li, T.; Weichenhan, D.; Lutsik, P.; Ciudad, L.; Álvarez-Prado, Á.F.; Rodríguez-Ubreva, J.; Klemann, C.; Speckmann, C.; et al. Activation-induced deaminase is critical for the establishment of DNA methylation patterns prior to the germinal center reaction. Nucleic Acids Res. 2021, 49, 5057–5073. [Google Scholar] [CrossRef]


| Enzyme Responsible for DNA Methylation | Enzyme Responsible for DNA Demethylation | |
|---|---|---|
| Memory CD4 T cells | De novo DNA methyltransferase 3a (DNMT3a) interacts with CREM, mediating the epigenetic remodelling of IL2 and IL17A during memory CD4 T cell differentiation [64]. | - |
| Memory CD8 T cells | The absence of DNMT3a promotes memory CD8 T cell differentiation in mice [86,91]. | The absence of ten-eleven translocation (TET)2 promotes memory CD8 T cell differentiation in mice [90]. |
| Memory B cells | DNMT3a expression is reduced in activated CG cells but is similar between naïve and memory B cells [111]. | TET mediates the demethylation during the transition from naïve to human memory B cells [117]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mittelstaedt, N.N.; Becker, A.L.; de Freitas, D.N.; Zanin, R.F.; Stein, R.T.; Duarte de Souza, A.P. DNA Methylation and Immune Memory Response. Cells 2021, 10, 2943. https://doi.org/10.3390/cells10112943
Mittelstaedt NN, Becker AL, de Freitas DN, Zanin RF, Stein RT, Duarte de Souza AP. DNA Methylation and Immune Memory Response. Cells. 2021; 10(11):2943. https://doi.org/10.3390/cells10112943
Chicago/Turabian StyleMittelstaedt, Nathalia Noschang, André Luiz Becker, Deise Nascimento de Freitas, Rafael F. Zanin, Renato T. Stein, and Ana Paula Duarte de Souza. 2021. "DNA Methylation and Immune Memory Response" Cells 10, no. 11: 2943. https://doi.org/10.3390/cells10112943
APA StyleMittelstaedt, N. N., Becker, A. L., de Freitas, D. N., Zanin, R. F., Stein, R. T., & Duarte de Souza, A. P. (2021). DNA Methylation and Immune Memory Response. Cells, 10(11), 2943. https://doi.org/10.3390/cells10112943





