Transcriptome Analysis of a Cotton Cultivar Provides Insights into the Differentially Expressed Genes Underlying Heightened Resistance to the Devastating Verticillium Wilt
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions and Fungal Pathogen Inoculations
2.2. RNA Extraction, Library Construction, and RNA Sequencing Analysis
2.3. Identification of Differentially Expressed Genes (DEGs)
2.4. Gene Ontology Enrichment and KEGG Pathway Analysis
2.5. Reverse Transcription and Quantitative PCR (RT-qPCR)
2.6. Statistical Analysis
3. Results
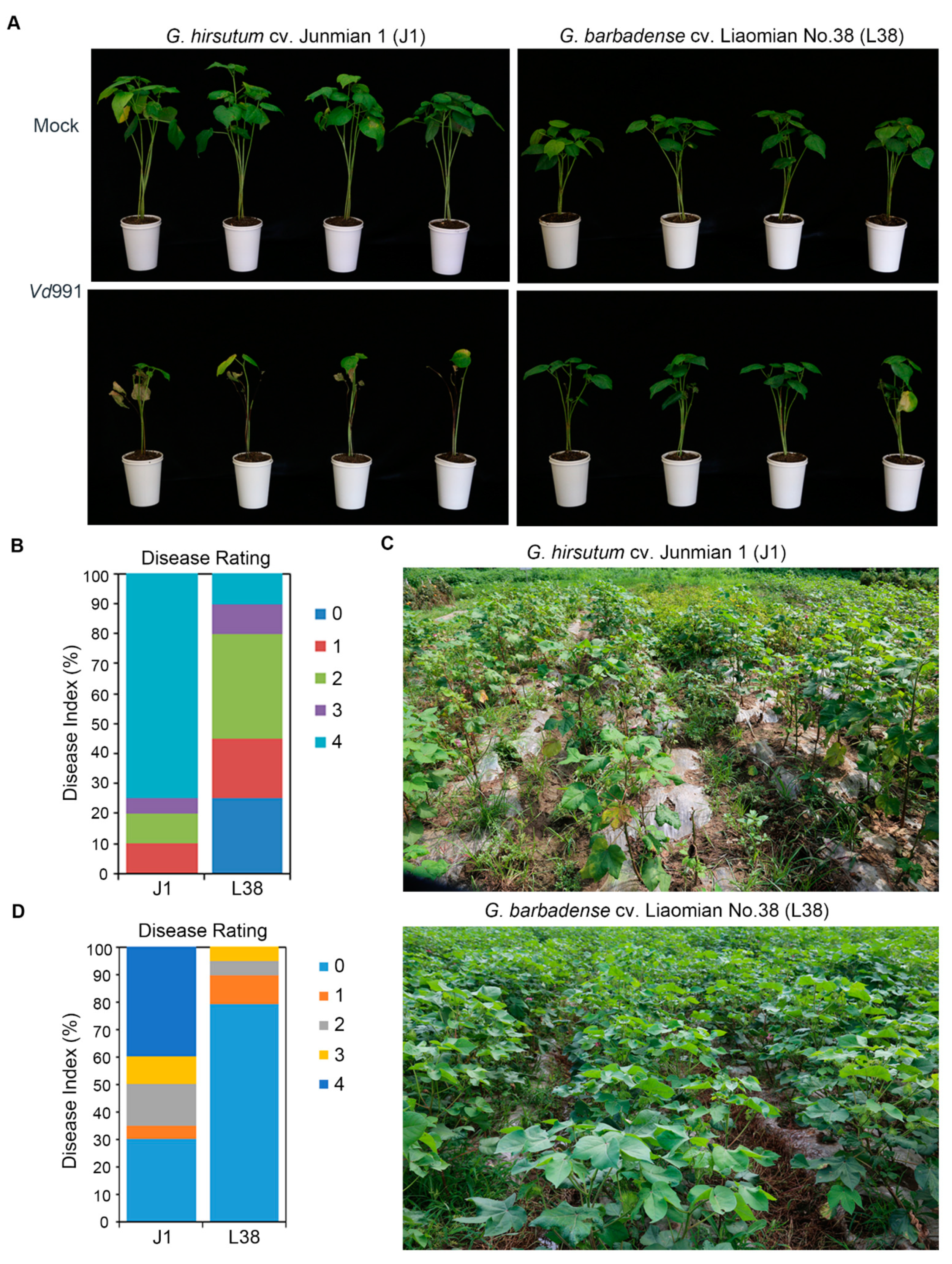
3.1. Resistant Phenotype of L38 Cotton
3.2. RNA-Seq Was Conducted to J1 and L38 Cotton
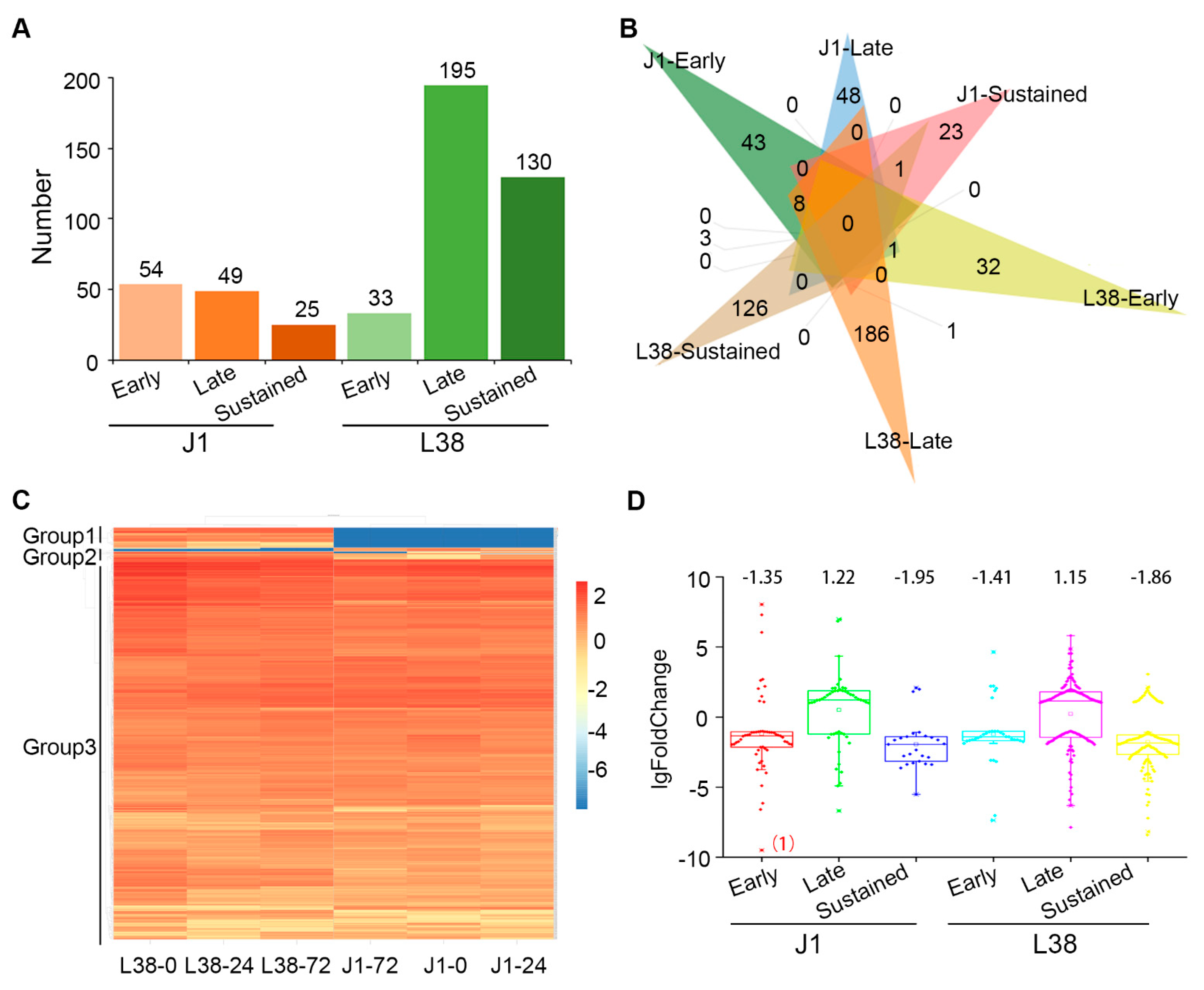
3.3. Identification of Differentially Expressed Genes in L38 during Pathogen Inoculation
3.4. Functional Analysis of Differentially Expressed Genes in L38 during V. dahliae Inoculation
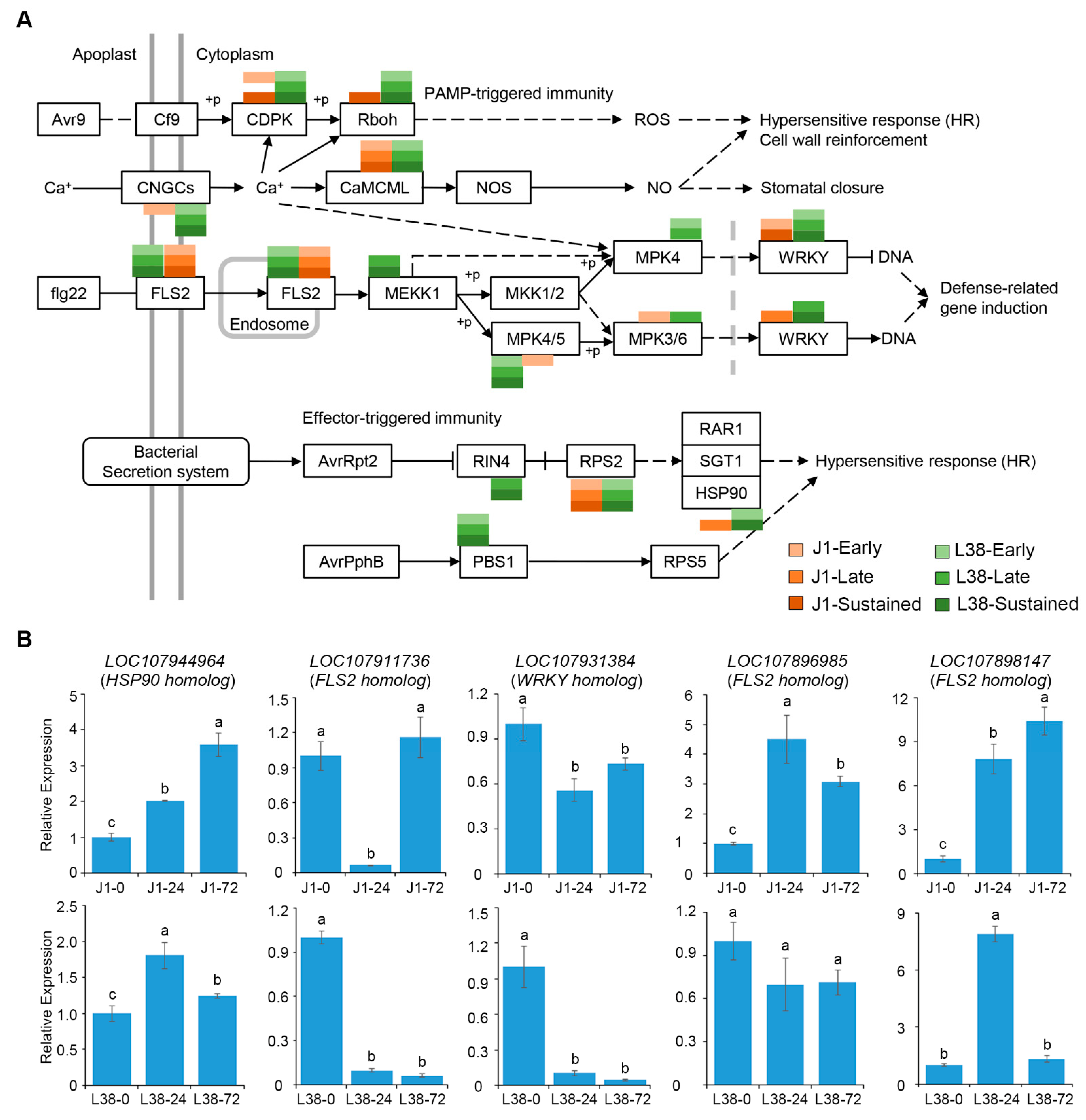
3.5. Resistance Related Pathways Analysis of DEGs
3.6. Discovering and Characteristic of Resistance Gene Analogues (RGAs) in J1 and L38
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurnberger, T.; Brunner, F.; Kemmerling, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- He, Y.; Zhou, J.; Shan, L.; Meng, X. Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 2018, 131, jcs209353. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor Kinases in Plant-Pathogen Interactions: More Than Pattern Recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.B.; Bogdanove, A.J.; Sessa, G. Understanding the Functions of Plant Disease Resistance Proteins. Annu. Rev. Plant Biol. 2003, 54, 23–61. [Google Scholar] [CrossRef] [Green Version]
- Thomma, B.P.; Penninckx, I.A.; Cammue, B.P.; Broekaert, W.F. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001, 13, 63–68. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting Mechanisms of Defense against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. alboatrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Wilhelm, S. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 1955, 45, 180–181. [Google Scholar]
- Wang, H.; Lin, Z.; Zhang, X.; Chen, W.; Guo, X.P.; Nie, Y.C.; Li, Y.H. Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J. Integr. Plant Biol. 2008, 50, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ma, X.; Li, N.; Zhou, L.; Liu, Z.; Han, H.; Gui, Y.; Bao, Y.; Chen, J.; Dai, X. Genome-wide association study discovered candidate genes of Verticillium wilt resistance in up-land cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2017, 15, 1520–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, H.; Chen, W.; Li, Y. Genetic Structure, Linkage Disequilibrium and Association Mapping of Verticillium Wilt Resistance in Elite Cotton (Gossypium hirsutum L.) Germplasm Population. PLoS ONE 2014, 9, e86308. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Wang, C.; Ma, Q.; Zhang, A.; Shi, C.; Liu, J.; Zhang, X.; Yang, D.; Ma, X. An RTM-GWAS procedure reveals the QTL alleles and candidate genes for three yield-related traits in upland cotton. BMC Plant Biol. 2020, 20, 416. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Huang, J.-Q.; Li, N.-Y.; Ma, X.-F.; Wang, J.-L.; Liu, C.; Liu, Y.-F.; Liang, Y.; Bao, Y.-M.; Dai, X.-F. Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 2015, 15, 148. [Google Scholar] [CrossRef] [Green Version]
- Li, T.-G.; Zhang, D.-D.; Zhou, L.; Kong, Z.-Q.; Hussaini, A.S.; Wang, D.; Li, J.-J.; Short, D.P.G.; Dhar, N.; Klosterman, S.J.; et al. Genome-Wide Identification and Functional Analyses of the CRK Gene Family in Cotton Reveals GbCRK18 Confers Verticillium Wilt Resistance in Gossypium barbadense. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, N.-Y.; Ma, X.-F.; Short, D.P.G.; Li, T.-G.; Zhou, L.; Gui, Y.-J.; Kong, Z.-Q.; Zhang, D.-D.; Zhang, W.-Q.; Li, J.-J.; et al. The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 2017, 19, 1466–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Wang, B.; Yin, C.; Zhang, D.; Wang, D.; Song, J.; Zhou, L.; Kong, Z.; Klosterman, S.J.; Li, J.; et al. The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol. Plant Pathol. 2019, 20, 857–876. [Google Scholar] [CrossRef] [Green Version]
- Dong, Q.; Magwanga, R.O.; Cai, X.; Lu, P.; Nyangasi Kirungu, J.; Zhou, Z.; Wang, X.; Wang, X.; Xu, Y.; Hou, Y.; et al. RNA-sequencing, physiological and RNAi analyses provide insights into the response mechanism of the ABC-mediated resistance to Verticillium dahliae infection in cotton. Genes 2019, 10, 110. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Sun, R.; Zhang, X.; Feng, Z.; Wei, F.; Zhao, L.; Zhang, Y.; Zhu, L.; Feng, H.; Zhu, H. Genome-wide analysis of OPR family genes in cotton identified a role for GhOPR9 in Verticillium dahliae resistance. Genes 2020, 11, 1134. [Google Scholar] [CrossRef]
- Hu, Q.; Zhu, L.; Zhang, X.; Guan, Q.; Xiao, S.; Min, L.; Zhang, X. GhCPK33 Negatively Regulates Defense against Verticillium dahliae by Phosphorylating GhOPR3. Plant Physiol. 2018, 178, 876–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, X.P.; Sun, S.C.; Zhang, X.Y.; Li, Y.J.; Liu, F.; Zhu, Q.H.; Xue, F.; Sun, J. GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front Plant Sci. 2020, 11, 69. [Google Scholar] [CrossRef]
- Meng, J.; Gao, H.; Zhai, W.; Shi, J.; Zhang, M.; Zhang, W.; Jian, G.; Zhang, M.; Qi, F. Subtle regulation of cotton resistance to Verticillium wilt mediated by MAPKK family members. Plant Sci. 2018, 272, 235–242. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, L.; Jamieson, P.; Zhang, L.; Zhao, Z.; Babilonia, K.; Shao, W.; Wu, L.; Mustafa, R.; Amin, I.; et al. The Cotton Wall-Associated Kinase GhWAK7A Mediates Responses to Fungal Wilt Pathogens by Complexing with the Chitin Sensory Receptors. Plant Cell 2020, 32, 3978–4001. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, Z.; Xiao, S.; Ye, Z.; Nie, X.; Zhang, X.; Kong, J.; Zhu, L. Phosphate deficiency enhances cotton resistance to Verticillium dahliae through activating jasmonic acid biosynthesis and phenylpropanoid pathway. Plant Sci. 2021, 302, 110724. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Jiang, H.; Zhu, X.; Wang, W.; He, X.; Shi, Y.; Yuan, Y.; Du, X.; Cai, Y. Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genom. 2013, 14, 852. [Google Scholar] [CrossRef] [Green Version]
- Xiang, L.; Cai, C.; Cheng, J.; Wang, L.; Wu, C.; Shi, Y.; Luo, J.; He, L.; Deng, Y.; Zhang, X.; et al. Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq. PeerJ 2018, 6, e4500. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Cheng, W.; Feng, Z.; Zhu, Q.; Sun, Y.; Li, Y.; Sun, J. Transcriptomic analysis of gene expression of Verticillium dahliae upon treatment of the cotton root exudates. BMC Genom. 2020, 21, 155. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Zhao, Z.; Ma, P.; Qu, Y.; Sun, G.; Chen, Q. Integrative transcriptomic and gene co-expression network analysis of host responses upon Verticillium dahliae infection in Gossypium hirsutum. Sci. Rep. 2021, 11, 20586. [Google Scholar] [CrossRef]
- Chen, J.Y.; Liu, C.; Gui, Y.J.; Si, K.W.; Zhang, D.D.; Wang, J.; Short, D.P.G.; Huang, J.-Q.; Li, N.-Y.; Liang, Y.; et al. Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dhar, N.; Chen, J.-Y.; Subbarao, K.V.; Klosterman, S.J. Hormone Signaling and Its Interplay With Development and Defense Responses in Verticillium-Plant Interactions. Front. Plant Sci. 2020, 11, 1. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Hermoso, A.; D’Alessandro, R.; Vlasova, A.; Andolfo, G.; Frusciante, L.; Lowy, E.; Roma, G.; Ercolano, M.R. PRGdb 2.0: Towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 2012, 41, D1167–D1171. [Google Scholar] [CrossRef]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Zhai, Y.; Li, L.; Yang, Z.; Ge, X.; Yang, Z.; Zhang, C.; Li, F.; Ren, M. BIN2 negatively regulates plant defence against Verticillium dahliae in Arabidopsis and cotton. Plant Biotechnol. J. 2021, 19, 2097–2112. [Google Scholar] [CrossRef] [PubMed]
- Sekhwal, M.K.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; You, F.M. Disease Resistance Gene Analogs (RGAs) in Plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Song, J.; Dhar, N.; Shan, Y.; Ma, X.-Y.; Wang, X.-L.; Chen, J.-Y.; Dai, X.-F.; Li, R.; Wang, Z.-S. Transcriptome Analysis of a Cotton Cultivar Provides Insights into the Differentially Expressed Genes Underlying Heightened Resistance to the Devastating Verticillium Wilt. Cells 2021, 10, 2961. https://doi.org/10.3390/cells10112961
Zhu H, Song J, Dhar N, Shan Y, Ma X-Y, Wang X-L, Chen J-Y, Dai X-F, Li R, Wang Z-S. Transcriptome Analysis of a Cotton Cultivar Provides Insights into the Differentially Expressed Genes Underlying Heightened Resistance to the Devastating Verticillium Wilt. Cells. 2021; 10(11):2961. https://doi.org/10.3390/cells10112961
Chicago/Turabian StyleZhu, He, Jian Song, Nikhilesh Dhar, Ying Shan, Xi-Yue Ma, Xiao-Lei Wang, Jie-Yin Chen, Xiao-Feng Dai, Ran Li, and Zi-Sheng Wang. 2021. "Transcriptome Analysis of a Cotton Cultivar Provides Insights into the Differentially Expressed Genes Underlying Heightened Resistance to the Devastating Verticillium Wilt" Cells 10, no. 11: 2961. https://doi.org/10.3390/cells10112961
APA StyleZhu, H., Song, J., Dhar, N., Shan, Y., Ma, X.-Y., Wang, X.-L., Chen, J.-Y., Dai, X.-F., Li, R., & Wang, Z.-S. (2021). Transcriptome Analysis of a Cotton Cultivar Provides Insights into the Differentially Expressed Genes Underlying Heightened Resistance to the Devastating Verticillium Wilt. Cells, 10(11), 2961. https://doi.org/10.3390/cells10112961





