The Motility and Mesenchymal Features of Breast Cancer Cells Correlate with the Levels and Intracellular Localization of Transglutaminase Type 2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Treatments
2.2. Parallel Artificial Membrane Permeability Assay (PAMPA)
2.3. Reverse Transcription and Real Time Quantitative PCR
2.4. Nuclear Protein Extracion, Immunoprecipitation and Immunochemical Analysis
2.5. Real-Time Assays of Cell Migration and Invasion
2.6. Immunocytochemical and Confocal Analysis
2.7. Statistical Analysis
3. Results
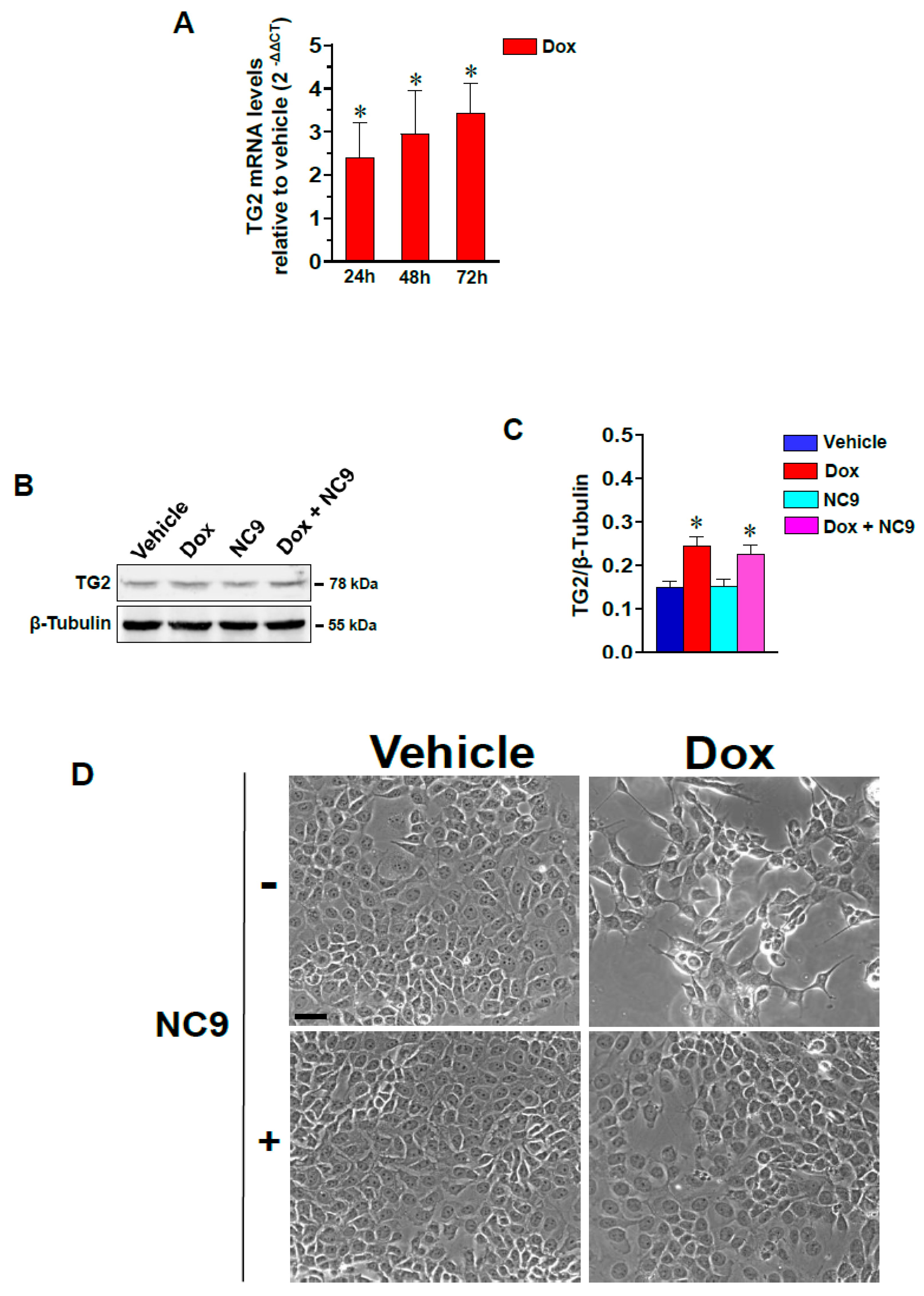
3.1. Dox-Induced Changes in Morphology, Motility and EMT Markers Involve TG2 in MCF-7 Cells
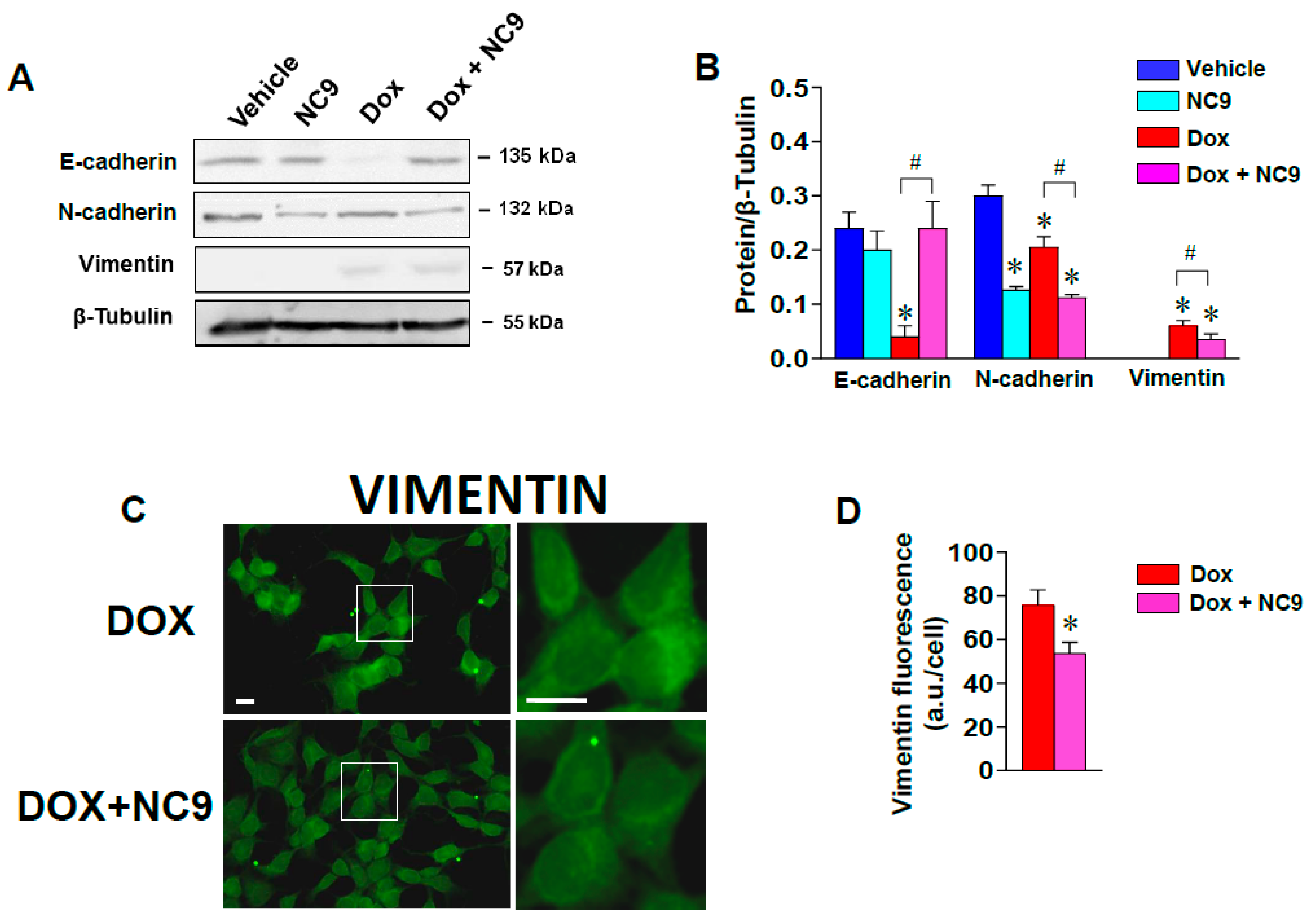
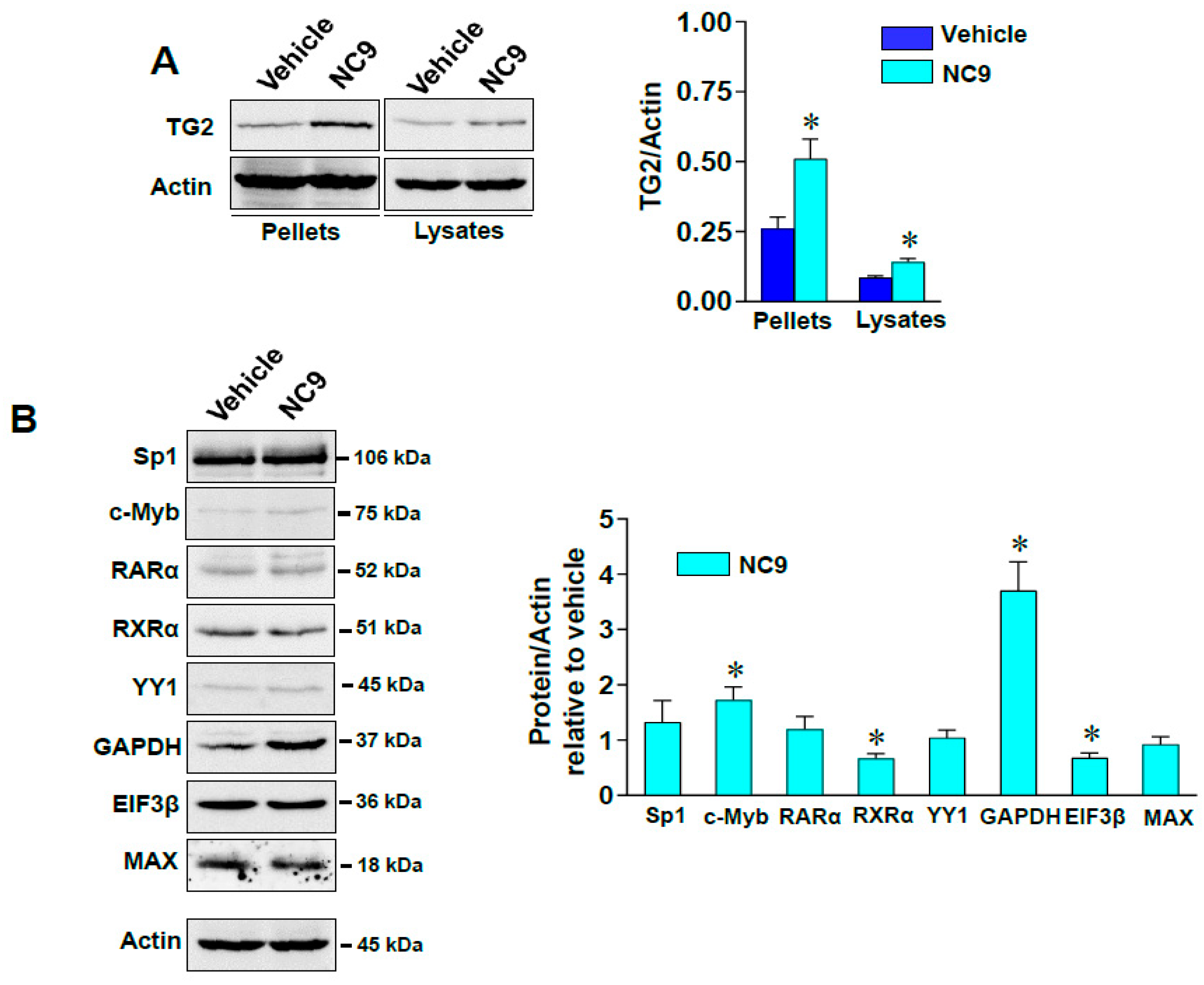
3.2. Impacts of TG2 on Cytoskeleton through Interactions with Vimentin and on Transcriptional Control in the Nucleus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehta, K.; Fok, J.; Miller, F.R.; Koul, D.; Sahin, A.A. Prognostic Significance of Tissue Transglutaminase in Drug Resistant and Metastatic Breast Cancer. Clin. Cancer Res. 2004, 10, 8068–8076. [Google Scholar] [CrossRef] [Green Version]
- Fisher, M.L.; Adhikary, G.; Xu, W.; Kerr, C.; Keillor, J.W.; Eckert, R.L. Type II transglutaminase stimulates epidermal cancer stem cell epithelial-mesenchymal transition. Oncotarget 2015, 6, 20525–20539. [Google Scholar] [CrossRef] [Green Version]
- Karicheva, O.; Rodriguez, J.M.; Wadier, N.; Martin-Hernandez, K.; Vauchelles, R.; Magroun, N.; Tissier, A.; Schreiber, V.; Dantzer, F. PARP3 controls TGFβ and ROS driven epithelial-to-mesenchymal transition and stemness by stimulating a TG2-Snail-E-cadherin axis. Oncotarget 2016, 7, 64109–64123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, K.D. Transglutaminase 2 and NF-κB: An odd couple that shapes breast cancer phenotype. Breast Cancer Res. Treat. 2012, 137, 329–336. [Google Scholar] [CrossRef]
- Ai, L.; Kim, W.-J.; Demircan, B.; Dyer, L.M.; Bray, K.J.; Skehan, R.R.; Massoll, N.A.; Brown, K.D. The transglutaminase 2 gene (TGM2), a potential molecular marker for chemotherapeutic drug sensitivity, is epigenetically silenced in breast cancer. Carcinogenesis 2008, 29, 510–518. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-S.; Park, S.-S.; Nam, B.-H.; Kim, I.-H.; Kim, S.-Y. Reversal of Drug Resistance in Breast Cancer Cells by Transglutaminase 2 Inhibition and Nuclear Factor-κB Inactivation. Cancer Res. 2006, 66, 10936–10943. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-S. Silencing of TGase 2 sensitizes breast cancer cells to apoptosis by regulation of survival factors. Front. Biosci. 2009, 14, 2514–2521. [Google Scholar] [CrossRef] [Green Version]
- Park, K.-S. Increase in transglutaminase 2 expression is associated with NF-kappaB activation in breast cancer tissues. Front. Biosci. 2009, 14, 1945–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, A.H.; O’Keefe, R.J.; Schwarz, E.M.; Rosier, R.N.; Puzas, J.E. Nuclear Factor-κB–Dependent Mechanisms in Breast Cancer Cells Regulate Tumor Burden and Osteolysis in Bone. Cancer Res. 2005, 65, 3209–3217. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, J.; Ying, X.; Lin, P.C.; Zhou, B.P. Twist-mediated Epithelial-mesenchymal Transition Promotes Breast Tumor Cell Invasion via Inhibition of Hippo Pathway. Sci. Rep. 2016, 6, 24606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Işeri, D.; Kars, M.D.; Arpaci, F.; Atalay, C.; Pak, I.; Gündüz, U. Drug resistant MCF-7 cells exhibit epithelial-mesenchymal transition gene expression pattern. Biomed. Pharmacother. 2011, 65, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-Y.; Jeong, E.M.; Lee, J.-H.; Kim, H.-J.; Lim, J.; Kim, C.-W.; Shin, N.-M.; Jeon, J.-H.; Choi, K.; Kim, I.-G. Doxorubicin Induces the Persistent Activation of Intracellular Transglutaminase 2 That Protects from Cell Death. Mol. Cells 2012, 33, 235–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eligula, L.; Chuang, L.; Phillips, M.L.; Motoki, M.; Seguro, K.; Muhlrad, A. Transglutaminase-Induced Cross-Linking between Subdomain 2 of G-Actin and the 636–642 Lysine-Rich Loop of Myosin Subfragment 1. Biophys. J. 1998, 74, 953–963. [Google Scholar] [CrossRef] [Green Version]
- Munsie, L.; Caron, N.; Atwal, R.S.; Marsden, I.; Wild, E.; Bamburg, J.R.; Tabrizi, S.; Truant, R. Mutant huntingtin causes defective actin remodeling during stress: Defining a new role for transglutaminase 2 in neurodegenerative disease. Hum. Mol. Genet. 2011, 20, 1937–1951. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Kirkpatrick, L.L.; Schilling, A.B.; Helseth, D.L.; Chabot, N.; Keillor, J.W.; Johnson, G.V.; Brady, S.T. Transglutaminase and Polyamination of Tubulin: Posttranslational Modification for Stabilizing Axonal Microtubules. Neuron 2013, 78, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Agnihotri, N.; Kumar, S.; Mehta, K. Tissue transglutaminase as a central mediator in inflammation-induced progression of breast cancer. Breast Cancer Res. 2013, 15, 202. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battaglia, R.; Delic, S.; Herrmann, H.; Snider, N.T. Vimentin on the move: New developments in cell migration. F1000Research 2018, 7, 1796. [Google Scholar] [CrossRef] [Green Version]
- Trejo-Skalli, A.V.; Velasco, P.T.; Murthy, S.N.; Lorand, L.; Goldman, R.D. Association of a transglutaminase-related antigen with intermediate filaments. Proc. Natl. Acad. Sci. USA 1995, 92, 8940–8944. [Google Scholar] [CrossRef] [Green Version]
- Gupta, M.; Greenberg, C.S.; Eckman, D.M.; Sane, D.C. Arterial Vimentin Is a Transglutaminase Substrate: A Link between Vasomotor Activity and Remodeling? J. Vasc. Res. 2007, 44, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Zhengkui, S.; Sun, Z.; Liu, Z. Silencing of TGM2 reverses epithelial to mesenchymal transition and modulates the chemosensitivity of breast cancer to docetaxel. Exp. Ther. Med. 2015, 10, 1413–1418. [Google Scholar] [CrossRef] [Green Version]
- Herman, J.F.; Mangala, L.S.; Mehta, K. Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene 2006, 25, 3049–3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangala, L.S.; Fok, J.Y.; Zorrilla-Calancha, I.R.; Verma, A.; Mehta, K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene 2006, 26, 2459–2470. [Google Scholar] [CrossRef] [Green Version]
- Patteson, A.E.; Vahabikashi, A.; Goldman, R.D.; Janmey, P.A. Mechanical and Non-Mechanical Functions of Filamentous and Non-Filamentous Vimentin. BioEssays 2020, 42, 2000078. [Google Scholar] [CrossRef]
- Serres, M.P.; Samwer, M.; Quang, B.A.T.; Lavoie, G.; Perera, U.; Görlich, D.; Charras, G.; Petronczki, M.; Roux, P.P.; Paluch, E.K. F-Actin Interactome Reveals Vimentin as a Key Regulator of Actin Organization and Cell Mechanics in Mitosis. Dev. Cell 2020, 52, 210–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Fang, Z.; Ma, J. Regulatory mechanisms and clinical significance of vimentin in breast cancer. Biomed. Pharmacother. 2020, 133, 111068. [Google Scholar] [CrossRef] [PubMed]
- Strouhalova, K.; Přechová, M.; Gandalovičová, A.; Brábek, J.; Gregor, M.; Rosel, D. Vimentin Intermediate Filaments as Potential Target for Cancer Treatment. Cancers 2020, 12, 184. [Google Scholar] [CrossRef] [Green Version]
- Kokkinos, M.I.; Wafai, R.; Wong, M.K.; Newgreen, D.F.; Thompson, E.W.; Waltham, M. Vimentin and Epithelial-Mesenchymal Transition in Human Breast Cancer–Observations in vitro and in vivo. Cells Tissues Organs 2007, 185, 191–203. [Google Scholar] [CrossRef]
- Kim, J.; Yang, C.; Kim, E.J.; Jang, J.; Kim, S.-J.; Kang, S.M.; Kim, M.G.; Jung, H.; Park, D.; Kim, C. Vimentin filaments regulate integrin-ligand interactions by binding to the cytoplasmic tail of integrin β3. J. Cell Sci. 2016, 129, 2030–2042. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Kaartinen, M.T. Transglutaminase activity regulates differentiation, migration and fusion of osteoclasts via affecting actin dynamics. J. Cell. Physiol. 2018, 233, 7497–7513. [Google Scholar] [CrossRef]
- Kerr, C.; Szmacinski, H.; Fisher, M.L.; Nance, B.; Lakowicz, J.R.; Akbar, A.; Keillor, J.W.; Wong, T.L.; Godoy-Ruiz, R.; A Toth, E.; et al. Transamidase site-targeted agents alter the conformation of the transglutaminase cancer stem cell survival protein to reduce GTP binding activity and cancer stem cell survival. Oncogene 2016, 36, 2981–2990. [Google Scholar] [CrossRef] [Green Version]
- Gundemir, S.; Monteagudo, A.; Akbar, A.; Keillor, J.W.; Johnson, G.V. The complex role of transglutaminase 2 in glioblastoma proliferation. Neuro-Oncology 2016, 19, 208–218. [Google Scholar] [CrossRef] [Green Version]
- Adhikary, G.; Grun, D.; Alexander, H.R.; Friedberg, J.S.; Xu, W.; Keillor, J.W.; Kandasamy, S.; Eckert, R.L. Transglutaminase is a mesothelioma cancer stem cell survival protein that is required for tumor formation. Oncotarget 2018, 9, 34495–34505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jambrovics, K.; Uray, I.P.; Keillor, J.W.; Fésüs, L.; Balajthy, Z. Benefits of Combined All-Trans Retinoic Acid and Arsenic Trioxide Treatment of Acute Promyelocytic Leukemia Cells and Further Enhancement by Inhibition of Atypically Expressed Transglutaminase 2. Cancers 2020, 12, 648. [Google Scholar] [CrossRef] [Green Version]
- Shinde, A.; Paez, J.S.; Libring, S.; Hopkins, K.; Solorio, L.; Wendt, M.K. Transglutaminase-2 facilitates extracellular vesicle-mediated establishment of the metastatic niche. Oncogenesis 2020, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgeois, K. Towards in vitro Pharmacokinetic Assessment of Novel Targeted Covalent Inhibitors for Human Tissue Transglutaminase. Doctoral Dissertation, University of Ottawa, Ottawa, Canada, April 2019. Available online: https://ruor.uottawa.ca/handle/10393/39472 (accessed on 1 October 2021).
- Wohnsland, F.; Faller, B. High-Throughput Permeability pH Profile and High-Throughput Alkane/Water log P with Artificial Membranes. J. Med. Chem. 2001, 44, 923–930. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H. Drug-Like Properties, 2nd ed.; Academic Press: Boston, MA, USA, 2016; pp. 325–337. [Google Scholar]
- Franzese, O.; Minotti, L.; Aguiari, G.; Corrà, F.; Cervellati, C.; Ferrari, C.; Volinia, S.; Bergamini, C.M.; Bianchi, N. Involvement of non-coding RNAs and transcription factors in the induction of Transglutaminase isoforms by ATRA. Amino Acids 2019, 51, 1273–1288. [Google Scholar] [CrossRef]
- De Kok, J.B.; Roelofs, R.W.; A Giesendorf, B.; Pennings, J.L.; Waas, E.T.; Feuth, T.; Swinkels, D.W.; Span, P. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab. Investig. 2004, 85, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolo, V.; Benedusi, M.; Brugnoli, F.; Lanuti, P.; Marchisio, M.; Querzoli, P.; Capitani, S. Phospholipase C-β2 promotes mitosis and migration of human breast cancer-derived cells. Carcinogenesis 2007, 28, 1638–1645. [Google Scholar] [CrossRef] [Green Version]
- Brugnoli, F.; Grassilli, S.; Al-Qassab, Y.; Capitani, S.; Bertagnolo, V. PLC-β2 is modulated by low oxygen availability in breast tumor cells and plays a phenotype dependent role in their hypoxia-related malignant potential. Mol. Carcinog. 2016, 55, 2210–2221. [Google Scholar] [CrossRef]
- Brugnoli, F.; Grassilli, S.; Piazzi, M.; Palomba, M.; Nika, E.; Bavelloni, A.; Capitani, S.; Bertagnolo, V. In triple negative breast tumor cells, PLC-β2 promotes the conversion of CD133high to CD133low phenotype and reduces the CD133-related invasiveness. Mol. Cancer 2013, 12, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brugnoli, F.; Grassilli, S.; Lanuti, P.; Marchisio, M.; Al-Qassab, Y.; Vezzali, F.; Capitani, S.; Bertagnolo, V. Up-modulation of PLC-β2 reduces the number and malignancy of triple-negative breast tumor cells with a CD133+/EpCAM+ phenotype: A promising target for preventing progression of TNBC. BMC Cancer 2017, 17, 617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckert, R.L. Transglutaminase 2 takes center stage as a cancer cell survival factor and therapy target. Mol. Carcinog. 2019, 58, 837–853. [Google Scholar] [CrossRef] [PubMed]
- Ballestar, E.; Franco, L. Use of the Transglutaminase Reaction To Study the Dissociation of Histone N-Terminal Tails from DNA in Nucleosome Core Particles†. Biochemistry 1997, 36, 5963–5969. [Google Scholar] [CrossRef] [PubMed]
- Lesort, M.; Attanavanich, K.; Zhang, J.; Johnson, G.V.W. Distinct Nuclear Localization and Activity of Tissue Transglutaminase. J. Biol. Chem. 1998, 273, 11991–11994. [Google Scholar] [CrossRef] [Green Version]
- Tarasewicz, E.; Oakes, R.S.; Aviles, M.O.; Straehla, J.; Chilton, K.M.; Decker, J.T.; Wu, J.; Shea, L.D.; Jeruss, J.S.; Straehla, J. Embryonic stem cell secreted factors decrease invasiveness of triple-negative breast cancer cells through regulome modulation. Cancer Biol. Ther. 2018, 19, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Tatsukawa, H.; Fukaya, Y.; Frampton, G.; Martinez–Fuentes, A.; Suzuki, K.; Kuo, T.; Nagatsuma, K.; Shimokado, K.; Okuno, M.; Wu, J.; et al. Role of Transglutaminase 2 in Liver Injury via Cross-linking and Silencing of Transcription Factor Sp1. Gastroenterology 2009, 136, 1783–1795. [Google Scholar] [CrossRef] [Green Version]
- Krig, S.R.; Chandraratna, R.A.S.; Chang, M.M.J.; Wu, R.; Rice, R.H. Gene-specific TCDD suppression of RARalpha- and RXR-mediated induction of tissue transglutaminase. Toxicol. Sci. 2002, 68, 102–108. [Google Scholar] [CrossRef] [Green Version]
- Nagy, L.; A Thomázy, V.; Saydak, M.M.; Stein, J.P.; Davies, P.J. The promoter of the mouse tissue transglutaminase gene directs tissue-specific, retinoid-regulated and apoptosis-linked expression. Cell Death Differ. 1997, 4, 534–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergamini, C.M.; Vischioni, C.; Aguiari, G.; Grandi, C.; Terrazzan, A.; Volinia, S.; Bianchi, N.; Taccioli, C. Inhibition of the lncRNA Coded within Transglutaminase 2 Gene Impacts Several Relevant Networks in MCF-7 Breast Cancer Cells. Non-Coding RNA 2021, 7, 49. [Google Scholar] [CrossRef]
- Zang, Y.; Zhang, X.; Yan, L.; Gu, G.; Li, D.; Zhang, Y.; Fang, L.; Fu, S.; Ren, J.; Xu, Z. Eukaryotic Translation Initiation Factor 3b is both a Promising Prognostic Biomarker and a Potential Therapeutic Target for Patients with Clear Cell Renal Cell Carcinoma. J. Cancer 2017, 8, 3049–3061. [Google Scholar] [CrossRef] [Green Version]
- Hugo, H.J.; Pereira, L.; Suryadinata, R.; Drabsch, Y.; Gonda, T.J.; Gunasinghe, N.P.A.D.; Pinto, C.; Soo, E.T.L.; Van Denderen, B.J.W.; Hill, P.; et al. Direct repression of MYB by ZEB1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res. 2013, 15, R113. [Google Scholar] [CrossRef] [Green Version]
- Orrù, S.; Ruoppolo, M.; Francese, S.; Vitagliano, L.; Marino, G.; Esposito, C. Identification of tissue transglutaminase-reactive lysine residues in glyceraldehyde-3-phosphate dehydrogenase. Protein Sci. 2009, 11, 137–146. [Google Scholar] [CrossRef]
- Beninati, S.; Piacentini, M.; Bergamini, C.M. Transglutaminase 2, a double face enzyme. Amino Acids 2017, 49, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Tatsukawa, H.; Furutani, Y.; Hitomi, K.; Kojima, S. Transglutaminase 2 has opposing roles in the regulation of cellular functions as well as cell growth and death. Cell Death Dis. 2016, 7, e2244. [Google Scholar] [CrossRef] [Green Version]
- Piacentini, M.; D’Eletto, M.; Farrace, M.G.; Rodolfo, C.; Del Nonno, F.; Ippolito, G.; Falasca, L. Characterization of distinct sub-cellular location of transglutaminase type II: Changes in intracellular distribution in physiological and pathological states. Cell Tissue Res. 2014, 358, 793–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deasey, S.; Shanmugasundaram, S.; Nurminskaya, M. Tissue-specific responses to loss of transglutaminase 2. Amino Acids 2011, 44, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Tempest, R.; Guarnerio, S.; Maani, R.; Cooper, J.; Peake, N. The Biological and Biomechanical Role of Transglutaminase-2 in the Tumour Microenvironment. Cancers 2021, 13, 2788. [Google Scholar] [CrossRef] [PubMed]
- Hettasch, J.M.; Bandarenko, N.; Burchette, J.L.; Lai, T.S.; Marks, J.R.; A Haroon, Z.; Peters, K.; Dewhirst, M.W.; Iglehart, J.D.; Greenberg, C.S. Tissue transglutaminase expression in human breast cancer. Lab. Investig. 1996, 75, 637–645. [Google Scholar]
- Budillon, A.; Carbone, C.; Di Gennaro, E. Tissue transglutaminase: A new target to reverse cancer drug resistance. Amino Acids 2011, 44, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phatak, V.M.; Croft, S.M.; Setty, S.G.R.; Scarpellini, A.; Hughes, D.C.; Rees, R.; McArdle, S.; Verderio, E.A.M. Expression of transglutaminase-2 isoforms in normal human tissues and cancer cell lines: Dysregulation of alternative splicing in cancer. Amino Acids 2011, 44, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-L.; Chen, M.-J.; Lin, J.-C.; Lin, C.-H.; Huang, W.-C.; Cheng, S.-P.; Chen, S.-N.; Chang, Y.-C. Doxorubicin Promotes Migration and Invasion of Breast Cancer Cells through the Upregulation of the RhoA/MLC Pathway. J. Breast Cancer 2019, 22, 185–195. [Google Scholar] [CrossRef]
- Kumar, A.; Hu, J.; A LaVoie, H.; Walsh, K.B.; DiPette, D.J.; Singh, U.S. Conformational changes and translocation of tissue-transglutaminase to the plasma membranes: Role in cancer cell migration. BMC Cancer 2014, 14, 256. [Google Scholar] [CrossRef] [Green Version]
- Cellura, D.; Pickard, K.; Quaratino, S.; Parker, H.; Strefford, P.J.C.; Thomas, G.; Mitter, R.; Mirnezami, A.; Peake, N. miR-19–Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer. Mol. Cancer Res. 2015, 13, 1095–1105. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Wang, X.-H.; Hua, Y.-T.; Zhang, Y.-Z.; Han, Y.; Yang, Z.-L. The tissue transglutaminase: A potential target regulating MDR in breast cancer. Eur. Rev. Med. Pharm. Sci. 2020, 24, 6175–6184. [Google Scholar]
- Clément, S.; Velasco, P.T.; Murthy, S.P.; Wilson, J.H.; Lukas, T.J.; Goldman, R.D.; Lorand, L. The Intermediate Filament Protein, Vimentin, in the Lens Is a Target for Cross-linking by Transglutaminase. J. Biol. Chem. 1998, 273, 7604–7609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, N.J.; Baker, P.; Jones, C.; Aplin, J.D. A Role for Tissue Transglutaminase in Stabilization of Membrane-Cytoskeletal Particles Shed from the Human Placenta. Biol. Reprod. 2007, 77, 648–657. [Google Scholar] [CrossRef]
- Caron, N.S.; Munsie, L.N.; Keillor, J.W.; Truant, R. Using FLIM-FRET to Measure Conformational Changes of Transglutaminase Type 2 in Live Cells. PLoS ONE 2012, 7, e44159. [Google Scholar] [CrossRef]
- Lowery, J.; Kuczmarski, E.R.; Herrmann, H.; Goldman, R.D. Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function. J. Biol. Chem. 2015, 290, 17145–17153. [Google Scholar] [CrossRef] [Green Version]
- Antonyak, M.A.; Miller, A.M.; Jansen, J.M.; Boehm, J.E.; Balkman, C.E.; Wakshlag, J.J.; Page, R.L.; Cerione, R.A. Augmentation of Tissue Transglutaminase Expression and Activation by Epidermal Growth Factor Inhibit Doxorubicin-induced Apoptosis in Human Breast Cancer Cells. J. Biol. Chem. 2004, 279, 41461–41467. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Hwang, H.; Im, C.Y.; Kim, S.-Y. Recent Progress in the Development of Transglutaminase 2 (TGase2) Inhibitors. J. Med. Chem. 2016, 60, 554–567. [Google Scholar] [CrossRef]
- Király, R.; Demény, M.; Fésüs, L. Protein transamidation by transglutaminase 2 in cells: A disputed Ca2+-dependent action of a multifunctional protein. FEBS J. 2011, 278, 4717–4739. [Google Scholar] [CrossRef]
- Sato, N.; Ohtake, Y.; Kato, H.; Abe, S.; Kohno, H.; Ohkubo, Y. Effects of polyamines on histone polymerization. Protein J. 2003, 22, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-S.; Kim, D.-S.; Park, K.-S.; Song, H.-J.; Kim, S.-Y. Proteomic analysis of high-molecular-weight protein polymers in a doxorubicin-resistant breast-cancer cell line. Proteom. Clin. Appl. 2007, 1, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.-F.; Tatsukawa, H.; Kojima, S. New insights into the functions and localization of nuclear transglutaminase 2. FEBS J. 2011, 278, 4756–4767. [Google Scholar] [CrossRef] [PubMed]
- Knopfova, L.; Biglieri, E.; Volodko, N.; Masarik, M.; Hermanová, M.; Garzón, J.G.; Dúcka, M.; Kučírková, T.; Souček, K.; Šmarda, J.; et al. Transcription factor c-Myb inhibits breast cancer lung metastasis by suppression of tumor cell seeding. Oncogene 2017, 37, 1020–1030. [Google Scholar] [CrossRef] [Green Version]
- Chuang, D.-M.; Hough, C.; Senatorov, V.V. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, C.; Pinto, A.R.; Li, H.; Li, L.; Wang, L.; Simpson, R.; Liu, J.-P. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) induces cancer cell senescence by interacting with telomerase RNA component. Proc. Natl. Acad. Sci. USA 2012, 109, 13308–13313. [Google Scholar] [CrossRef] [Green Version]
- Kosova, A.; Khodyreva, S.N.; Lavrik, O.I. Role of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in DNA repair. Biochem. 2017, 82, 643–654. [Google Scholar] [CrossRef]
- Ma, F.; Li, X.; Ren, J.; Guo, R.; Li, Y.; Liu, J.; Sun, Y.; Liu, Z.; Jia, J.; Li, W. Downregulation of eukaryotic translation initiation factor 3b inhibited proliferation and metastasis of gastric cancer. Cell Death Dis. 2019, 10, 623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brtko, J. Role of Retinoids and their Cognate Nuclear Receptors in Breast Cancer Chemoprevention. Central Eur. J. Public Health 2007, 15, 3–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchi, N.; Brugnoli, F.; Grassilli, S.; Bourgeois, K.; Keillor, J.W.; Bergamini, C.M.; Aguiari, G.; Volinia, S.; Bertagnolo, V. The Motility and Mesenchymal Features of Breast Cancer Cells Correlate with the Levels and Intracellular Localization of Transglutaminase Type 2. Cells 2021, 10, 3059. https://doi.org/10.3390/cells10113059
Bianchi N, Brugnoli F, Grassilli S, Bourgeois K, Keillor JW, Bergamini CM, Aguiari G, Volinia S, Bertagnolo V. The Motility and Mesenchymal Features of Breast Cancer Cells Correlate with the Levels and Intracellular Localization of Transglutaminase Type 2. Cells. 2021; 10(11):3059. https://doi.org/10.3390/cells10113059
Chicago/Turabian StyleBianchi, Nicoletta, Federica Brugnoli, Silvia Grassilli, Karine Bourgeois, Jeffrey W. Keillor, Carlo M. Bergamini, Gianluca Aguiari, Stefano Volinia, and Valeria Bertagnolo. 2021. "The Motility and Mesenchymal Features of Breast Cancer Cells Correlate with the Levels and Intracellular Localization of Transglutaminase Type 2" Cells 10, no. 11: 3059. https://doi.org/10.3390/cells10113059






