Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine
Abstract
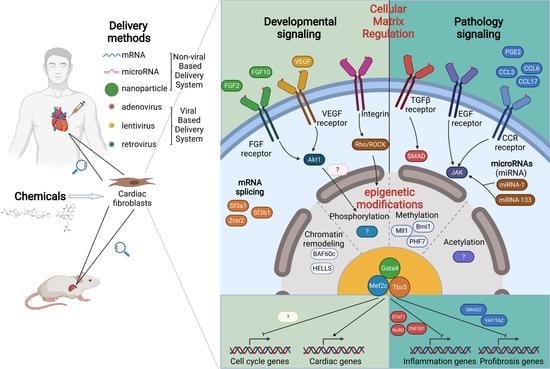
:1. Introduction
2. Transcriptional Regulation in Cardiac Reprogramming
2.1. Additional Transcriptional Factors Screening
2.2. Transcriptional Factors Optimization
2.3. Mechanisms of the Transcriptional Network in Cardiac Reprogramming
3. Epigenetic Regulations in Reprogramming
3.1. MicroRNAs
3.2. Chromatin Remodeling Factors
3.3. Epigenetic Factors Related to Methylation
4. Cellular Microenvironment in Cardiac Reprogramming
4.1. Growth Factor Regulation
4.2. Signaling Pathways
4.3. Inflammation Regulation
4.4. Cellular Matrix Regulation
5. Cell Cycling Regulation in Cardiac Reprogramming
6. Clinical Translational Progress for Cardiac Reprogramming
6.1. Non-Exogenous Genes Induced Cardiac Reprogramming
6.2. Viral Gene Based Delivery System Optimization
6.3. Non-Viral Gene Based Delivery System Optimization
6.4. Reprogramming Human Fibroblasts into iCMs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protze, S.; Khattak, S.; Poulet, C.; Lindemann, D.; Tanaka, E.M.; Ravens, U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 2012, 53, 323–332. [Google Scholar] [CrossRef]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE 2013, 8, e63577. [Google Scholar] [CrossRef] [Green Version]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Zhang, Y.; Xu, X.; Sun, Q.; Yang, C.; Wang, H.; Yang, J.; Yang, Y.; Yang, X.; Liu, Y.; et al. Sall4 and Myocd Empower Direct Cardiac Reprogramming from Adult Cardiac Fibroblasts After Injury. Front. Cell Dev. Biol. 2021, 9, 608367. [Google Scholar] [CrossRef]
- Hirai, H.; Tani, T.; Kikyo, N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 2010, 54, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Tani, T.; Katoku-Kikyo, N.; Kellner, S.; Karian, P.; Firpo, M.; Kikyo, N. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells 2011, 29, 1349–1361. [Google Scholar] [CrossRef] [Green Version]
- Hirai, H.; Katoku-Kikyo, N.; Karian, P.; Firpo, M.; Kikyo, N. Efficient iPS cell production with the MyoD transactivation domain in serum-free culture. PLoS ONE 2012, 7, e34149. [Google Scholar] [CrossRef]
- Wang, L.; Huang, P.; Near, D.; Ravi, K.; Xu, Y.; Liu, J.; Qian, L. Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells 2020, 9, 268. [Google Scholar] [CrossRef] [Green Version]
- Mathison, M.; Singh, V.P.; Gersch, R.P.; Ramirez, M.O.; Cooney, A.; Kaminsky, S.M.; Chiuchiolo, M.J.; Nasser, A.; Yang, J.; Crystal, R.G.; et al. “Triplet” polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. J. Thorac. Cardiovasc. Surg. 2014, 148, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, W.; Nam, Y.J. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Sci. Rep. 2019, 9, 14970. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, A.D.; Kim, L.J.; Nam, Y.J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019, 9, 6362. [Google Scholar] [CrossRef] [PubMed]
- Mathison, M.; Singh, V.P.; Sanagasetti, D.; Yang, L.; Pinnamaneni, J.P.; Yang, J.; Rosengart, T.K. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J. Thorac. Cardiovasc. Surg. 2017, 154, 1601–1610. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Perez, A.; Sathe, A.A.; Bhakta, M.; Leggett, K.; Xing, C.; Munshi, N.V. Hand2 Selectively Reorganizes Chromatin Accessibility to Induce Pacemaker-like Transcriptional Reprogramming. Cell Rep. 2019, 27, 2354–2369. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Wang, Z.; Garry, G.A.; Malladi, V.S.; Botten, G.A.; Ye, W.; Zhou, H.; Osterwalder, M.; Dickel, D.E.; Visel, A.; et al. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell 2019, 25, 69–86. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Welch, J.D.; Ma, H.; Zhou, Y.; Vaseghi, H.R.; Yu, S.; Wall, J.B.; Alimohamadi, S.; Zheng, M.; et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 2017, 551, 100–104. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Alimohamadi, S.; Wang, L.; Liu, Z.; Wall, J.B.; Yin, C.; Liu, J.; Qian, L. A Loss of Function Screen of Epigenetic Modifiers and Splicing Factors during Early Stage of Cardiac Reprogramming. Stem Cells Int. 2018, 2018, 3814747. [Google Scholar] [CrossRef]
- Stone, N.R.; Gifford, C.A.; Thomas, R.; Pratt, K.J.B.; Samse-Knapp, K.; Mohamed, T.M.A.; Radzinsky, E.M.; Schricker, A.; Ye, L.; Yu, P.Z.; et al. Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. Cell Stem Cell 2019, 25, 87–102. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Hodgkinson, C.P.; Pratt, R.E.; Lee, J.; Sullenger, B.A.; Dzau, V.J. Enhancing cardiac reprogramming via synthetic RNA oligonucleotides. Mol. Ther. Nucleic Acids 2021, 23, 55–62. [Google Scholar] [CrossRef]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [Green Version]
- Wamstad, J.A.; Alexander, J.M.; Truty, R.M.; Shrikumar, A.; Li, F.; Eilertson, K.E.; Ding, H.; Wylie, J.N.; Pico, A.R.; Capra, J.A.; et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 2012, 151, 206–220. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [Green Version]
- Testa, G.; Russo, M.; Di Benedetto, G.; Barbato, M.; Parisi, S.; Pirozzi, F.; Tocchetti, C.G.; Abete, P.; Bonaduce, D.; Russo, T.; et al. Bmi1 inhibitor PTC-209 promotes Chemically-induced Direct Cardiac Reprogramming of cardiac fibroblasts into cardiomyocytes. Sci. Rep. 2020, 10, 7129. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lei, I.; Karatas, H.; Li, Y.; Wang, L.; Gnatovskiy, L.; Dou, Y.; Wang, S.; Qian, L.; Wang, Z. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2016, 2, 16036. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Heo, J.S.; Kim, J.S.; Jun, E.K.; Lee, J.H.; Kim, A.; Kim, J.; Whang, K.Y.; Kang, Y.K.; Yeo, S.; et al. Reprogramming fibroblasts into induced pluripotent stem cells with Bmi1. Cell Res. 2011, 21, 1305–1315. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gayen, S.; Xiong, J.; Zhou, B.; Shanmugam, A.K.; Sun, Y.Q.; Karatas, H.; Liu, L.; Rao, R.C.; Wang, S.M.; et al. MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell 2016, 18, 481–494. [Google Scholar] [CrossRef] [Green Version]
- Garry, G.A.; Bezprozvannaya, S.; Chen, K.; Zhou, H.; Hashimoto, H.; Morales, M.G.; Liu, N.; Bassel-Duby, R.; Olson, E.N. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat. Cell Biol. 2021, 23, 467–475. [Google Scholar] [CrossRef]
- Eroglu, E.; Schell, J.P.; Chien, K.R. PHF7 directs cardiac reprogramming. Nat. Cell Biol. 2021, 23, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Mathison, M.; Gersch, R.P.; Nasser, A.; Lilo, S.; Korman, M.; Fourman, M.; Hackett, N.; Shroyer, K.; Yang, J.; Ma, Y.; et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J. Am. Heart Assoc. 2012, 1, e005652. [Google Scholar] [CrossRef] [Green Version]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [Green Version]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2016, 135, 978–995. [Google Scholar] [CrossRef]
- Guo, Y.; Lei, I.; Tian, S.; Gao, W.; Hacer, K.; Li, Y.; Wang, S.; Liu, L.; Wang, Z. Chemical suppression of specific C-C chemokine signaling pathways enhances cardiac reprogramming. J. Biol. Chem. 2019, 294, 9134–9146. [Google Scholar] [CrossRef]
- Muraoka, N.; Nara, K.; Tamura, F.; Kojima, H.; Yamakawa, H.; Sadahiro, T.; Miyamoto, K.; Isomi, M.; Haginiwa, S.; Tani, H.; et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 2019, 10, 674. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fidalgo, M.; Faiola, F.; Pereira, C.F.; Ding, J.J.; Saunders, A.; Gingold, J.; Schaniel, C.; Lemischka, I.R.; Silva, J.C.R.; Wang, J.L. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl. Acad. Sci. USA 2012, 109, 16202–16207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fidalgo, M.; Huang, X.; Guallar, D.; Sanchez-Priego, C.; Valdes, V.J.; Saunders, A.; Ding, J.; Wu, W.S.; Clavel, C.; Wang, J.L. Zfp281 Coordinates Opposing Functions of Tet1 and Tet2 in Pluripotent States. Cell Stem Cell 2016, 19, 355–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurotsu, S.; Sadahiro, T.; Fujita, R.; Tani, H.; Yamakawa, H.; Tamura, F.; Isomi, M.; Kojima, H.; Yamada, Y.; Abe, Y.; et al. Soft Matrix Promotes Cardiac Reprogramming via Inhibition of YAP/TAZ and Suppression of Fibroblast Signatures. Stem Cell Rep. 2020, 15, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.B.; Ratcliffe, M.B.; Fallert, M.A.; Edmunds, L.H.; Bogen, D.K. Changes In Passive Mechanical Stiffness Of Myocardial Tissue with Aneurysm Formation. Circulation 1994, 89, 2315–2326. [Google Scholar] [CrossRef] [Green Version]
- Kouskoff, V.; Lacaud, G.; Schwantz, S.; Fehling, H.J.; Keller, G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 13170–13175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamura, T.; Suzuki, J.; Wang, Y.V.; Menendez, S.; Morera, L.B.; Raya, A.; Wahl, G.M.; Izpisua Belmonte, J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009, 460, 1140–1144. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, L.; Liu, Z.; Alimohamadi, S.; Yin, C.; Liu, J.; Qian, L. Comparative Gene Expression Analyses Reveal Distinct Molecular Signatures between Differentially Reprogrammed Cardiomyocytes. Cell Rep. 2017, 20, 3014–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bektik, E.; Dennis, A.; Prasanna, P.; Madabhushi, A.; Fu, J.D. Single cell qPCR reveals that additional HAND2 and microRNA-1 facilitate the early reprogramming progress of seven-factor-induced human myocytes. PLoS ONE 2017, 12, e0183000. [Google Scholar] [CrossRef] [Green Version]
- Bektik, E.; Dennis, A.; Pawlowski, G.; Zhou, C.; Maleski, D.; Takahashi, S.; Laurita, K.R.; Deschenes, I.; Fu, J.D. S-phase Synchronization Facilitates the Early Progression of Induced-Cardiomyocyte Reprogramming through Enhanced Cell-Cycle Exit. Int. J. Mol. Sci. 2018, 19, 1364. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Huang, J.; Yang, X.; Liu, B.; Zhang, W.; Huang, L.; Deng, F.; Ma, J.; Bai, Y.; Lu, R.; et al. Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PLoS ONE 2012, 7, e28203. [Google Scholar] [CrossRef] [Green Version]
- Umei, T.C.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Kurotsu, S.; Tamura, F.; Osakabe, R.; et al. Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. Int. J. Mol. Sci. 2017, 18, 1805. [Google Scholar] [CrossRef] [Green Version]
- Abad, M.; Hashimoto, H.; Zhou, H.; Morales, M.G.; Chen, B.; Bassel-Duby, R.; Olson, E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017, 8, 548–560. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Naya, F.J. Antagonistic regulation of cell-cycle and differentiation gene programs in neonatal cardiomyocytes by homologous MEF2 transcription factors. J. Biol. Chem. 2017, 292, 10613–10629. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X.; et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef] [Green Version]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015, 25, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Yu, P.; Lingampalli, N.; Kim, H.J.; Tang, R.; Murthy, N. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomed. 2015, 10, 1841–1854. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, D.; Ku, S.H.; Kim, K.; Kim, S.H.; Kwon, I.C. MicroRNA-mediated non-viral direct conversion of embryonic fibroblasts to cardiomyocytes: Comparison of commercial and synthetic non-viral vectors. J. Biomater. Sci. Polym. Ed. 2017, 28, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Finch, E.A.; Zhang, L.; Zhang, H.; Hodgkinson, C.P.; Pratt, R.E.; Rosenberg, P.B.; Mirotsou, M.; Dzau, V.J. MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015, 116, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Mathison, M.; Singh, V.P.; Chiuchiolo, M.J.; Sanagasetti, D.; Mao, Y.; Patel, V.B.; Yang, J.; Kaminsky, S.M.; Crystal, R.G.; Rosengart, T.K. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J. Thorac. Cardiovasc. Surg. 2017, 153, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Isomi, M.; Sadahiro, T.; Yamakawa, H.; Fujita, R.; Yamada, Y.; Abe, Y.; Murakata, Y.; Akiyama, T.; Shu, T.; Mizukami, H.; et al. Overexpression of Gata4, Mef2c, and Tbx5 Generates Induced Cardiomyocytes Via Direct Reprogramming and Rare Fusion in the Heart. Circulation 2021, 143, 2123–2125. [Google Scholar] [CrossRef]
- Yang, L.; Xue, S.; Du, M.J.; Lian, F. Highly Efficient MicroRNA Delivery Using Functionalized Carbon Dots for Enhanced Conversion of Fibroblasts to Cardiomyocytes. Int. J. Nanomed. 2021, 16, 3741–3754. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Z.; Song, Y.A.; Chen, J.; Li, Q.Y.; Gao, J.F.; Tan, H.P.; Zhu, Y.F.; Wang, Z.M.; Li, M.H.; Yang, H.B.; et al. Direct in vivo reprogramming with non-viral sequential targeting nanoparticles promotes cardiac regeneration. Biomaterials 2021, 276, 121028. [Google Scholar] [CrossRef]
- Chang, Y.; Lee, E.; Kim, J.; Kwon, Y.W.; Kwon, Y.; Kim, J. Efficient in vivo direct conversion of fibroblasts into cardiomyocytes using a nanoparticle-based gene carrier. Biomaterials 2019, 192, 500–509. [Google Scholar] [CrossRef]
- Passaro, F.; Testa, G.; Ambrosone, L.; Costagliola, C.; Tocchetti, C.G.; di Nezza, F.; Russo, M.; Pirozzi, F.; Abete, P.; Russo, T.; et al. Nanotechnology-Based Cardiac Targeting and Direct Cardiac Reprogramming: The Betrothed. Stem Cells Int. 2017, 2017, 4940397. [Google Scholar] [CrossRef] [Green Version]
- England, C.G.; Im, H.J.; Feng, L.Z.; Chen, F.; Graves, S.A.; Hernandez, R.; Orbay, H.; Xu, C.; Cho, S.Y.; Nickles, R.J.; et al. Re-assessing the enhanced permeability and retention effect in peripheral arterial disease using radiolabeled long circulating nanoparticles. Biomaterials 2016, 100, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [Green Version]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Oh, H.J.; Park, J.S.; Lee, J.S.; Kim, J.H.; Park, K.H. Direct Conversion of Human Dermal Fibroblasts into Cardiomyocyte-Like Cells Using CiCMC Nanogels Coupled with Cardiac Transcription Factors and a Nucleoside Drug. Adv. Sci. 2020, 7, 1901818. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [Green Version]
- Christoforou, N.; Chakraborty, S.; Kirkton, R.D.; Adler, A.F.; Addis, R.C.; Leong, K.W. Core Transcription Factors, MicroRNAs, and Small Molecules Drive Transdifferentiation of Human Fibroblasts towards the Cardiac Cell Lineage. Sci. Rep. 2017, 7, 40285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Liu, Z.; Welch, J.D.; Gao, X.; Wang, L.; Garbutt, T.; Keepers, B.; Ma, H.; Prins, J.F.; Shen, W.; et al. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell 2019, 25, 149–164.e149. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Guo, Y.; Li, Z.; Wang, Z. Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine. Cells 2021, 10, 3297. https://doi.org/10.3390/cells10123297
Liu L, Guo Y, Li Z, Wang Z. Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine. Cells. 2021; 10(12):3297. https://doi.org/10.3390/cells10123297
Chicago/Turabian StyleLiu, Liu, Yijing Guo, Zhaokai Li, and Zhong Wang. 2021. "Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine" Cells 10, no. 12: 3297. https://doi.org/10.3390/cells10123297
APA StyleLiu, L., Guo, Y., Li, Z., & Wang, Z. (2021). Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine. Cells, 10(12), 3297. https://doi.org/10.3390/cells10123297