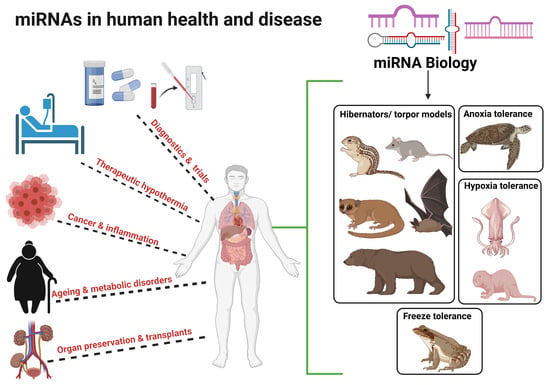
MicroRNA Cues from Nature: A Roadmap to Decipher and Combat Challenges in Human Health and Disease?
Abstract
:1. Introduction
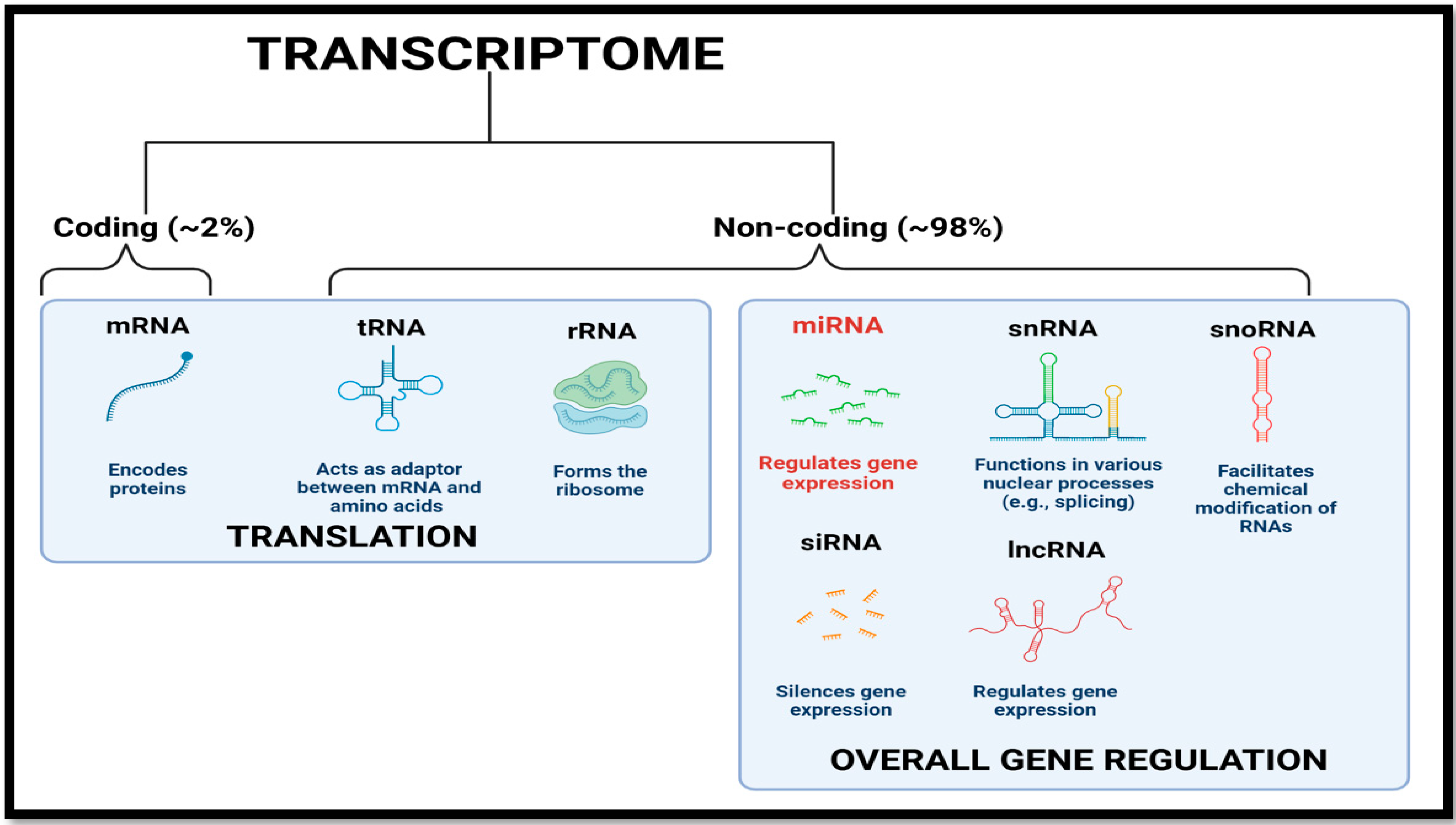
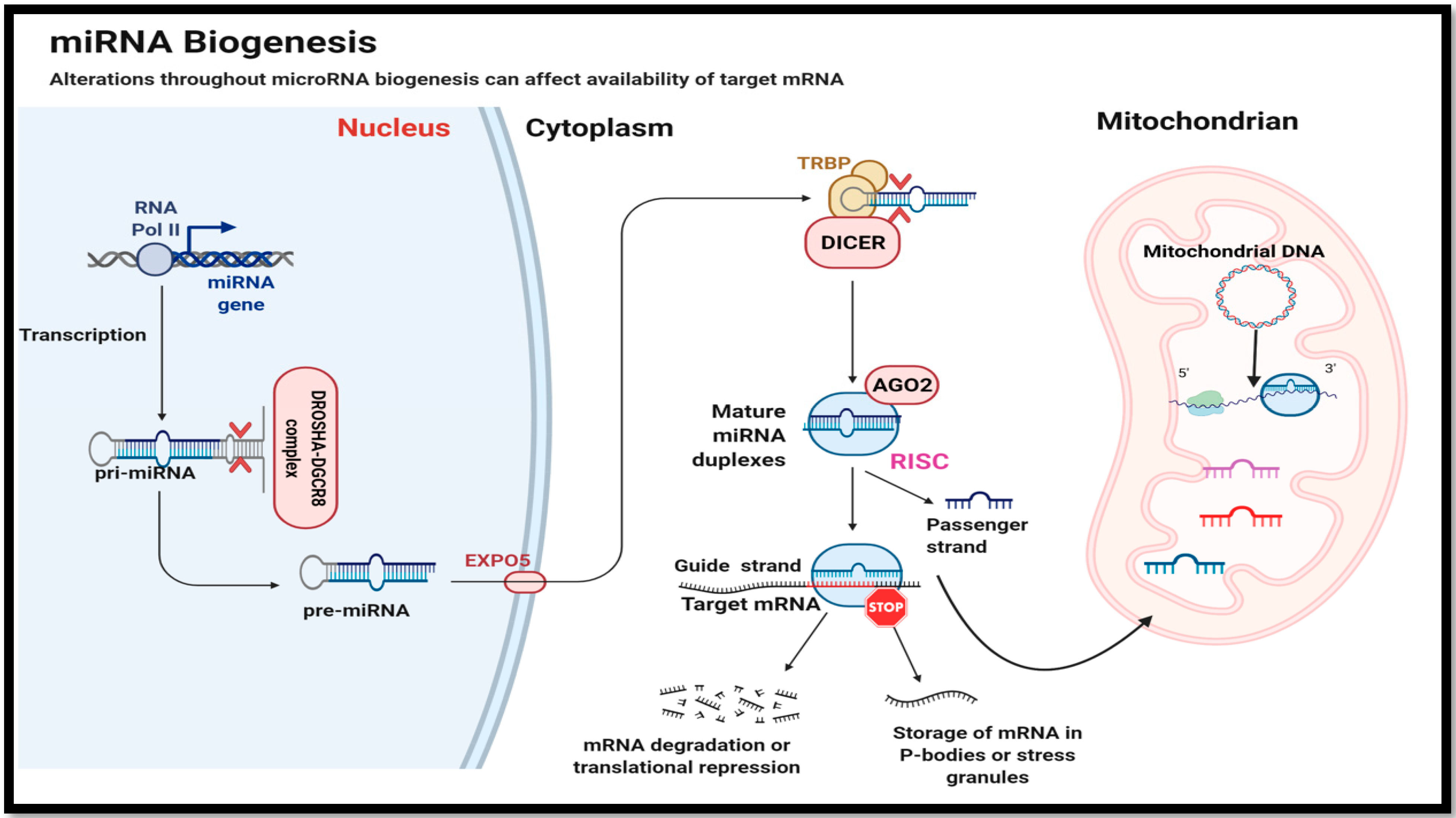
2. MicroRNA Biogenesis and Mode of Action
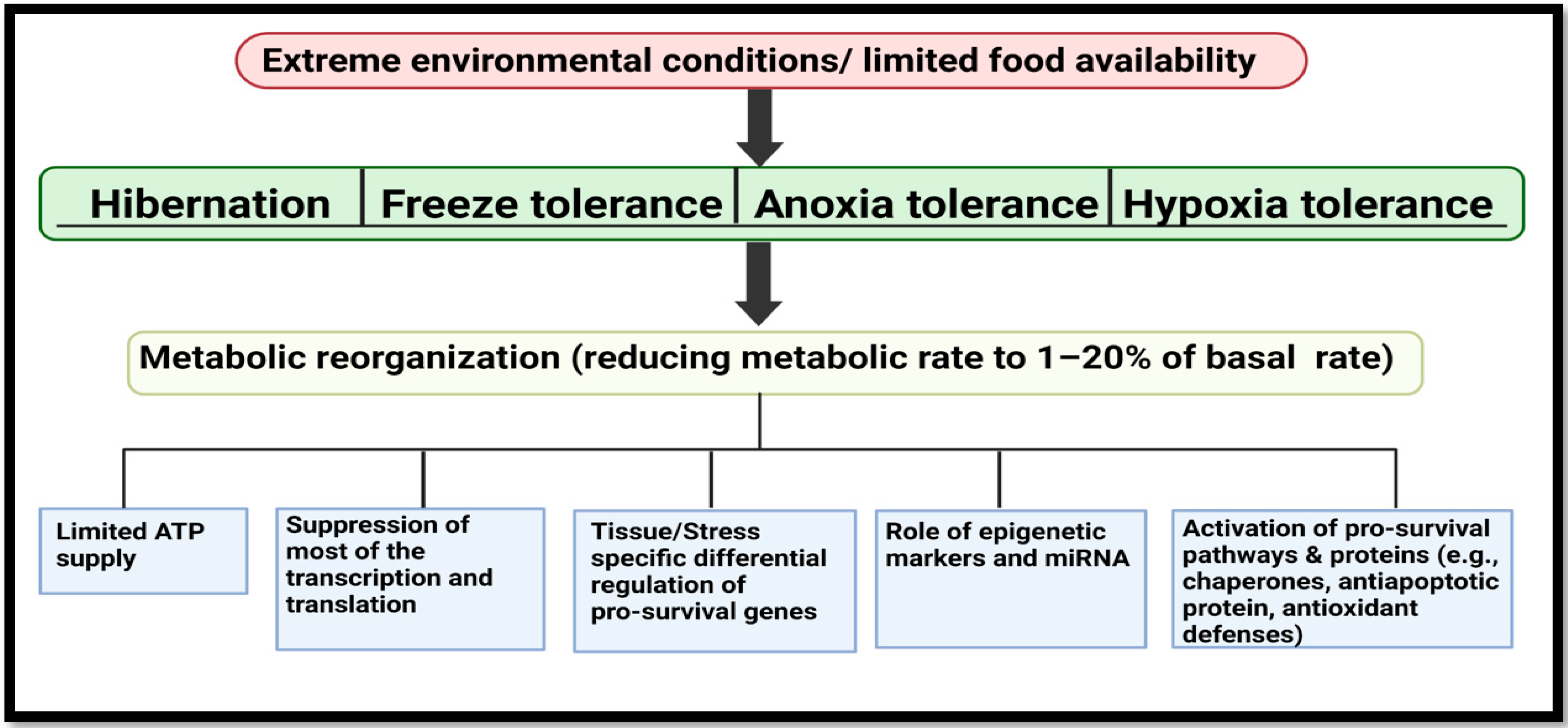
3. MicroRNA Biology from Extreme Animal Survivalists to Human Health and Disease
3.1. Ageing, Oxidative Stress and Related Disorders
3.2. MiRNA: Cancer, Inflammation and Other Diseases
3.3. Mitochondria Specific MiRNA, MitoMirs and MDPs: Dark Horses Involved in Mitochondrial Dysfunction?
3.4. MicroRNAs in Organ Preservation and Transplantation
3.5. Low Temperature MiRNA Target Selection: A Possible Role in Inducing Therapeutic Hypothermia?
| miRNA Species | Animal/Cells | Putative Target/Pathways | Reference |
|---|---|---|---|
| miR-21, miR-1, miR-29b, miR-23a, miR-181b, miR-15a, miR-20a, miR-128, miR-206 | Myotis lucifugis | Muscle atrophy | [23] |
| miR-29b | Myotis ricketti | Neuroprotection | [49] |
| miR-1 family | Dromiciops Gliroides | MEF-2 signaling in myogenesis and muscle maintenance | [26] |
| miR-1, miR-31, miR-23a, miR-29b, miR-206 | Ursus arctos | MEF-2 signaling in myogenesis and muscle maintenance | [30] |
| miR-365, miR-99, miR-92a, miR-103, miR-107 | Ictidomys tridecemlineatus | Affects adipogenesis, levels of FGF-21 during torpor and BAT mitochondrial regulation respectively | [13,102] |
| miR-200a, miR-15b, miR-25 | Ictidomys tridecemlineatus | Antioxidant response (NRF-2), suppress cell proliferation and mitochondrial apoptosis respectively | [38,105] |
| miR-195 | Ictidomys tridecemlineatus | Fatty acid synthase regulation | [77] |
| miR-200b, miR-200c, miR-141, miR-429, miR-182, miR-183, miR-96 | Ictidomys tridecemlineatus | Targets SUMOlyation and Ubiquitin like identifiers (ULMs) | [89] |
| miR-24 | Heterocephalus glaber | Hypoxia induced reduction in mitochondrial protein, cytochrome c | [67] |
| miR-335, miR-155 | Heterocephalus glaber | Regulating HIF signaling and NF-ĸB respectively | [69] |
| miR-92a, miR-193b, miR-218, miR-222, miR-874 | Microcebus murinus | P53 signaling and cell survival pathway | [71] |
| miR-2 family, miR-133 | Dosidicus gigas | Suppressing pro-apoptotic genes and Ischemic injury recovery in cardiomyopathy respectively | [80] |
| miR-34, miR-15a, miR-16 | T.s elegans | Suppress Cell cycle and P53 signaling | [87,88] |
| miR-20a, miR-21 | T.s elegans | Induce anti-apoptotic response | [162] |
| miR-21 | Rana sylvatica | Targets mRNA for inflammatory caspases, casp-3 | [157] |
| miR-451, miR-181a | Rana sylvatica | Reduces anoxia/reperfusion injury via restricting apoptosis | [158] |
| miR-145 | Rana sylvatica | Reduces ischemic injury | [161] |
| hsa-mir-mit3, hsa-mir-mit4 | Homo sapiens | Humanin gene regulation in the mitochondria | [119] |
| Let-7b, miR-146a, miR-19b, miR-34a, miR-221 | Homo sapiens | Cell senescence | [124] |
| miR-128-2, miR-205 | Homo sapiens | P53 signaling and anti-apoptotic response | [57,58] |
| miR-874, MIR-497, miR-290 | Rattus norvegicus | Increasing transport of cytosolic and membrane proteins | [150] |
| miR-9 | Rattus norvegicus | Reduced levels promote transcription and translation in generating proteins for cell and cytoskeleton integrity | [150] |
| miR-274 | Rattus norvegicus | Inhibit Mex3B and affecting genes in apoptotic cascade | [151] |
| mitomiR-181c | Rattus norvegicus | Decrease COX-1 in ETC chain | [120] |
| mitomiR-696, -532, -690 | Mus musculus | Mitochondrial energetics in heart failure | [122] |
| mitomiR-378a | Mus musculus | Suppress mitochondrial ATP6 and induce apoptosis | [123] |
4. MiRNA Therapeutics and Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Moussa, H.; Hawkins, L.J.; Storey, K.B. Role of MicroRNAs in extreme animal survival strategies. In miRNomics; Allmer, J., Yousef, M., Eds.; Humana: New York, NY, USA, 2022; pp. 311–347. [Google Scholar] [CrossRef]
- Shu, J.; e Silva, B.V.R.; Gao, T.; Xu, Z.; Cui, J. Dynamic and modularized MicroRNA regulation and its implication in human cancers. Sci. Rep. 2017, 7, 13356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. (Lond.) 2018, 15, 68. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.-P.; Slack, F.J. MicroRNA-mediated silencing inside P bodies. RNA Biol. 2006, 3, 97–100. [Google Scholar] [CrossRef]
- Ha, T.-Y. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune Netw. 2011, 11, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Long, B.; Han, W.; Yuan, S.; Wang, K. MicroRNAs: Important regulators of stem cells. Stem Cell Res. Ther. 2017, 8, 110. [Google Scholar] [CrossRef]
- Hwang, H.-W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776–780. [Google Scholar] [CrossRef]
- English, S.G.; Hadj-Moussa, H.; Storey, K.B. MicroRNAs regulate survival in oxygen-deprived environments. J. Exp. Biol. 2018. [Google Scholar] [CrossRef] [Green Version]
- Morin, P.; Storey, K.B. Mammalian hibernation: Differential gene expression and novel application of epigenetic controls. Int. J. Dev. Biol. 2009, 53, 433–442. [Google Scholar] [CrossRef]
- Logan, S.M.; Storey, K.B. MicroRNA expression patterns in the brown fat of hibernating 13-lined ground squirrels. Genomics 2021, 113, 769–781. [Google Scholar] [CrossRef]
- Luu, B.E.; Storey, K.B. Solving donor organ shortage with insights from freeze tolerance in nature. BioEssays 2018, 40, 1800092. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Molecular physiology of freeze tolerance in vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular basis for the recognition of primary MicroRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microrna biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Graves, P.; Zeng, Y. Biogenesis of mammalian micrornas: A global view. Genom. Proteom. Bioinform. 2012, 10, 239–245. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of micrornas: A review. J. Physiol. Biochem. 2011, 67, 129–139. [Google Scholar] [CrossRef]
- da Costa, J.P.; Vitorino, R.; Silva, G.M.; Vogel, C.; Duarte, A.C.; Rocha-Santos, T. A synopsis on aging—Theories, mechanisms and future prospects. Ageing Res. Rev. 2016, 29, 90–112. [Google Scholar] [CrossRef]
- Demetrius, L. Aging in mouse and human systems: A comparative study. Ann. N. Y. Acad. Sci. 2006, 1067, 66–82. [Google Scholar] [CrossRef]
- Smith-Vikos, T.; Slack, F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012, 125, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Kornfeld, S.F.; Biggar, K.K.; Storey, K.B. Differential expression of mature MicroRNAs involved in muscle maintenance of hibernating little brown bats, Myotis lucifugus: A model of muscle atrophy resistance. Genom. Proteom. Bioinform. 2012, 10, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Kollias, H.D.; Perry, R.L.S.; Miyake, T.; Aziz, A.; McDermott, J.C. Smad7 promotes and enhances skeletal muscle differentiation. Mol. Cell. Biol. 2006, 26, 6248–6260. [Google Scholar] [CrossRef] [Green Version]
- Bozinovic, F.; Ruiz, G.; Rosenmann, M. Energetics and torpor of a South American “living fossil”, the microbiotheriid Dromiciops gliroides. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2004, 174, 293–297. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Moggridge, J.A.; Luu, B.E.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. The hibernating south american marsupial, Dromiciops gliroides, displays torpor-sensitive MicroRNA expression patterns. Sci. Rep. 2016, 6, 24627. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The role of MicroRNA-1 and MicroRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Opazo, J.C.; Nespolo, R.F.; Bozinovic, F. Arousal from torpor in the Chilean mouse-opposum (Thylamys elegans): Does non-shivering thermogenesis play a role? Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1999, 123, 393–397. [Google Scholar] [CrossRef]
- Rose, R.W.; West, A.K.; Ye, J.; McCormack, G.H.; Colquhoun, E.Q. Nonshivering thermogenesis in a marsupial (the Tasmanian Bettong Bettongia gaimardi ) is not attributable to brown adipose tissue. Physiol. Biochem. Zool. 1999, 72, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Luu, B.E.; Lefai, E.; Giroud, S.; Swenson, J.E.; Chazarin, B.; Gauquelin-Koch, G.; Arnemo, J.M.; Evans, A.L.; Bertile, F.; Storey, K.B. MicroRNAs facilitate skeletal muscle maintenance and metabolic suppression in hibernating brown bears. J. Cell. Physiol. 2020, 235, 3984–3993. [Google Scholar] [CrossRef]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [Green Version]
- Burton, G.J.; Jauniaux, E. Oxidative stress. Best Pract. Res. Clin. Obstet. Gynaecol. 2011, 25, 287–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trist, B.G.; Hare, D.J.; Double, K.L. Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell 2019, 18, e13031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Heid, J.; Cencioni, C.; Ripa, R.; Baumgart, M.; Atlante, S.; Milano, G.; Scopece, A.; Kuenne, C.; Guenther, S.; Azzimato, V.; et al. Age-dependent increase of oxidative stress regulates MicroRNA-29 family preserving cardiac health. Sci. Rep. 2017, 7, 16839. [Google Scholar] [CrossRef] [PubMed]
- Tong, N.; Jin, R.; Zhou, Z.; Wu, X. Involvement of MicroRNA-34a in age-related susceptibility to oxidative stress in ARPE-19 cells by targeting the silent mating type information regulation 2 Homolog 1/P66shc pathway: Implications for age-related macular degeneration. Front. Aging Neurosci. 2019, 11, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiles, J.M.; Pepin, M.E.; Sunny, S.; Shelar, S.B.; Challa, A.K.; Dalley, B.; Hoidal, J.R.; Pogwizd, S.M.; Wende, A.R.; Rajasekaran, N.S. Identification of Nrf2-responsive MicroRNA networks as putative mediators of myocardial reductive stress. Sci. Rep. 2021, 11, 11977. [Google Scholar] [CrossRef]
- Frigault, J.J.; Gaudet, J.D.; Morin, P.J. Investigating Nrf2-associated non-coding RNAs in the hibernating ground squirrel, Ictidomys tridecemlineatus. J. Therm. Biol. 2018, 75, 38–44. [Google Scholar] [CrossRef]
- Izzo, C.; Vitillo, P.; Di Pietro, P.; Visco, V.; Strianese, A.; Virtuoso, N.; Ciccarelli, M.; Galasso, G.; Carrizzo, A.; Vecchione, C. The role of oxidative stress in cardiovascular aging and cardiovascular diseases. Life 2021, 11, 60. [Google Scholar] [CrossRef]
- Wu, C.W.; Storey, K.B. Hibernation and aging molecular mechanisms of mammalian hypometabolism and its links to longevity. In Aging: Exploring a Complex Phenomenon; Ahmad, S.I., Ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 617–633. [Google Scholar] [CrossRef]
- Al-Attar, R.; Storey, K.B. Suspended in time: Molecular responses to hibernation also promote longevity. Exp. Gerontol. 2020, 134, 110889. [Google Scholar] [CrossRef]
- Turbill, C.; Bieber, C.; Ruf, T. Hibernation is associated with increased survival and the evolution of slow life histories among mammals. Proc. R. Soc. B Biol. Sci. 2011, 278, 3355–3363. [Google Scholar] [CrossRef] [Green Version]
- Camandola, S.; Mattson, M.P. Brain Metabolism in Health, Aging, and Neurodegeneration. EMBO J. 2017, 36, 1474–1492. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Aulston, B.; Liu, Q.; Mante, M.; Florio, J.; Rissman, R.A.; Yuan, S.H. Extracellular vesicles isolated from familial alzheimer’s disease neuronal cultures induce aberrant Tau phosphorylation in the wild-type mouse brain. J. Alzheimer’s Dis. 2019, 72, 575–585. [Google Scholar] [CrossRef]
- Logan, S.; Storey, K. Regrowth and neuronal protection are key for mammalian hibernation: Roles for metabolic suppression. Neural Regen. Res. 2020, 15, 2027. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, Y.-H.; Yin, Q.; Yang, T.; Dong, D.; Liao, C.-C.; Zhang, S. Critical roles of mitochondria in brain activities of torpid myotis ricketti bats revealed by a proteomic approach. J. Proteom. 2014, 105, 266–284. [Google Scholar] [CrossRef]
- Biggar, K.K.; Storey, K.B. Identification and expression of microrna in the brain of hibernating bats, Myotis lucifugus. Gene 2014, 544, 67–74. [Google Scholar] [CrossRef]
- Yuan, L.; Geiser, F.; Lin, B.; Sun, H.; Chen, J.; Zhang, S. Down but Not Out: The role of MicroRNAs in hibernating bats. PLoS ONE 2015, 10, e0135064. [Google Scholar] [CrossRef] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Manikandan, J.; Aarthi, J.J.; Kumar, S.D.; Pushparaj, P.N. Oncomirs: The potential role of non-coding MicroRNAs in understanding cancer. Bioinformation 2008, 2, 330–334. [Google Scholar] [CrossRef] [Green Version]
- Cho, W.C.S. OncomiRs: The discovery and progress of MicroRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [Green Version]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sültmann, H. Serum MicroRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Adams, B.D.; Kasinski, A.L.; Slack, F.J. Aberrant regulation and function of MicroRNAs in cancer. Curr. Biol. 2014, 24, R762–R776. [Google Scholar] [CrossRef] [Green Version]
- Hoey, C.; Ahmed, M.; Fotouhi Ghiam, A.; Vesprini, D.; Huang, X.; Commisso, K.; Commisso, A.; Ray, J.; Fokas, E.; Loblaw, D.A.; et al. Circulating MiRNAs as non-invasive biomarkers to predict aggressive prostate cancer after radical prostatectomy. J. Transl. Med. 2019, 17, 173. [Google Scholar] [CrossRef]
- Kashyap, D.; Kaur, H. Cell-free MiRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 2020, 246, 117417. [Google Scholar] [CrossRef]
- Donzelli, S.; Fontemaggi, G.; Fazi, F.; Di Agostino, S.; Padula, F.; Biagioni, F.; Muti, P.; Strano, S.; Blandino, G. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant P53 gain of function. Cell Death Differ. 2012, 19, 1038–1048. [Google Scholar] [CrossRef] [Green Version]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of MicroRNA biogenesis in cancer: The Impact of mutant P53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef] [Green Version]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Geörg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 MiRNA microprocessor complex underlie high-risk blastemal type wilms tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [Green Version]
- Lupo, M.A.; Walts, A.E.; Sistrunk, J.W.; Giordano, T.J.; Sadow, P.M.; Massoll, N.; Campbell, R.; Jackson, S.A.; Toney, N.; Narick, C.M.; et al. Multiplatform molecular test performance in indeterminate thyroid nodules. Diagn. Cytopathol. 2020, 48, 1254–1264. [Google Scholar] [CrossRef]
- Barré-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Futur. Sci. OA 2015, 1. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yin, T.; Feng, Y.; Cona, M.M.; Huang, G.; Liu, J.; Song, S.; Jiang, Y.; Xia, Q.; Swinnen, J.V.; et al. Mammalian models of chemically induced primary malignancies exploitable for imaging-based preclinical theragnostic research. Quant. Imaging Med. Surg. 2015, 5, 708–729. [Google Scholar] [CrossRef] [PubMed]
- Willyard, C. The mice with human tumours: Growing pains for a popular cancer model. Nature 2018, 560, 156–157. [Google Scholar] [CrossRef] [PubMed]
- Buffenstein, R.; Amoroso, V.; Andziak, B.; Avdieiev, S.; Azpurua, J.; Barker, A.J.; Bennett, N.C.; Brieño-Enríquez, M.A.; Bronner, G.N.; Coen, C.; et al. The naked truth: A comprehensive clarification and classification of current ‘myths’ in naked mole-rat biology. Biol. Rev. 2021, brv.12791. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [Green Version]
- Logan, S.M.; Szereszewski, K.E.; Bennett, N.C.; Hart, D.W.; van Jaarsveld, B.; Pamenter, M.E.; Storey, K.B. The brains of six african mole-rat species show divergent responses to hypoxia. J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef]
- Liu, F.J.; Kaur, P.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Wong, P.T.H.; Jeyaseelan, K. MiR-335 regulates Hif-1α to reduce cell death in both mouse cell line and rat ischemic models. PLoS ONE 2015, 10, e0128432. [Google Scholar] [CrossRef] [Green Version]
- Hadj-Moussa, H.; Pamenter, M.E.; Storey, K.B. Hypoxic naked mole–rat brains use MicroRNA to coordinate hypometabolic fuels and neuroprotective defenses. J. Cell. Physiol. 2021, 236, 5080–5097. [Google Scholar] [CrossRef]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef]
- Biggar, K.K.; Luu, B.E.; Wu, C.W.; Pifferi, F.; Perret, M.; Storey, K.B. Identification of novel and conserved MicroRNA and their expression in the gray mouse lemur, Microcebus murinus, a primate capable of daily torpor. Gene 2018, 677, 332–339. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, J.; Sun, B.F.; Hou, G.Z.; Yu, Z.J.; Luo, H. Microrna-92a targets Sertad3 and regulates the growth, invasion, and migration of prostate cancer cells via the P53 pathway. OncoTargets Ther. 2020. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Cao, B.; Shang, H. Downregulation of MicroRNA-193b-3p promotes autophagy and cell survival by targeting TSC1/MTOR signaling in NSC-34 cells. Front. Mol. Neurosci. 2017, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Torres-Berrío, A.; Nouel, D.; Cuesta, S.; Parise, E.M.; Restrepo-Lozano, J.M.; Larochelle, P.; Nestler, E.J.; Flores, C. MiR-218: A molecular switch and potential biomarker of susceptibility to stress. Mol. Psychiatry 2020, 25, 951–964. [Google Scholar] [CrossRef]
- Fhu, C.W.; Ali, A. Fatty acid synthase: An emerging target in cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef]
- Syed-Abdul, M.M.; Parks, E.J.; Gaballah, A.H.; Bingham, K.; Hammoud, G.M.; Kemble, G.; Buckley, D.; McCulloch, W.; Manrique-Acevedo, C. Fatty acid synthase inhibitor TVB-2640 reduces hepatic de novo lipogenesis in males with metabolic abnormalities. Hepatology 2020, 72, 103–118. [Google Scholar] [CrossRef]
- Lang-Ouellette, D.; Morin, P.J. Differential expression of MiRNAs with metabolic implications in hibernating thirteen-lined ground squirrels, Ictidomys tridecemlineatus. Mol. Cell. Biochem. 2014, 394, 291–298. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Storey, K.B. The OxymiR response to oxygen limitation: A comparative MicroRNA perspective. J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef]
- Rosa, R.; Seibel, B.A. Metabolic physiology of the humboldt squid, Dosidicus gigas: Implications for vertical migration in a pronounced oxygen minimum zone. Prog. Oceanogr. 2010, 86, 72–80. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Logan, S.M.; Seibel, B.A.; Storey, K.B. Potential role for MicroRNA in regulating hypoxia-induced metabolic suppression in jumbo squids. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 586–593. [Google Scholar] [CrossRef]
- Teixeira, A.F.; ten Dijke, P.; Zhu, H.-J. On-target anti-TGF-β therapies are not succeeding in clinical cancer treatments: What are remaining challenges? Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Ouyang, Y.-B.; Stary, C.M.; Yang, G.-Y.; Giffard, R. MicroRNAs: Innovative targets for cerebral ischemia and stroke. Curr. Drug Targets 2013, 14, 90–101. [Google Scholar] [CrossRef]
- Abdellatif, M. The role of MicroRNA-133 in cardiac hypertrophy uncovered. Circ. Res. 2010, 106, 16–18. [Google Scholar] [CrossRef]
- Storey, K.B. Anoxia tolerance in turtles: Metabolic regulation and gene expression. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 263–276. [Google Scholar] [CrossRef]
- Reese, S.A.; Ultsch, G.R.; Jackson, D.C. Lactate accumulation, glycogen depletion, and shell composition of hatchling turtles during simulated aquatic hibernation. J. Exp. Biol. 2004, 207, 2889–2895. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, M.L.; Gladden, L.B.; Nijsten, M.W.N.; Jones, K.B. Lactate and cancer: Revisiting the Warburg Effect in an era of lactate shuttling. Front. Nutr. 2015, 1, 27. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Biggar, K.K.; Storey, K.B. Regulation of P53 by reversible post-transcriptional and post-translational mechanisms in liver and skeletal muscle of an anoxia tolerant turtle, Trachemys scripta elegans. Gene 2013, 513, 147–155. [Google Scholar] [CrossRef]
- Biggar, K.K.; Storey, K.B. Evidence for cell cycle suppression and MicroRNA regulation of Cyclin D1 during anoxia exposure in turtles. Cell Cycle 2012, 11, 1705–1713. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Johnson, K.R.; Hallenbeck, J.M. Global protein conjugation by Ubiquitin-like-modifiers during ischemic stress is regulated by MicroRNAs and confers robust tolerance to ischemia. PLoS ONE 2012, 7, e47787. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Lin, H.-K. The key role of Ubiquitination and Sumoylation in signaling and cancer: A research topic. Front. Oncol. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Miyake, S.; Wakita, H.; McMullen, D.C.; Azuma, Y.; Auh, S.; Hallenbeck, J.M. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J. Cereb. Blood Flow Metab. 2007, 27, 950–962. [Google Scholar] [CrossRef] [Green Version]
- Humphries, B.; Yang, C. The MicroRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [Green Version]
- Spitschak, A.; Meier, C.; Kowtharapu, B.; Engelmann, D.; Pützer, B.M. MiR-182 promotes cancer invasion by linking RET oncogene activated NF-ΚB to loss of the HES1/Notch1 regulatory circuit. Mol. Cancer 2017, 16, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyfried, T.N.; Mukherjee, P. Targeting energy metabolism in brain cancer: Review and hypothesis. Nutr. Metab. (Lond.) 2005, 2, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chondronikola, M.; Sidossis, L.S. The Physiological Significance of Brown Adipose Tissue and the Beiging of White Adipose Tissue in People. In Adipose Tissue Biology; Symonds, M., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 201–227. [Google Scholar] [CrossRef]
- Kajimura, S.; Saito, M. A new era in brown adipose tissue biology: Molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 2014, 76, 225–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omran, F.; Christian, M. Inflammatory signaling and brown fat activity. Front. Endocrinol. (Lausanne) 2020, 11, 156. [Google Scholar] [CrossRef]
- Ballinger, M.A.; Andrews, M.T. Nature’s fat-burning machine: Brown adipose tissue in a hibernating mammal. J. Exp. Biol. 2018, 221 (Suppl. 1). [Google Scholar] [CrossRef] [Green Version]
- Golden, R.J.; Chen, B.; Li, T.; Braun, J.; Manjunath, H.; Chen, X.; Wu, J.; Schmid, V.; Chang, T.-C.; Kopp, F.; et al. An Argonaute phosphorylation cycle promotes MicroRNA-mediated silencing. Nature 2017, 542, 197–202. [Google Scholar] [CrossRef] [Green Version]
- de Jong, J.M.A.; Larsson, O.; Cannon, B.; Nedergaard, J. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Metab. 2015, 308, E1085–E1105. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Jiang, H.; Li, X.; Chen, X.; Huang, Y. MiR-92a regulates brown adipocytes differentiation, mitochondrial oxidative respiration, and heat generation by targeting SMAD7. J. Cell. Biochem. 2020, 121, 3825–3836. [Google Scholar] [CrossRef]
- Wu, C.-W.; Biggar, K.K.; Storey, K.B. Expression profiling and structural characterization of MicroRNAs in adipose tissues of hibernating ground squirrels. Genom. Proteom. Bioinform. 2014, 12, 284–291. [Google Scholar] [CrossRef]
- Ruiz, J.R.; Martinez-Tellez, B.; Sanchez-Delgado, G.; Osuna-Prieto, F.J.; Rensen, P.C.N.; Boon, M.R. Role of human brown fat in obesity, metabolism and cardiovascular disease: Strategies to turn up the heat. Prog. Cardiovasc. Dis. 2018, 61, 232–245. [Google Scholar] [CrossRef]
- Javadov, S.; Kozlov, A.V.; Camara, A.K.S. Mitochondria in health and diseases. Cells 2020, 9, 1177. [Google Scholar] [CrossRef]
- Morin, M.D.; Lang-Ouellette, D.; Lyons, P.J.; Crapoulet, N.; Morin, P. Characterization of MiRNAs modulated by torpor in the hibernating ground squirrel, Ictidomys tridecemlineatus liver by next-generation sequencing. Cryo Lett. 2017, 38, 269–277. [Google Scholar]
- Zhang, Y.; Huang, F.; Wang, J.; Peng, L.; Luo, H. MiR-15b mediates liver cancer cells proliferation through targeting BCL-2. Int. J. Clin. Exp. Pathol. 2015, 8, 15677–15683. [Google Scholar]
- Feng, X.; Jiang, J.; Shi, S.; Xie, H.; Zhou, L.; Zheng, S. Knockdown of MiR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int. J. Oncol. 2016, 49, 2600–2610. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; Ito, Y.; Niikura, T.; Shao, Z.; Hata, M.; Oyama, F.; Nishimoto, I. Mechanisms of neuroprotection by a novel rescue factor humanin from swedish mutant amyloid precursor protein. Biochem. Biophys. Res. Commun. 2001, 283, 460–468. [Google Scholar] [CrossRef]
- Szereszewski, K.E.; Storey, K.B. Identification of a prosurvival neuroprotective mitochondrial peptide in a mammalian hibernator. Cell Biochem. Funct. 2019, 37, 494–503. [Google Scholar] [CrossRef]
- Wijenayake, S.; Storey, K.B. Oxidative Damage? not a problem! the characterization of Humanin-like mitochondrial peptide in anoxia tolerant freshwater turtles. Protein J. 2021, 40, 87–107. [Google Scholar] [CrossRef]
- Yen, K.; Wan, J.; Mehta, H.H.; Miller, B.; Christensen, A.; Levine, M.E.; Salomon, M.P.; Brandhorst, S.; Xiao, J.; Kim, S.-J.; et al. Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans. Sci. Rep., 2018, 8, 14212. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P. New role for the mitochondrial peptide Humanin: Protective agent against chemotherapy-induced side effects. JNCI J. Natl. Cancer Inst. 2014, 106, dju006. [Google Scholar] [CrossRef] [Green Version]
- Sreekumar, P.G.; Ishikawa, K.; Spee, C.; Mehta, H.H.; Wan, J.; Yen, K.; Cohen, P.; Kannan, R.; Hinton, D.R. The mitochondrial-derived peptide Humanin protects RPE cells from oxidative stress, senescence, and mitochondrial dysfunction. Investig. Opthalmology Vis. Sci. 2016, 57, 1238. [Google Scholar] [CrossRef]
- Guo, B.; Zhai, D.; Cabezas, E.; Welsh, K.; Nouraini, S.; Satterthwait, A.C.; Reed, J.C. Humanin peptide suppresses apoptosis by interfering with bax activation. Nature 2003, 423, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Moreno Ayala, M.A.; Gottardo, M.F.; Zuccato, C.F.; Pidre, M.L.; Nicola Candia, A.J.; Asad, A.S.; Imsen, M.; Romanowski, V.; Creton, A.; Isla Larrain, M.; et al. Humanin promotes tumor progression in experimental triple negative breast cancer. Sci. Rep. 2020, 10, 8542. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Wang, M.; Liu, Z.; Wang, S.; Xie, Y. [Gly14]-Humanin ameliorates high glucose-induced apoptosis by inhibiting the expression of MicroRNA-155 in endothelial microparticles. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 2335–2347. [Google Scholar] [CrossRef]
- Niikura, T.; Chiba, T.; Aiso, S.; Matsuoka, M.; Nishimoto, I. Humanin: After the discovery. Mol. Neurobiol. 2004, 30, 327–340. [Google Scholar] [CrossRef]
- Borralho, P.M.; Rodrigues, C.M.P.; Steer, C.J. MicroRNAs in mitochondria: An unexplored niche. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2015; pp. 31–51. [Google Scholar] [CrossRef]
- Shinde, S.; Bhadra, U. A complex genome-MicroRNA interplay in human mitochondria. BioMed Res. Int. 2015, 2015, 206382. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear MiRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef] [Green Version]
- Arranz, E.; Peña, A.S.; Bernardo, D. Mediators of inflammation and immune responses in the human gastrointestinal tract. Mediat. Inflamm. 2013, 2013, 865638. [Google Scholar] [CrossRef]
- Wang, X.; Song, C.; Zhou, X.; Han, X.; Li, J.; Wang, Z.; Shang, H.; Liu, Y.; Cao, H. Mitochondria associated MicroRNA expression profiling of heart failure. BioMed Res. Int. 2017, 2017, 4042509. [Google Scholar] [CrossRef]
- Machado, I.F.; Teodoro, J.S.; Palmeira, C.M.; Rolo, A.P. MiR-378a: A new emerging microrna in metabolism. Cell. Mol. Life Sci. 2020, 77, 1947–1958. [Google Scholar] [CrossRef]
- Rippo, M.R.; Olivieri, F.; Monsurrò, V.; Prattichizzo, F.; Albertini, M.C.; Procopio, A.D. MitomiRs in human inflamm-aging: A hypothesis involving MiR-181a, MiR-34a and MiR-146a. Exp. Gerontol. 2014, 56, 154–163. [Google Scholar] [CrossRef]
- Olivieri, F.; Rippo, M.R.; Procopio, A.D.; Fazioli, F. Circulating Inflamma-MiRs in aging and age-related diseases. Front. Genet. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Luna, C.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Modulation of inflammatory markers by MiR-146a during replicative senescence in trabecular meshwork cells. Investig. Opthalmol. Vis. Sci. 2010, 51, 2976. [Google Scholar] [CrossRef]
- Li, H.; Dai, B.; Fan, J.; Chen, C.; Nie, X.; Yin, Z.; Zhao, Y.; Zhang, X.; Wang, D.W. The different roles of MiRNA-92a-2-5p and Let-7b-5p in mitochondrial translation in Db/Db mice. Mol. Ther. Nucleic Acids 2019, 17, 424–435. [Google Scholar] [CrossRef]
- Yan, K.; An, T.; Zhai, M.; Huang, Y.; Wang, Q.; Wang, Y.; Zhang, R.; Wang, T.; Liu, J.; Zhang, Y.; et al. Mitochondrial MiR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 2019, 10, 500. [Google Scholar] [CrossRef] [Green Version]
- Siasos, G.; Tsigkou, V.; Kosmopoulos, M.; Theodosiadis, D.; Simantiris, S.; Tagkou, N.M.; Tsimpiktsioglou, A.; Stampouloglou, P.K.; Oikonomou, E.; Mourouzis, K.; et al. Mitochondria and cardiovascular diseases—from pathophysiology to treatment. Ann. Transl. Med. 2018, 6, 256. [Google Scholar] [CrossRef]
- Tan, S.; Merchant, J. Joseph Murray (1919–2012): First transplant surgeon. Singapore Med. J. 2019, 60, 162–163. [Google Scholar] [CrossRef]
- Beyar, R. Challenges in organ transplantation. Rambam Maimonides Med. J. 2011, 2. [Google Scholar] [CrossRef]
- Giwa, S.; Lewis, J.K.; Alvarez, L.; Langer, R.; Roth, A.E.; Church, G.M.; Markmann, J.F.; Sachs, D.H.; Chandraker, A.; Wertheim, J.A.; et al. The promise of organ and tissue preservation to transform medicine. Nat. Biotechnol. 2017, 35, 530–542. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Storey, K.B. Bringing nature back: Using hibernation to reboot organ preservation. FEBS J. 2019, 286, 1094–1100. [Google Scholar] [CrossRef]
- Jing, L.; Yao, L.; Zhao, M.; Peng, L.-P.; Liu, M. Organ preservation: From the past to the future. Acta Pharmacol. Sin. 2018, 39, 845–857. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic rate depression. In Advances in Clinical Chemistry; Elsevier/Academic Press: Amsterdam, The Netherlands, 2010; pp. 77–108. [Google Scholar] [CrossRef]
- Storey, K.B. The gray mouse lemur: A model for studies of primate metabolic rate depression. Genom. Proteom Bioinform. 2015, 13, 77–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giroud, S.; Habold, C.; Nespolo, R.F.; Mejías, C.; Terrien, J.; Logan, S.M.; Henning, R.H.; Storey, K.B. The torpid state: Recent advances in metabolic adaptations and protective mechanisms. Front. Physiol. 2021, 11, 1824. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-W.; Biggar, K.K.; Luu, B.E.; Szereszewski, K.E.; Storey, K.B. Analysis of MicroRNA expression during the torpor-arousal cycle of a mammalian hibernator, the 13-lined ground squirrel. Physiol. Genomics 2016, 48, 388–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, G.; Weir, M.R.; Li, X.C.; Mandelbrot, D.A. The evolving role of MTOR inhibition in transplantation tolerance. J. Am. Soc. Nephrol. 2011, 22, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-W.; Storey, K.B. Regulation of the MTOR signaling network in hibernating thirteen-lined ground squirrels. J. Exp. Biol. 2012, 215, 1720–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, T.; Zha, D.; Gao, P.; Shui, H.; Wu, X. MiR-874 alleviates renal injury and inflammatory response in diabetic nephropathy through targeting toll-like receptor-4. J. Cell. Physiol. 2019, 234, 871–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, S.; Choi, J.; Lim, S.; Choi, H.J.; Park, J.; Hong, S.H.; Park, C.S.; Choi, J.H.; Chae, M.S. Stress burden related to postreperfusion syndrome may aggravate hyperglycemia with insulin resistance during living donor liver transplantation: A propensity score-matching analysis. PLoS ONE 2020, 15, e0243873. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, J. Normothermic preservation poised to change organ transplants. JAMA. 2018, 319, 2263. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, N.M.; Cunningham, A.J. Mild therapeutic hypothermia to improve the neurologic outcome after cardiac arrest. Surv. Anesthesiol. 2003, 47, 219–220. [Google Scholar] [CrossRef] [Green Version]
- Benson, D.W.; Williams, G.R.; Spencer, F.C.; Yates, A.J. The use of hypothermia after cardiac arrest. Anesth. Analg. 1959, 38, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Peberdy, M.A.; Callaway, C.W.; Neumar, R.W.; Geocadin, R.G.; Zimmerman, J.L.; Donnino, M.; Gabrielli, A.; Silvers, S.M.; Zaritsky, A.L.; Merchant, R.; et al. Part 9: Post-cardiac arrest care: 2010 American Heart Association guidelines for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation 2010, 122 (Suppl. 3). [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Sobel, R.A.; Cheng, D.; Steinberg, G.K.; Yenari, M.A. Mild hypothermia increases Bcl-2 protein expression following global cerebral ischemia. Mol. Brain Res. 2001, 95, 75–85. [Google Scholar] [CrossRef]
- Whalen, M.J.; Carlos, T.M.; Clark, R.S.B.; Marion, D.W.; Dekosky, S.T.; Heineman, S.; Schiding, J.K.; Memarzadeh, F.; Kochanek, P.M. The effect of brain temperature on acute inflammation after traumatic brain injury in rats. J. Neurotrauma 1997, 14, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Brodhun, M.; Fritz, H.; Walter, B.; Antonow-Schlorke, I.; Reinhart, K.; Zwiener, U.; Bauer, R.; Patt, S. Immunomorphological sequelae of severe brain injury induced by fluid-percussion in juvenile pigs—Effects of mild hypothermia. Acta Neuropathol. 2001, 101, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Truettner, J.S.; Alonso, O.F.; Bramlett, H.M.; Dietrich, W.D. Therapeutic hypothermia alters microrna responses to traumatic brain injury in rats. J. Cereb. Blood Flow Metab. 2011, 31, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Umehara, T.; Kagawa, S.; Tomida, A.; Murase, T.; Abe, Y.; Shingu, K.; Ikematsu, K. Body temperature-dependent MicroRNA expression analysis in rats: Rno-MiR-374-5p regulates apoptosis in skeletal muscle cells via Mex3B under hypothermia. Sci. Rep. 2020, 10, 15432. [Google Scholar] [CrossRef]
- Biggar, K.K.; Storey, K.B. Insight into post-transcriptional gene regulation: Stress-responsive MicroRNAs and their role in the environmental stress survival of tolerant animals. J. Exp. Biol. 2015, 218, 1281–1289. [Google Scholar] [CrossRef] [Green Version]
- Biggar, K.K.; Storey, K.B. Functional impact of MicroRNA regulation in models of extreme stress adaptation. J. Mol. Cell Biol. 2018, 10, 93–101. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Molecular biology of freezing tolerance. In Comprehensive Physiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Rubinsky, B.; Wong, S.T.; Hong, J.S.; Gilbert, J.; Roos, M.; Storey, K.B. 1H magnetic resonance imaging of freezing and thawing in freeze-tolerant frogs. Am. J. Physiol. Integr. Comp. Physiol. 1994, 266, R1771–R1777. [Google Scholar] [CrossRef]
- Singh, G.; Storey, K.B. MondoA:MLX complex regulates glucose-dependent gene expression and links to circadian rhythm in liver and brain of the freeze-tolerant wood frog, Rana sylvatica. Mol. Cell. Biochem., 2020, 473, 203–216. [Google Scholar] [CrossRef]
- Biggar, K.K.; Dubuc, A.; Storey, K. MicroRNA regulation below zero: Differential expression of MiRNA-21 and MiRNA-16 during freezing in wood frogs. Cryobiology 2009, 59, 317–321. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Storey, K.B. Micromanaging freeze tolerance: The biogenesis and regulation of neuroprotective MicroRNAs in frozen brains. Cell. Mol. Life Sci. 2018, 75, 3635–3647. [Google Scholar] [CrossRef]
- Moon, J.; Xu, L.; Giffard, R.G. Inhibition of MicroRNA-181 reduces forebrain ischemia-induced neuronal loss. J. Cereb. Blood Flow Metab. 2013, 33, 1976–1982. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.-J.; Ouyang, Y.-B.; Xiong, X.; Stary, C.M.; Giffard, R.G. Post-Stroke treatment with MiR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp. Neurol. 2015, 264, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Bansal, S.; Luu, B.E.; Storey, K.B. MicroRNA regulation in heart and skeletal muscle over the freeze–thaw cycle in the freeze tolerant wood frog. J. Comp. Physiol. B 2016, 186, 229–241. [Google Scholar] [CrossRef]
- Biggar, K.K.; Storey, K.B. Exploration of low temperature MicroRNA function in an anoxia tolerant vertebrate ectotherm, the red eared slider turtle (Trachemys scripta elegans). J. Therm. Biol. 2017, 68, 139–146. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Gabriely, G. MiR-21: A small multi-faceted rna. J. Cell. Mol. Med. 2008, 13, 39–53. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating high confidence MicroRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA Therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal MiR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated MicroRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biggar, K.K.; Storey, K.B. The emerging roles of micrornas in the molecular responses of metabolic rate depression. J. Mol. Cell Biol. 2011, 3, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, M.; Slack, F.J. Challenges identifying efficacious MiRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, G.; Storey, K.B. MicroRNA Cues from Nature: A Roadmap to Decipher and Combat Challenges in Human Health and Disease? Cells 2021, 10, 3374. https://doi.org/10.3390/cells10123374
Singh G, Storey KB. MicroRNA Cues from Nature: A Roadmap to Decipher and Combat Challenges in Human Health and Disease? Cells. 2021; 10(12):3374. https://doi.org/10.3390/cells10123374
Chicago/Turabian StyleSingh, Gurjit, and Kenneth B. Storey. 2021. "MicroRNA Cues from Nature: A Roadmap to Decipher and Combat Challenges in Human Health and Disease?" Cells 10, no. 12: 3374. https://doi.org/10.3390/cells10123374