Functional Conservation and Genetic Divergence of Chordate Glycinergic Neurotransmission: Insights from Amphioxus Glycine Transporters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Identification and Cloning of Glycine Transporter Genes from B. lanceolatum
2.3. Phylogenetic Analyses
2.4. Whole Mount In Situ Hybridization
3. Results
3.1. Identification and Phylogenetic Analysis of Amphioxus Glycine Transporter Genes
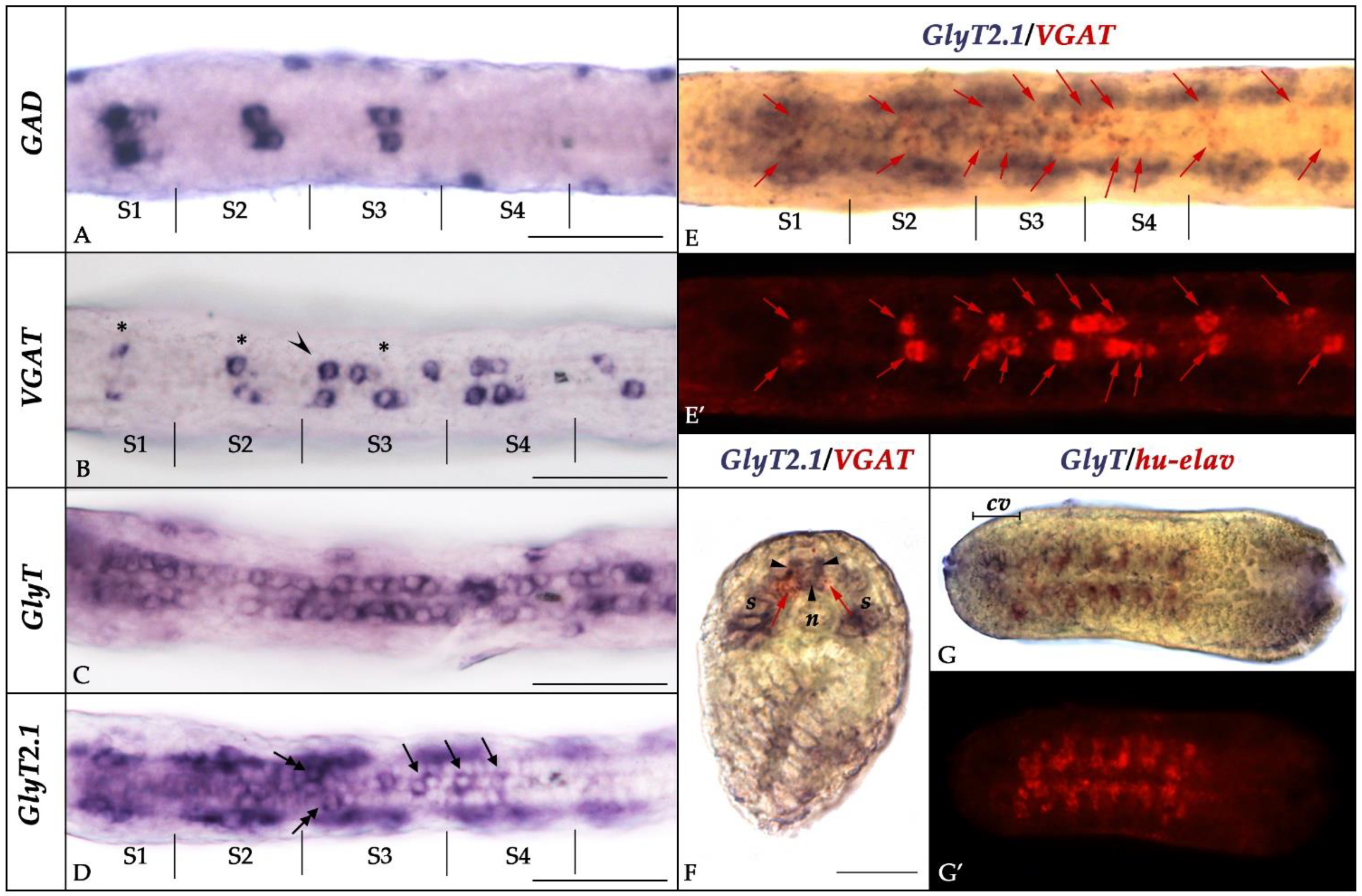
3.2. Expression of GlyT, GlyT2.1, and GlyT2.2 in Developing Amphioxus
3.3. Glycine Transporter Expression and the Neurochemistry of the Developing Amphioxus Nervous System
4. Discussion
4.1. Evolution of Glycine Neurotransmission in Metazoan Animals
4.2. Evolution of Glycine Transporters in Deuterostomes: Competing Scenarios
4.3. Amphioxus Glycine Transporter Paralogs Are Differentially Expressed in Glia and Neurons
4.4. Role of Glycine and Glycine Transporters in Larval Swimming
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coleman, W.L.; Fischl, M.J.; Weimann, S.R.; Burger, R.M. GABAergic and glycinergic inhibition modulate monaural auditory response properties in the avian superior olivary nucleus. J. Neurophysiol. 2011, 105, 2405–2420. [Google Scholar] [CrossRef]
- Wässle, H.; Koulen, P.; Brandstätter, J.H.; Fletcher, E.L.; Becker, C.M. Glycine and GABA receptors in the mammalian retina. Vis. Res. 1998, 38, 1411–1430. [Google Scholar] [CrossRef] [Green Version]
- Pow, D.V.; Hendrickson, A.E. Expression of glycine and the glycine transporter Glyt-1 in the developing rat retina. Vis. Neurosci. 2000, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Marques, B.L.; Oliveira-Lima, O.C.; Carvalho, G.A.; de Almeida Chiarelli, R.; Ribeiro, R.I.; Parreira, R.C.; da Madeira Freitas, E.M.; Resende, R.R.; Klempin, F.; Ulrich, H.; et al. Neurobiology of glycine transporters: From molecules to behavior. Neurosci. Biobehav. Rev. 2020, 118, 97–110. [Google Scholar] [CrossRef]
- Harvey, R.J.; Yee, B.K. Glycine transporters as novel therapeutic targets in schizophrenia, alcohol dependence and pain. Nat. Rev. Drug Discov. 2013, 12, 866–885. [Google Scholar] [CrossRef]
- Zafra, F.; Ibáñez, I.; Bartolomé-Martín, D.; Piniella, D.; Arribas-Blázquez, M.; Giménez, C. Glycine transporters and its coupling with NMDA receptors. Adv. Neurobiol. 2017, 16, 55–83. [Google Scholar]
- Eulenburg, V.; Armsen, W.; Betz, H.; Gomeza, J. Glycine transporters: Essential regulators of neurotransmission. Trends Biochem. Sci. 2005, 30, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Höglund, P.J.; Adzic, D.; Scicluna, S.J.; Lindblom, J.; Fredriksson, R. The repertoire of solute carriers of family 6: Identification of new human and rodent genes. Biochem. Biophys. Res. Commun. 2005, 336, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Kingsmore, S.F.; Han, H.; Yang-Feng, T.L.; Godinot, N.; Seldin, M.F.; Caron, M.G.; Giros, B. Cloning of the human glycine transporter type 1: Molecular and pharmacological characterization of novel isoform variants and chromosomal localization of the gene in the human and mouse genomes. Mol. Pharmacol. 1994, 45, 608–617. [Google Scholar]
- Liu, Q.R.; Lopez-Corcuera, B.; Mandiyan, S.; Nelson, H.; Nelson, N. Cloning and expression of a spinal cord- and brain-specific glycine transporter with novel structural features. J. Biol. Chem. 1993, 268, 22802–22808. [Google Scholar] [CrossRef]
- Gallagher, M.J.; Burgess, L.H.; Brunden, K.R. Characterization of multiple forms of the human glycine transporter type-2. Mol. Brain Res. 1999, 70, 101–115. [Google Scholar] [CrossRef]
- Raiteri, L.; Raiteri, M.; Bonanno, G. Glycine is taken up through GLYT1 and GLYT2 transporters into mouse spinal cord axon terminals and causes vesicular and carrier-mediated release of its proposed co-transmitter GABA. J. Neurochem. 2001, 76, 1823–1832. [Google Scholar] [CrossRef] [Green Version]
- Adams, R.H.; Sato, K.; Shimada, S.; Tohyama, M.; Püschel, A.W.; Betz, H. Gene structure and glial expression of the glycine transporter GlyT1 in embryonic and adult rodents. J. Neurosci. 1995, 15, 2524–2532. [Google Scholar] [CrossRef]
- Hanley, J.G.; Jones, E.M.C.; Moss, S.J. GABA receptor ρ1 subunit interacts with a novel splice variant of the glycine transporter, GLYT-1. J. Biol. Chem. 2000, 275, 840–846. [Google Scholar] [CrossRef] [Green Version]
- Borowsky, B.; Hoffman, B.J. Analysis of a gene encoding two glycine transporter variants reveals alternative promoter usage and a novel gene structure. J. Biol. Chem. 1998, 273, 29077–29085. [Google Scholar] [CrossRef] [Green Version]
- Ebihara, S.; Yamamoto, T.; Obata, K.; Yanagawa, Y. Gene structure and alternative splicing of the mouse glycine transporter type-2. Biochem. Biophys. Res. Commun. 2004, 317, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Ponce, J.; Poyatos, I.; Aragón, C.; Giménez, C.; Zafra, F. Characterization of the 5′region of the rat brain glycine transporter GLYT2 gene: Identification of a novel isoform. Neurosci. Lett. 1998, 242, 25–28. [Google Scholar] [CrossRef]
- Erdem, F.A.; Ilic, M.; Koppensteiner, P.; Gołacki, J.; Lubec, G.; Freissmuth, M.; Sandtner, W. A comparison of the transport kinetics of glycine transporter 1 and glycine transporter 2. J. Gen. Physiol. 2019, 151, 1035–1050. [Google Scholar] [CrossRef] [PubMed]
- Jursky, F.; Tamura, S.; Tamura, A.; Mandiyan, S.; Nelson, H.; Nelson, N. Structure, function and brain localization of neurotransmitter transporters. J. Exp. Biol. 1994, 196, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Zafra, F.; Gomeza, J.; Olivares, L.; Aragón, C.; Giménez, C. Regional distribution and developmental variation of the glycine transporters GLYT1 and GLYT2 in the rat CNS. Eur. J. Neurosci. 1995, 7, 1342–1352. [Google Scholar] [CrossRef]
- Cubelos, B.; Giménez, C.; Zafra, F. Localization of the GLYT1 glycine transporter at glutamatergic synapses in the rat brain. Cereb. Cortex 2005, 15, 448–459. [Google Scholar] [CrossRef]
- Gomeza, J.; Hülsmann, S.; Ohno, K.; Eulenburg, V.; Szöke, K.; Richter, D.; Betz, H. Inactivation of the glycine transporter 1 gene discloses vital role of glial glycine uptake in glycinergic inhibition. Neuron 2003, 40, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Gomeza, J.; Ohno, K.; Hülsmann, S.; Armsen, W.; Eulenburg, V.; Richter, D.W.; Laube, B.; Betz, H. Deletion of the mouse glycine transporter 2 results in a hyperekplexia phenotype and postnatal lethality. Neuron 2003, 40, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Sawada, M.; McAdoo, D.J.; Blankenship, J.E.; Price, C.H. Modulation of arterial muscle contraction in Aplysia by glycine and neuron R14. Brain Res. 1981, 207, 486–490. [Google Scholar] [CrossRef]
- Sawada, M.; McAdoo, D.J.; Ichinose, M.; Price, C.H. Influences of glycine and neuron R14 on contraction of the anterior aorta of Aplysia. Jpn. J. Physiol. 1984, 34, 747–767. [Google Scholar] [CrossRef] [Green Version]
- Frenkel, L.; Muraro, N.I.; Beltrán González, A.N.; Marcora, M.S.; Bernabó, G.; Hermann-Luibl, C.; Romero, J.I.; Helfrich-Förster, C.; Castaño, E.M.; Marino-Busjle, C.; et al. Organization of circadian behavior relies on glycinergic transmission. Cell Rep. 2017, 19, 72–85. [Google Scholar] [CrossRef] [Green Version]
- Nishino, A.; Okamura, Y.; Piscopo, S.; Brown, E.R. A glycine receptor is involved in the organization of swimming movements in an invertebrate chordate. BMC Neurosci. 2010, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Aubrey, K.R.; Rossi, F.M.; Ruivo, R.; Alboni, S.; Bellenchi, G.C.; Le Goff, A.; Gasnier, B.; Supplisson, S. The transporters GlyT2 and VIAAT cooperate to determine the vesicular glycinergic phenotype. J. Neurosci. 2007, 27, 6273–6281. [Google Scholar] [CrossRef] [PubMed]
- Pierobon, P.; Minei, R.; Porcu, P.; Sogliano, C.; Tino, A.; Marino, G.; Biggio, G.; Concas, A. Putative glycine receptors in Hydra: A biochemical and behavioural study. Eur. J. Neurosci. 2001, 14, 1659–1666. [Google Scholar] [CrossRef] [PubMed]
- Thimgan, M.S.; Berg, J.S.; Stuart, A.E. Comparative sequence analysis and tissue localization of members of the SLC6 family of transporters in adult Drosophila melanogaster. J. Exp. Biol. 2006, 209, 3383–3404. [Google Scholar] [CrossRef] [Green Version]
- Shpak, M.; Gentil, L.G.; Miranda, M. The origin and evolution of vertebrate glycine transporters. J. Mol. Evol. 2014, 78, 188–193. [Google Scholar] [CrossRef]
- Holland, L.Z.; Holland, N.D. Cephalochordates: A window into vertebrate origins. Curr. Top. Dev. Biol. 2021, 141, 119–147. [Google Scholar] [PubMed]
- Wicht, H.; Lacalli, T.C. The nervous system of amphioxus: Structure, development, and evolutionary significance. Can. J. Zool. 2005, 83, 122–150. [Google Scholar] [CrossRef]
- Albuixech-Crespo, B.; Lo, L.; Moreno-Bravo, J.A.; Maeso, I.; Sa, L.; Somorjai, I.; Pascual-Anaya, J.; Puelles, E.; Bovolenta, P. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS Biol. 2017, 15, e2001573. [Google Scholar] [CrossRef] [Green Version]
- Candiani, S.; Moronti, L.; Ramoino, P.; Schubert, M.; Pestarino, M. A neurochemical map of the developing amphioxus nervous system. BMC Neurosci. 2012, 13, 59. [Google Scholar] [CrossRef] [Green Version]
- Lacalli, T.; Candiani, S. Locomotory control in amphioxus larvae: New insights from neurotransmitter data. Evodevo 2017, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bozzo, M.; Lacalli, T.C.; Obino, V.; Caicci, F.; Marcenaro, E.; Bachetti, T.; Manni, L.; Pestarino, M.; Schubert, M.; Candiani, S. Amphioxus neuroglia: Molecular characterization and evidence for early compartmentalization of the developing nerve cord. Glia 2021, 69, 1654–1678. [Google Scholar] [CrossRef]
- Telford, M.J.; Budd, G.E.; Philippe, H. Phylogenomic insights into animal evolution. Curr. Biol. 2015, 25, R876–R887. [Google Scholar] [CrossRef] [Green Version]
- Marlétaz, F.; Peijnenburg, K.T.C.A.; Goto, T.; Satoh, N.; Rokhsar, D.S. A new spiralian phylogeny places the enigmatic arrow worms among gnathiferans. Curr. Biol. 2019, 29, 312–318.e3. [Google Scholar] [CrossRef] [Green Version]
- Cannon, J.T.; Vellutini, B.C.; Smith, J.; Ronquist, F.; Jondelius, U.; Hejnol, A. Xenacoelomorpha is the sister group to Nephrozoa. Nature 2016, 530, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Rouse, G.W.; Wilson, N.G.; Carvajal, J.I.; Vrijenhoek, R.C. New deep-sea species of Xenoturbella and the position of Xenacoelomorpha. Nature 2016, 530, 94–97. [Google Scholar] [CrossRef]
- Philippe, H.; Poustka, A.J.; Chiodin, M.; Hoff, K.J.; Dessimoz, C.; Tomiczek, B.; Schiffer, P.H.; Müller, S.; Domman, D.; Horn, M.; et al. Mitigating anticipated effects of systematic errors supports sister-group relationship between Xenacoelomorpha and Ambulacraria. Curr. Biol. 2019, 29, 1818–1826.e6. [Google Scholar] [CrossRef] [Green Version]
- Kapli, P.; Natsidis, P.; Leite, D.J.; Fursman, M.; Jeffrie, N.; Rahman, I.A.; Philippe, H.; Copley, R.R.; Telford, M.J. Lack of support for Deuterostomia prompts reinterpretation of the first Bilateria. Sci. Adv. 2021, 7, eabe2741. [Google Scholar] [CrossRef]
- Simion, P.; Philippe, H.; Baurain, D.; Jager, M.; Richter, D.J.; Di Franco, A.; Roure, B.; Satoh, N.; Quéinnec, E.; Ereskovsky, A.; et al. A Large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 2017, 27, 958–967. [Google Scholar] [CrossRef] [Green Version]
- Whelan, N.V.; Kocot, K.M.; Moroz, T.P.; Mukherjee, K.; Williams, P.; Paulay, G.; Moroz, L.L.; Halanych, K.M. Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 2017, 1, 1737–1746. [Google Scholar] [CrossRef] [Green Version]
- Kapli, P.; Telford, M.J. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 2020, 6, eabc5162. [Google Scholar] [CrossRef]
- Bozzo, M.; Candiani, S.; Schubert, M. Whole mount in situ hybridization and immunohistochemistry for studying retinoic acid signaling in developing amphioxus. Methods Enzymol. 2020, 637, 419–452. [Google Scholar]
- Fuentes, M.; Schubert, M.; Dalfo, D.; Candiani, S.; Benito, E.; Gardenyes, J.; Godoy, L.; Moret, F.; Illas, M.; Patten, I.; et al. Preliminary observations on the spawning conditions of the European amphioxus (Branchiostoma lanceolatum) in captivity. J. Exp. Zool. Part B Mol. Dev. Evol. 2004, 302, 384–391. [Google Scholar] [CrossRef]
- Carvalho, J.E.; Lahaye, F.; Yong, L.W.; Croce, J.C.; Escrivá, H.; Yu, J.-K.; Schubert, M. An updated staging system for cephalochordate development: One table suits them all. Front. Cell Dev. Biol. 2021, 9, 668006. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [Green Version]
- Bi, C.; Lu, N.; Han, T.; Huang, Z.; Chen, J.Y.; He, C.; Lu, Z. Whole-genome resequencing of twenty Branchiostoma belcheri individuals provides a brand-new variant dataset for Branchiostoma. BioMed Res. Int. 2020, 2020, 3697342. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. 1), 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zieger, E.; Garbarino, G.; Robert, N.S.M.; Yu, J.K.; Croce, J.C.; Candiani, S.; Schubert, M. Retinoic acid signaling and neurogenic niche regulation in the developing peripheral nervous system of the cephalochordate amphioxus. Cell. Mol. Life Sci. 2018, 75, 2407–2429. [Google Scholar] [CrossRef]
- Zieger, E.; Candiani, S.; Garbarino, G.; Croce, J.C.; Schubert, M. Roles of retinoic acid signaling in shaping the neuronal architecture of the developing amphioxus nervous system. Mol. Neurobiol. 2018, 55, 5210–5229. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 12, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Chevenet, F.; Brun, C.; Bañuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 493. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Candiani, S.; Garbarino, G.; Pestarino, M. Detection of mRNA and microRNA expression in basal chordates, amphioxus, and ascidians. In In Situ Hybridization Methods; Hauptmann, G., Ed.; Humana Press: New York, NY, USA, 2015; Volume 99, pp. 279–292. [Google Scholar]
- Bozzo, M.; Pergner, J.; Kozmik, Z.; Kozmikova, I. Novel polyclonal antibodies as a useful tool for expression studies in amphioxus embryos. Int. J. Dev. Biol. 2017, 61, 793–800. [Google Scholar] [CrossRef] [Green Version]
- Pergner, J.; Vavrova, A.; Kozmikova, I.; Kozmik, Z. Molecular fingerprint of amphioxus frontal eye illuminates the evolution of homologous cell types in the chordate retina. Front. Cell Dev. Biol. 2020, 8, 705. [Google Scholar] [CrossRef]
- Satoh, G.; Wang, Y.; Zhang, P.; Satoh, N. Early development of amphioxus nervous system with special reference to segmental cell organization and putative sensory cell precursors: A study based on the expression of pan-neuronal marker gene Hu/elav. J. Exp. Zool. 2001, 291, 354–364. [Google Scholar] [CrossRef]
- Villar-Cerviño, V.; Barreiro-Iglesias, A.; Anadón, R.; Rodicio, M.C. Distribution of glycine immunoreactivity in the brain of adult sea lamprey (Petromyzon marinus). Comparison with γ-aminobutyric acid. J. Comp. Neurol. 2008, 507, 1441–1463. [Google Scholar] [CrossRef]
- Imboden, M.; Devignot, V.; Korn, H.; Goblet, C. Regional distribution of glycine receptor messenger RNA in the central nervous system of zebrafish. Neuroscience 2001, 103, 811–830. [Google Scholar] [CrossRef]
- Berki, A.C.; O’Donovan, M.J.; Antal, M. Developmental expression of glycine immunoreactivity and its colocalization with GABA in the embryonic chick lumbosacral spinal cord. J. Comp. Neurol. 1995, 362, 583–596. [Google Scholar] [CrossRef]
- Vitanova, L. Immunocytochemical study of glycine receptors in the retina of the frog Xenopus laevis. Anat. Embryol. 2006, 211, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Anadón, R.; Rodríguez-Moldes, I.; Adrio, F. Glycine-immunoreactive neurons in the brain of a shark (Scyliorhinus canicula L.). J. Comp. Neurol. 2013, 521, 3057–3082. [Google Scholar] [CrossRef]
- Sakima, A.; Yamazato, M.; Sesoko, S.; Muratani, H.; Fukiyama, K. Cardiovascular and sympathetic effects of L-glutamate and glycine injected into the rostral ventrolateral medulla of conscious rats. Hypertens. Res. 2000, 23, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Barreiro-Iglesias, A.; Mysiak, K.S.; Adrio, F.; Rodicio, M.C.; Becker, C.G.; Becker, T.; Anadón, R. Distribution of glycinergic neurons in the brain of glycine transporter-2 transgenic Tg(glyt2:Gfp) adult zebrafish: Relationship to brain–spinal descending systems. J. Comp. Neurol. 2013, 521, 389–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yáñez, J.; Busch, J.; Anadón, R.; Meissl, H. Pineal projections in the zebrafish (Danio rerio): Overlap with retinal and cerebellar projections. Neuroscience 2009, 164, 1712–1720. [Google Scholar] [CrossRef]
- Moly, P.K.; Ikenaga, T.; Kamihagi, C.; Islam, A.F.M.T.; Hatta, K. Identification of initially appearing glycine-immunoreactive neurons in the embryonic zebrafish brain. Dev. Neurobiol. 2014, 74, 616–632. [Google Scholar] [CrossRef]
- Redecker, P.; Pabst, H.; Löscher, W.; Steinlechner, S. Evidence for microvesicular storage and release of glycine in rodent pinealocytes. Neurosci. Lett. 2001, 299, 93–96. [Google Scholar] [CrossRef]
- Sato, K.; Kiyama, H.; Shimada, S.; Tohyama, M. Gene expression of KA type and NMDA receptors and of a glycine transporter in the rat pineal gland. Neuroendocrinology 1993, 58, 77–79. [Google Scholar] [CrossRef]
- Villar-Cerviño, V.; Barreiro-Iglesias, A.; Anadón, R.; Rodicio, M.C. Development of glycine immunoreactivity in the brain of the sea lamprey: Comparison with γ-aminobutyric acid immunoreactivity. J. Comp. Neurol. 2009, 512, 747–767. [Google Scholar] [CrossRef]
- Adrio, F.; Rodriguez-Moldes, I.; Anadon, R. Distribution of glycine immunoreactivity in the brain of the Siberian sturgeon (Acipenser baeri): Comparison with γ-aminobutyric acid. J. Comp. Neurol. 2011, 519, 1115–1142. [Google Scholar] [CrossRef]
- Lacalli, T.C.; Holland, N.D.; West, J.E. Landmarks in the anterior central nervous system of amphioxus larvae. Philos. Trans. R. Soc. B Biol. Sci. 1994, 344, 165–185. [Google Scholar]
- Wickstead, J.H.; Bone, Q. Ecology of acraniate larvæ. Nature 1959, 184, 1849–1851. [Google Scholar] [CrossRef]
- Pergner, J.; Kozmik, Z. Amphioxus photoreceptors—Insights into the evolution of vertebrate opsins, vision, and circadian rhythmicity. Int. J. Dev. Biol. 2017, 61, 665–681. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, S.; Anadón, R. The fine structure of lamellate cells in the brain of amphioxus (Branchiostoma lanceolatum, Cephalochordata). Cell Tissue Res. 1991, 263, 597–600. [Google Scholar] [CrossRef]
- Glardon, S.; Holland, L.Z.; Gehring, W.J.; Holland, N.D. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): Insights into eye and photoreceptor evolution. Development 1998, 125, 2701–2710. [Google Scholar] [CrossRef]
- Bozzo, M.; Macrì, S.; Calzia, D.; Sgarra, R.; Manfioletti, G.; Ramoino, P.; Lacalli, T.; Vignali, R.; Pestarino, M.; Candiani, S. The HMGA gene family in chordates: Evolutionary perspectives from amphioxus. Dev. Genes Evol. 2017, 227, 201–211. [Google Scholar] [CrossRef]
- Hobert, O. The neuronal genome of Caenorhabditis elegans. In WormBook; The C. elegans Research Community, Ed.; WormBase: Pasadena, CA, USA, 2013; pp. 1–106. [Google Scholar]
- Gendrel, M.; Atlas, E.G.; Hobert, O. A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife 2016, 5, e17686. [Google Scholar] [CrossRef]
- Serrano-Saiz, E.; Pereira, L.; Gendrel, M.; Aghayeva, U.; Battacharya, A.; Howell, K.; Garcia, L.R.; Hobert, O. A neurotransmitter atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics 2017, 206, 1251–1269. [Google Scholar] [CrossRef] [Green Version]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef] [Green Version]
- Wester, M.R.; Teasley, D.C.; Byers, S.L.; Saha, M.S. Expression patterns of glycine transporters (xGlyT1, xGlyT2, and xVIAAT) in Xenopus laevis during early development. Gene Expr. Patterns 2008, 8, 261–270. [Google Scholar] [CrossRef]
- Cui, W.W.; Low, S.E.; Hirata, H.; Saint-Amant, L.; Geisler, R.; Hume, R.I.; Kuwada, J.Y. The zebrafish shocked gene encodes a glycine transporter and is essential for the function of early neural circuits in the CNS. J. Neurosci. 2005, 25, 6610–6620. [Google Scholar] [CrossRef] [Green Version]
- Raiteri, L.; Raiteri, M. Functional “glial” GLYT1 glycine transporters expressed in neurons. J. Neurochem. 2010, 114, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Jursky, F.; Nelson, N. Developmental expression of the glycine transporters GLYT1 and GLYT2 in mouse brain. J. Neurochem. 1996, 67, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.H. Hyperekplexia. In The Curated Reference Collection in Neuroscience and Biobehavioral Psychology; Stein, J., Bennett, D., Coen, C., Dunbar, R., Goodwin, G., Husain, M., Mann, E., Morris, J., Rolls, E., Taylor, J.S.H., et al., Eds.; Elsevier Science Ltd.: London, UK, 2016; pp. 52–56. [Google Scholar]
- Hirata, H.; Carta, E.; Yamanaka, I.; Harvey, R.J.; Kuwada, J.Y. Defective glycinergic synaptic transmission in zebrafish motility mutants. Front. Mol. Neurosci. 2010, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grillner, S. The motor infrastructure: From ion channels to neuronal networks. Nat. Rev. Neurosci. 2003, 4, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.; Lu, Z.; Meinertzhagen, I.A. Circuit homology between decussating pathways in the Ciona larval CNS and the vertebrate startle-response pathway. Curr. Biol. 2017, 27, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Grillner, S.; Ekeberg, O.; El Manira, A.; Lansner, A.; Parker, D.; Tegnér, J.; Wallén, P. Intrinsic function of a neuronal network—A vertebrate central pattern generator. Brain Res. Rev. 1998, 26, 184–197. [Google Scholar] [CrossRef]
- Goulding, M. Circuits controlling vertebrate locomotion: Moving in a new direction. Nat. Rev. Neurosci. 2009, 10, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.H.; Harris-Warrick, R.M. Strychnine eliminates alternating motor output during fictive locomotion in the lamprey. Brain Res. 1984, 293, 164–167. [Google Scholar] [CrossRef]
- Ryan, K.; Lu, Z.; Meinertzhagen, I.A. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 2016, 5, e16962. [Google Scholar] [CrossRef] [PubMed]
- Lacalli, T.C.; Kelly, S.J. Ventral neurons in the anterior nerve cord of amphioxus larvae. I. An inventory of cell types and synaptic patterns. J. Morphol. 2003, 257, 190–211. [Google Scholar] [CrossRef]
- Lacalli, T.C. Ventral neurons in the anterior nerve cord of amphioxus larvae. II. Further data on the pacemaker circuit. J. Morphol. 2003, 257, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Lacalli, T.C.; Kelly, S.J. Somatic motoneurones in amphioxus larvae: Cell types, cell position, and innervation patterns. Acta Zool. 1999, 80, 113–124. [Google Scholar] [CrossRef]
- Lacalli, T.C. Sensory systems in amphioxus: A window on the ancestral chordate condition. Brain. Behav. Evol. 2004, 64, 148–162. [Google Scholar] [CrossRef] [PubMed]







| Name | Species | Genomic Position | GenBank Accession Number | Transcriptome/EST Data |
|---|---|---|---|---|
| GlyT | B. belcheri | Unplaced scaffold | XP_019646809.1 (isoform X1) XP_019646811.1 (isoform X2) XP_019646812.1 (isoform X3) | n.a. |
| B. floridae* | Chr 7:197986 | XP_035682443 | GETA01030290.1 | |
| B. lanceolatum | scf42:456448 | n.a. | JT884165.1 | |
| GlyT2.1 | B. belcheri | Unplaced scaffold | XP_019644968.1 (isoform X1) XP_019644969.1 (isoform X2) | n.a. |
| B. floridae* | Chr 12: 3077852 | XP_035694152.1 (isoform X1) XP_035694153.1 (isoform X2) XP_035694154.1 (isoform X3) | n.a. | |
| B. lanceolatum | scf11:2848268 | n.a. | JT866905.1 JT890693.1 | |
| B. belcheri | Unplaced scaffold | XP_019644970.1 (isoform X1) XP_019644972.1 (isoform X2) XP_019644973.1 (isoform X3) | n.a. | |
| GlyT2.2 | B. floridae | Chr 12: 2983693 | XP_035694157.1 | FE595048 |
| B. lanceolatum | scf11:1893629 | n.a. | JT855933.1 JT862870.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bozzo, M.; Costa, S.; Obino, V.; Bachetti, T.; Marcenaro, E.; Pestarino, M.; Schubert, M.; Candiani, S. Functional Conservation and Genetic Divergence of Chordate Glycinergic Neurotransmission: Insights from Amphioxus Glycine Transporters. Cells 2021, 10, 3392. https://doi.org/10.3390/cells10123392
Bozzo M, Costa S, Obino V, Bachetti T, Marcenaro E, Pestarino M, Schubert M, Candiani S. Functional Conservation and Genetic Divergence of Chordate Glycinergic Neurotransmission: Insights from Amphioxus Glycine Transporters. Cells. 2021; 10(12):3392. https://doi.org/10.3390/cells10123392
Chicago/Turabian StyleBozzo, Matteo, Simone Costa, Valentina Obino, Tiziana Bachetti, Emanuela Marcenaro, Mario Pestarino, Michael Schubert, and Simona Candiani. 2021. "Functional Conservation and Genetic Divergence of Chordate Glycinergic Neurotransmission: Insights from Amphioxus Glycine Transporters" Cells 10, no. 12: 3392. https://doi.org/10.3390/cells10123392








