The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks
Abstract
:1. Introduction
2. Replisome Assembly and DNA Replication
Nucleosomes and Chromatin Remodeling
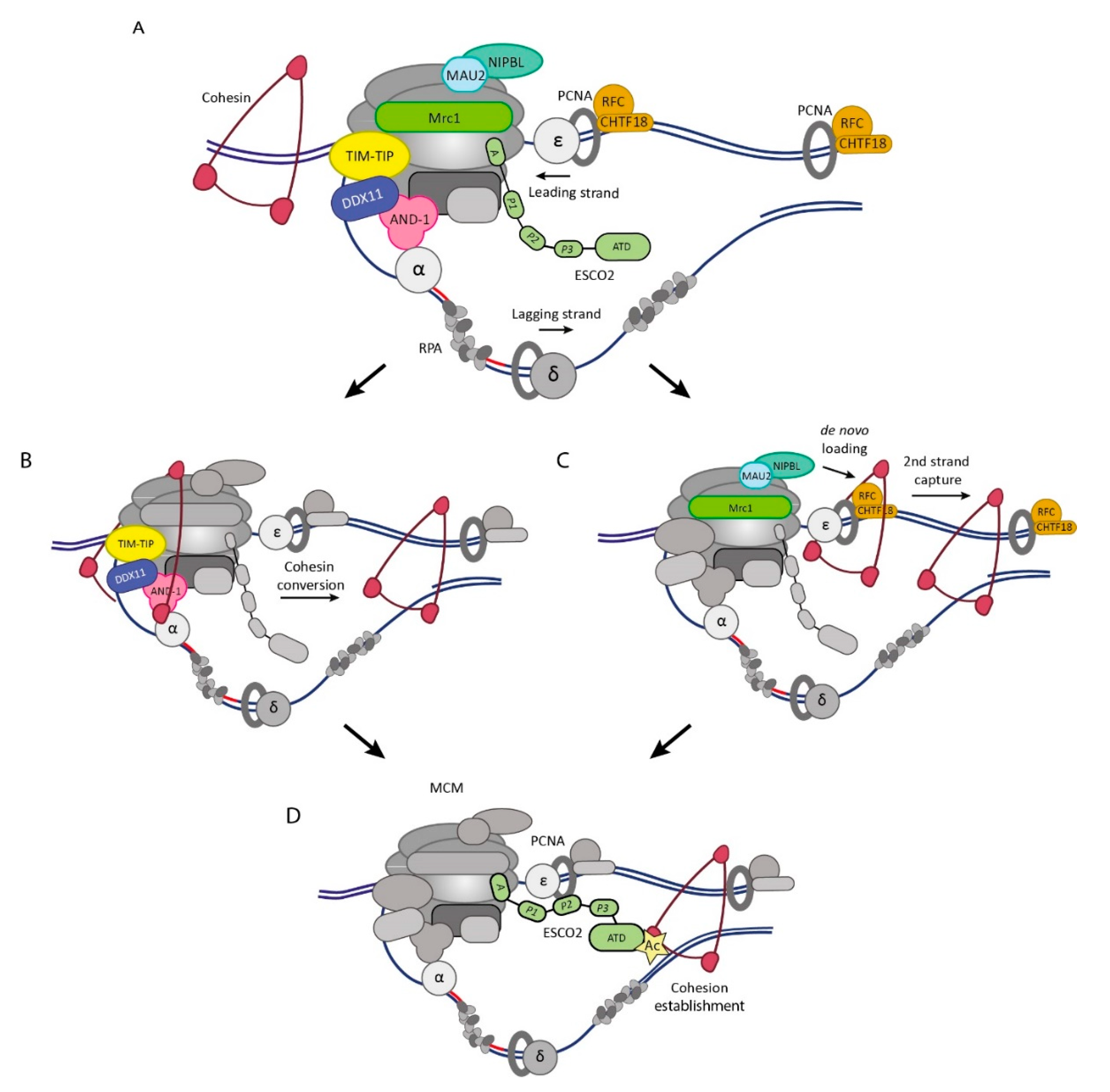
3. Sister DNA Entrapment at Processive Replication Forks
3.1. Conversion Pathway
3.2. De Novo Pathway
4. Establishment of Sister Chromatid Cohesion
4.1. SMC3 Acetyltransferases
4.2. Interactions of Eco1/ESCO2 with the Replisome
4.3. SORORIN
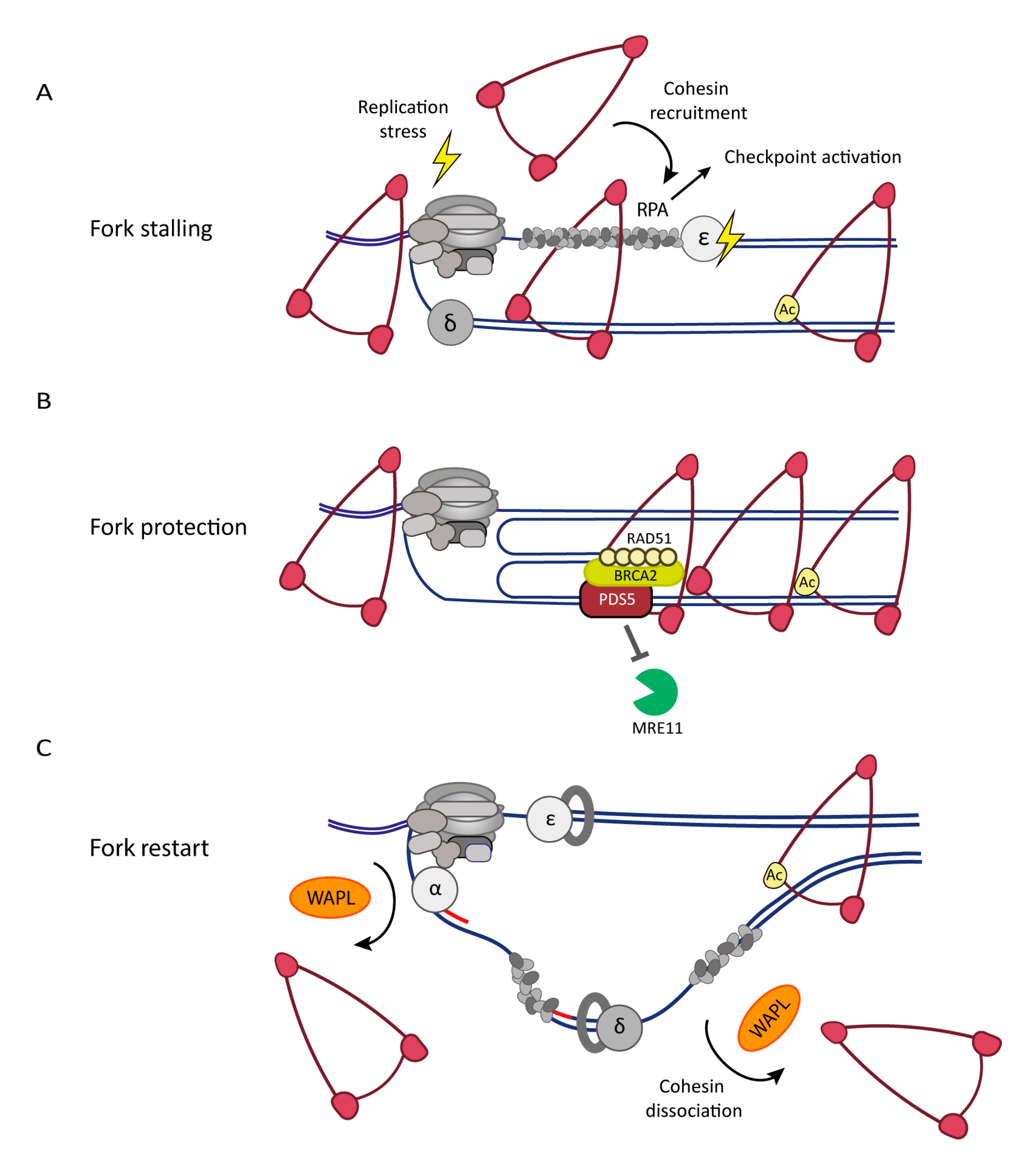
5. The Cohesin Complex and Associated Factors in the DNA Replication Stress Response
5.1. Cohesin Dynamics and Replication Stress
5.2. DNA Replication Stress Affects Sister Chromatid Cohesion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Haarhuis, J.H.; Elbatsh, A.M.; Rowland, B.D. Cohesin and Its Regulation: On the Logic of X-Shaped Chromosomes. Dev. Cell 2014, 31, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, M.; Scheinost, J.C.; Petela, N.J.; Gligoris, T.G.; Wissler, M.; Ogushi, S.; Collier, J.E.; Voulgaris, M.; Kurze, A.; Chan, K.-L.; et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-topological as well as Topological Mechanisms. Cell 2018, 173, 1508–1519.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, S.; Minamino, M.; Casas-Delucchi, C.S.; Patel, H.; Uhlmann, F. A Role for Chromatin Remodeling in Cohesin Loading onto Chromosomes. Mol. Cell 2019, 74, 664–673.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakimi, M.-A.; Bochar, D.A.; Schmiesing, J.A.; Dong, Y.; Barak, O.G.; Speicher, D.W.; Yokomori, K.; Shiekhattar, R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 2002, 418, 994–998. [Google Scholar] [CrossRef]
- Garcia-Luis, J.; Lazar-Stefanita, L.; Gutierrez-Escribano, P.; Thierry, A.; Cournac, A.; García, A.; González, S.; Sánchez, M.; Jarmuz, A.; Montoya, A.; et al. FACT mediates cohesin function on chromatin. Nat. Struct. Mol. Biol. 2019, 26, 970–979. [Google Scholar] [CrossRef]
- Lopez-Serra, L.; Kelly, G.; Patel, H.; Stewart, A.; Uhlmann, F. The Scc2–Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014, 46, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busslinger, G.; Stocsits, R.R.; van der Lelij, P.; Axelsson, E.; Tedeschi, A.; Galjart, N.; Peters, J.-M. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 2017, 544, 503–507. [Google Scholar] [CrossRef]
- Heinz, S.; Texari, L.; Hayes, M.G.; Urbanowski, M.; Chang, M.; Givarkes, N.; Rialdi, A.; White, K.M.; Albrecht, R.A.; Pache, L.; et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018, 174, 1522–1536.e22. [Google Scholar] [CrossRef] [Green Version]
- Lengronne, A.; Katou, Y.; Mori, S.; Yokobayashi, S.; Kelly, G.; Itoh, T.; Watanabe, Y.; Shirahige, K.; Uhlmann, F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 2004, 430, 573–578. [Google Scholar] [CrossRef]
- Davidson, I.F.; Goetz, D.; Zaczek, M.P.; Molodtsov, M.; Veld, P.J.H.I.; Weissmann, F.; Litos, G.; Cisneros, D.; Ocampo-Hafalla, M.; Ladurner, R.; et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 2016, 35, 2671–2685. [Google Scholar] [CrossRef]
- Stigler, J.; Çamdere, G.; Koshland, D.E.; Greene, E.C. Single-Molecule Imaging Reveals a Collapsed Conformational State for DNA-Bound Cohesin. Cell Rep. 2016, 15, 988–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dequeker, B.J.H.; Brandão, H.B.; Scherr, M.J.; Gassler, J.; Powell, S.; Gaspar, I.; Flyamer, I.M.; Tang, W.; Stocsits, R.; Davidson, I.F.; et al. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kueng, S.; Hegemann, B.; Peters, B.H.; Lipp, J.J.; Schleiffer, A.; Mechtler, K.; Peters, J.-M. Wapl Controls the Dynamic Association of Cohesin with Chromatin. Cell 2006, 127, 955–967. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, R.; Gillespie, P.J.; Hirano, T. Human Wapl Is a Cohesin-Binding Protein that Promotes Sister-Chromatid Resolution in Mitotic Prophase. Curr. Biol. 2006, 16, 2406–2417. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, T.; Ladurner, R.; Schmitz, J.; Kreidl, E.; Schleiffer, A.; Bhaskara, V.; Bando, M.; Shirahige, K.; Hyman, A.A.; Mechtler, K.; et al. Sororin Mediates Sister Chromatid Cohesion by Antagonizing Wapl. Cell 2010, 143, 737–749. [Google Scholar] [CrossRef] [Green Version]
- Gerlich, D.W.; Koch, B.; Dupeux, F.; Peters, J.-M.; Ellenberg, J. Live-Cell Imaging Reveals a Stable Cohesin-Chromatin Interaction after but Not before DNA Replication. Curr. Biol. 2006, 16, 1571–1578. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.S.; Pustova, I.; Cattoglio, C.; Tjian, R.; Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 2017, 6, e25776. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Peters, J.-M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 445–464. [Google Scholar] [CrossRef]
- Litwin, I.; Pilarczyk, E.; Wysocki, R. The Emerging Role of Cohesin in the DNA Damage Response. Genes 2018, 9, 581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted Loading of Mcm2–7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, T.; Locke, J.; Greiwe, J.F.; Diffley, J.F.X.; Costa, A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 2019, 575, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Fragkos, M.; Ganier, O.; Coulombe, P.; Méchali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef]
- Masai, H.; Matsumoto, S.; You, Z.; Yoshizawa-Sugata, N.; Oda, M. Eukaryotic Chromosome DNA Replication: Where, When, and How? Annu. Rev. Biochem. 2010, 79, 89–130. [Google Scholar] [CrossRef]
- Fu, Y.; Yardimci, H.; Long, D.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Schärer, O.D.; Walter, J.C. Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Ali, F.A.; Renault, L.; Gannon, J.; Gahlon, H.; Kotecha, A.; Zhou, J.C.; Rueda, D.; Costa, A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun. 2016, 7, 10708. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Shi, Y.; Georgescu, R.E.; Yuan, Z.; Chait, B.T.; Li, H.; O’Donnell, M. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015, 22, 976–982. [Google Scholar] [CrossRef] [Green Version]
- Goswami, P.; Ali, F.A.; Douglas, M.E.; Locke, J.; Purkiss, A.; Janska, A.; Eickhoff, P.; Early, A.; Nans, A.; Cheung, A.M.C.; et al. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Georgescu, R.; Schauer, G.D.; O’Donnell, M.E.; Li, H. Structure of the polymerase ε holoenzyme and atomic model of the leading strand replisome. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Choe, K.N.; Moldovan, G.-L. Forging Ahead through Darkness: PCNA, Still the Principal Conductor at the Replication Fork. Mol. Cell 2017, 65, 380–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, A.C.; Zhou, J.C.; Perera, R.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinković, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.-H.; Farina, A.; Bermudez, V.P.; Tappin, I.; Du, F.; Galal, W.C.; Hurwitz, J. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc. Natl. Acad. Sci. USA 2013, 110, 19760–19765. [Google Scholar] [CrossRef] [Green Version]
- Kilkenny, M.L.; Simon, A.C.; Mainwaring, J.; Wirthensohn, D.; Holzer, S.; Pellegrini, L. The human CTF4-orthologue AND-1 interacts with DNA polymerase α/primase via its unique C-terminal HMG box. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Lancey, C.; Tehseen, M.; Raducanu, V.-S.; Rashid, F.; Merino, N.; Ragan, T.J.; Savva, C.G.; Zaher, M.S.; Shirbini, A.; Blanco, F.J.; et al. Structure of the processive human Pol δ holoenzyme. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Stodola, J.L.; Burgers, P.M. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat. Struct. Mol. Biol. 2016, 23, 402–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, L.; Bambara, R.A. Okazaki Fragment Metabolism. Cold Spring Harb. Perspect. Biol. 2013, 5, a010173. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Kanchwala, M.; Xing, C.; Yu, H. MCM2–7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. eLife 2018, 7. [Google Scholar] [CrossRef]
- Guillou, E.; Ibarra, A.; Coulon, V.; Casado-Vela, J.; Rico, D.; Casal, I.; Schwob, E.; Losada, A.; Méndez, J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010, 24, 2812–2822. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.S.; Basu, A.; Bermudez, V.; Hurwitz, J.; Walter, J.C. Cdc7–Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008, 22, 1894–1905. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.S.; Yiu, P.; Chou, M.F.; Gygi, S.; Walter, J.C. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nature 2004, 6, 991–996. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Hirano, T. Scc2 Couples Replication Licensing to Sister Chromatid Cohesion in Xenopus Egg Extracts. Curr. Biol. 2004, 14, 1598–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlmann, F.; Nasmyth, K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998, 8, 1095–1102. [Google Scholar] [CrossRef] [Green Version]
- MacAlpine, H.K.; Gordân, R.; Powell, S.K.; Hartemink, A.J.; MacAlpine, D.M. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2009, 20, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, M.P.; Ladurner, R.; Poser, I.; Beveridge, R.; Rampler, E.; Hudecz, O.; Novatchkova, M.; Hériché, J.; Wutz, G.; van der Lelij, P.; et al. The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Minamino, M.; Tei, S.; Negishi, L.; Kanemaki, M.; Yoshimura, A.; Sutani, T.; Bando, M.; Shirahige, K. Temporal Regulation of ESCO2 Degradation by the MCM Complex, the CUL4-DDB1-VPRBP Complex, and the Anaphase-Promoting Complex. Curr. Biol. 2018, 28, 2665–2672.e5. [Google Scholar] [CrossRef]
- Cucco, F.; Palumbo, E.; Camerini, S.; D’Alessio, B.; Quarantotti, V.; Casella, M.L.; Rizzo, I.M.; Cukrov, D.; Delia, M.; Russo, A.; et al. Separase prevents genomic instability by controlling replication fork speed. Nucleic Acids Res. 2017, 46, 267–278. [Google Scholar] [CrossRef] [Green Version]
- Tittel-Elmer, M.; Lengronne, A.; Davidson, M.B.; Bacal, J.; François, P.; Hohl, M.; Petrini, J.; Pasero, P.; Cobb, J.A. Cohesin Association to Replication Sites Depends on Rad50 and Promotes Fork Restart. Mol. Cell 2012, 48, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Baetz, K.K.; Krogan, N.J.; Emili, A.; Greenblatt, J.; Hieter, P. The ctf13-30/CTF13 Genomic Haploinsufficiency Modifier Screen Identifies the Yeast Chromatin Remodeling Complex RSC, which Is Required for the Establishment of Sister Chromatid Cohesion. Mol. Cell. Biol. 2004, 24, 1232–1244. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Hsu, J.-M.; Laurent, B.C. The RSC Nucleosome-Remodeling Complex Is Required for Cohesin’s Association with Chromosome Arms. Mol. Cell 2004, 13, 739–750. [Google Scholar] [CrossRef]
- Brownlee, P.M.; Chambers, A.; Cloney, R.; Bianchi, A.; Downs, J.A. BAF180 Promotes Cohesion and Prevents Genome Instability and Aneuploidy. Cell Rep. 2014, 6, 973–981. [Google Scholar] [CrossRef] [Green Version]
- Devaiah, B.N.; Case-Borden, C.; Gegonne, A.; Hsu, C.H.; Chen, Q.; Meerzaman, D.; Dey, A.; Ozato, K.; Singer, D.S. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016, 23, 540–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olley, G.; Ansari, M.; Bengani, H.; Grimes, G.R.; Rhodes, J.; von Kriegsheim, A.; Blatnik, A.; Stewart, F.J.; Wakeling, E.; Carroll, N.; et al. BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange–like syndrome. Nat. Genet. 2018, 50, 329–332. [Google Scholar] [CrossRef]
- Boginya, A.; Detroja, R.; Matityahu, A.; Frenkel-Morgenstern, M.; Onn, I. The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungrawala, H.; Rose, K.L.; Bhat, K.; Mohni, K.N.; Glick, G.G.; Couch, F.B.; Cortez, D. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol. Cell 2015, 59, 998–1010. [Google Scholar] [CrossRef] [Green Version]
- MacAlpine, D.M.; Almouzni, G. Chromatin and DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010207. [Google Scholar] [CrossRef]
- Kurat, C.; Yeeles, J.; Patel, H.; Early, A.; Diffley, J.F. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol. Cell 2016, 65, 117–130. [Google Scholar] [CrossRef]
- Delamarre, A.; Barthe, A.; Saint-André, C.D.L.R.; Luciano, P.; Forey, R.; Padioleau, I.; Skrzypczak, M.; Ginalski, K.; Géli, V.; Pasero, P.; et al. MRX Increases Chromatin Accessibility at Stalled Replication Forks to Promote Nascent DNA Resection and Cohesin Loading. Mol. Cell 2019, 77, 395–410.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, D.; Li, X.; Ding, L.; Tang, J.; Liu, C.; Shirahige, K.; Cao, Q.; Lou, H. Rtt101-Mms1-Mms22 coordinates replication-coupled sister chromatid cohesion and nucleosome assembly. EMBO Rep. 2017, 18, 1294–1305. [Google Scholar] [CrossRef]
- Haering, C.; Löwe, J.; Hochwagen, A.; Nasmyth, K. Molecular Architecture of SMC Proteins and the Yeast Cohesin Complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef]
- Veld, P.J.H.I.; Herzog, F.; Ladurner, R.; Davidson, I.F.; Piric, S.; Kreidl, E.; Bhaskara, V.; Aebersold, R.; Peters, J.-M. Characterization of a DNA exit gate in the human cohesin ring. Science 2014, 346, 968–972. [Google Scholar] [CrossRef]
- Pradhan, B.; Barth, R.; Kim, E.; Davidson, I.F.; Bauer, B.; Laar, T.v.; Yang, W.; Ryu, J.K.; Torre, J.v.d.; Peters, J.M.; et al. SMC complexes can traverse physical roadblocks bigger than their ring size. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lengronne, A.; McIntyre, J.; Katou, Y.; Kanoh, Y.; Hopfner, K.-P.; Shirahige, K.; Uhlmann, F. Establishment of Sister Chromatid Cohesion at the S. cerevisiae Replication Fork. Mol. Cell 2006, 23, 787–799. [Google Scholar] [CrossRef]
- Srinivasan, M.; Petela, N.J.; Scheinost, J.C.; Collier, J.; Voulgaris, M.; Roig, M.B.; Beckouet, F.; Hu, B.; A Nasmyth, K. Scc2 counteracts a Wapl-independent mechanism that releases cohesin from chromosomes during G1. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.D.; Haarhuis, J.H.; Grimm, J.B.; Rowland, B.D.; Lavis, L.D.; Nasmyth, K.A. Cohesin Can Remain Associated with Chromosomes during DNA Replication. Cell Rep. 2017, 20, 2749–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murayama, Y.; Samora, C.P.; Kurokawa, Y.; Iwasaki, H.; Uhlmann, F. Establishment of DNA-DNA Interactions by the Cohesin Ring. Cell 2018, 172, 465–477.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasmyth, K.A. Scc2-mediated loading of cohesin onto chromosomes in G1 yeast cells is insufficient to build cohesion during S phase. bioRxiv 2017, 123596. [Google Scholar] [CrossRef] [Green Version]
- Borges, V.; Smith, D.J.; Whitehouse, I.; Uhlmann, F. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma 2013, 122, 121–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Boone, C.; Brown, G.W. Genetic Dissection of Parallel Sister-Chromatid Cohesion Pathways. Genetics 2007, 176, 1417–1429. [Google Scholar] [CrossRef] [Green Version]
- Kawasumi, R.; Abe, T.; Psakhye, I.; Miyata, K.; Hirota, K.; Branzei, D. Vertebrate CTF18 and DDX11 essential function in cohesion is bypassed by preventing WAPL-mediated cohesin release. Genes Dev. 2021, 35, 1368–1382. [Google Scholar] [CrossRef]
- Srinivasan, M.; Fumasoni, M.; Petela, N.J.; Murray, A.; Nasmyth, K.A. Cohesion is established during DNA replication utilising chromosome associated cohesin rings as well as those loaded de novo onto nascent DNAs. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yokoyama, M.; Matsumoto, S.; Fukatsu, R.; You, Z.; Masai, H. Fission Yeast Swi1-Swi3 Complex Facilitates DNA Binding of Mrc1. J. Biol. Chem. 2010, 285, 39609–39622. [Google Scholar] [CrossRef] [Green Version]
- Witosch, J.; Wolf, E.; Mizuno, N. Architecture and ssDNA interaction of the Timeless-Tipin-RPA complex. Nucleic Acids Res. 2014, 42, 12912–12927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eickhoff, P.; Kose, H.B.; Martino, F.; Petojevic, T.; Ali, F.A.; Locke, J.; Tamberg, N.; Nans, A.; Berger, J.M.; Botchan, M.R.; et al. Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome. Cell Rep. 2019, 28, 2673–2688.e8. [Google Scholar] [CrossRef] [PubMed]
- Grabarczyk, D.B. Crystal structure and interactions of the Tof1–Csm3 (Timeless–Tipin) fork protection complex. Nucleic Acids Res. 2020, 48, 6996–7004. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Akan, Z.; Yilmaz, S.; Grillo, M.; Smith-Roe, S.L.; Kang, T.-H.; Cordeiro-Stone, M.; Kaufmann, W.K.; Abraham, R.T.; Sancar, A.; et al. Tipin-Replication Protein A Interaction Mediates Chk1 Phosphorylation by ATR in Response to Genotoxic Stress. J. Biol. Chem. 2010, 285, 16562–16571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef]
- Cho, W.-H.; Kang, Y.-H.; An, Y.-Y.; Tappin, I.; Hurwitz, J.; Lee, J.-K. Human Tim-Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc. Natl. Acad. Sci. USA 2013, 110, 2523–2527. [Google Scholar] [CrossRef] [Green Version]
- Leman, A.R.; Noguchi, C.; Lee, C.Y.; Noguchi, E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J. Cell Sci. 2010, 123, 660–670. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.L.; Pot, I.; Chang, M.; Xu, H.; Aneliunas, V.; Kwok, T.; Newitt, R.; Aebersold, R.; Boone, C.; Brown, G.W.; et al. Identification of Protein Complexes Required for Efficient Sister Chromatid Cohesion. Mol. Biol. Cell 2004, 15, 1736–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortone, G.; Zheng, G.; Pensieri, P.; Chiappetta, V.; Tatè, R.; Malacaria, E.; Pichierri, P.; Yu, H.; Pisani, F.M. Interaction of the Warsaw breakage syndrome DNA helicase DDX11 with the replication fork-protection factor Timeless promotes sister chromatid cohesion. PLoS Genet. 2018, 14, e1007622. [Google Scholar] [CrossRef]
- Samora, C.P.; Saksouk, J.; Goswami, P.; Wade, B.O.; Singleton, M.; Bates, P.; Lengronne, A.; Costa, A.; Uhlmann, F. Ctf4 Links DNA Replication with Sister Chromatid Cohesion Establishment by Recruiting the Chl1 Helicase to the Replisome. Mol. Cell 2016, 63, 371–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, T.; Kawasumi, R.; Arakawa, H.; Hori, T.; Shirahige, K.; Losada, A.; Fukagawa, T.; Branzei, D. Chromatin determinants of the inner-centromere rely on replication factors with functions that impart cohesion. Oncotarget 2016, 7, 67934–67947. [Google Scholar] [CrossRef] [PubMed]
- van Schie, J.J.M.; Faramarz, A.; Balk, J.A.; Stewart, G.S.; Cantelli, E.; Oostra, A.B.; Rooimans, M.A.; Parish, J.L.; de Almeida Estéves, C.; Dumic, K.; et al. Warsaw breakage syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat. Commun. 2020, 11, 4287. [Google Scholar] [CrossRef]
- Simon, A.K.; Kummer, S.; Wild, S.; Lezaja, A.; Teloni, F.; Jozwiakowski, S.K.; Altmeyer, M.; Gari, K. The iron–sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation. Life Sci. Alliance 2020, 3, e201900547. [Google Scholar] [CrossRef]
- Farina, A.; Shin, J.-H.; Kim, D.-H.; Bermudez, V.P.; Kelman, Z.; Seo, Y.-S.; Hurwitz, J. Studies with the Human Cohesin Establishment Factor, ChlR1. J. Biol. Chem. 2008, 283, 20925–20936. [Google Scholar] [CrossRef] [Green Version]
- Fumasoni, M.; Zwicky, K.; Vanoli, F.; Lopes, M.; Branzei, D. Error-Free DNA Damage Tolerance and Sister Chromatid Proximity during DNA Replication Rely on the Polα/Primase/Ctf4 Complex. Mol. Cell 2015, 57, 812–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.-Y.; Park, S.H. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 2020, 52, 1948–1958. [Google Scholar] [CrossRef]
- Liu, H.W.; Bouchoux, C.; Panarotto, M.; Kakui, Y.; Patel, H.; Uhlmann, F. Division of Labor between PCNA Loaders in DNA Replication and Sister Chromatid Cohesion Establishment. Mol. Cell 2020, 78, 725–738.e4. [Google Scholar] [CrossRef]
- Mayer, M.L.; Gygi, S.P.; Aebersold, R.; Hieter, P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): An Alternative RFC Complex Required for Sister Chromatid Cohesion in S. cerevisiae. Mol. Cell 2001, 7, 959–970. [Google Scholar] [CrossRef]
- Terret, M.-E.; Sherwood, R.; Rahman, S.; Qin, J.; Jallepalli, P.V. Cohesin acetylation speeds the replication fork. Nature 2009, 462, 231–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, J.S.; Kroll, E.S.; Lundblad, V.; Spencer, F.A. Saccharomyces cerevisiae CTF18 and CTF4 Are Required for Sister Chromatid Cohesion. Mol. Cell. Biol. 2001, 21, 3144–3158. [Google Scholar] [CrossRef] [Green Version]
- Stokes, K.; Winczura, A.; Song, B.; de Piccoli, G.; Grabarczyk, D.B. Ctf18-RFC and DNA Pol ϵ form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Nucleic Acids Res. 2020, 48, 8128–8145. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Han, J.; Zhou, Z.-X.; Jia, S.; Chabes, A.; Farrugia, G.; Ordog, T.; Zhang, Z. Strand-Specific Analysis Shows Protein Binding at Replication Forks and PCNA Unloading from Lagging Strands when Forks Stall. Mol. Cell 2014, 56, 551–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, M.-S.; Ryu, E.; Lee, S.-W.; Park, J.; Ha, N.Y.; Ra, J.S.; Kim, Y.J.; Kim, J.; Abdel-Rahman, M.; Park, S.H.; et al. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Kanellis, P.; Agyei, R.; Durocher, D. Elg1 Forms an Alternative PCNA-Interacting RFC Complex Required to Maintain Genome Stability. Curr. Biol. 2003, 13, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Faramarz, A.; Balk, J.A.; van Schie, J.J.M.; Oostra, A.B.; Ghandour, C.A.; Rooimans, M.A.; Wolthuis, R.M.F.; de Lange, J. Non-redundant roles in sister chromatid cohesion of the DNA helicase DDX11 and the SMC3 acetyl transferases ESCO1 and ESCO2. PLoS ONE 2020, 15, e0220348. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Boone, C.; Klein, H.L. Mrc1 Is Required for Sister Chromatid Cohesion to Aid in Recombination Repair of Spontaneous Damage. Mol. Cell. Biol. 2004, 24, 7082–7090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef]
- Naylor, M.L.; Li, J.-M.; Osborn, A.J.; Elledge, S.J. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc. Natl. Acad. Sci. USA 2009, 106, 12765–12770. [Google Scholar] [CrossRef] [Green Version]
- Osborn, A.J.; Elledge, S.J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003, 17, 1755–1767. [Google Scholar] [CrossRef] [Green Version]
- Tourrière, H.; Versini, G.; Cordón-Preciado, V.; Alabert, C.; Pasero, P. Mrc1 and Tof1 Promote Replication Fork Progression and Recovery Independently of Rad53. Mol. Cell 2005, 19, 699–706. [Google Scholar] [CrossRef]
- Tanaka, H.; Kubota, Y.; Tsujimura, T.; Kumano, M.; Masai, H.; Takisawa, H. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells 2009, 14, 949–963. [Google Scholar] [CrossRef] [Green Version]
- Smith-Roe, S.L.; Patel, S.S.; Simpson, D.A.; Zhou, Y.C.; Rao, S.; Ibrahim, J.G.; Kaiser-Rogers, K.A.; Cordeiro-Stone, M.; Kaufmann, W.K. Timeless functions independently of the Tim-Tipin complex to promote sister chromatid cohesion in normal human fibroblasts. Cell Cycle 2011, 10, 1618–1624. [Google Scholar] [CrossRef]
- Gutierrez-Escribano, P.; Newton, M.D.; Llauró, A.; Huber, J.; Tanasie, L.; Davy, J.; Aly, I.; Aramayo, R.; Montoya, A.; Kramer, H.; et al. A conserved ATP- and Scc2/4-dependent activity for cohesin in tethering DNA molecules. Sci. Adv. 2019, 5, eaay6804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Rodríguez, L.J.; de Piccoli, G.; Marchesi, V.; Jones, R.C.; Edmondson, R.D.; Labib, K. A conserved Polϵ binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res. 2015, 43, 8830–8838. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.; Takano, R.; Takeo, S.; Taniguchi, R.; Ogawa, K.; Ohashi, E.; Tsurimoto, T. Stable Interaction between the Human Proliferating Cell Nuclear Antigen Loader Complex Ctf18-Replication Factor C (RFC) and DNA Polymerase ϵ Is Mediated by the Cohesion-specific Subunits, Ctf18, Dcc1, and Ctf8. J. Biol. Chem. 2010, 285, 34608–34615. [Google Scholar] [CrossRef] [Green Version]
- Grabarczyk, D.B.; Silkenat, S.; Kisker, C. Structural Basis for the Recruitment of Ctf18-RFC to the Replisome. Structure 2018, 26, 137–144.e3. [Google Scholar] [CrossRef] [Green Version]
- Wade, B.O.; Liu, H.W.; Samora, C.P.; Uhlmann, F.; Singleton, M.R. Structural studies of RFC C tf18 reveal a novel chromatin recruitment role for Dcc1. EMBO Rep. 2017, 18, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, M.; Cho, T.; Álvarez-Quilón, A.; Li, K.; Schellenberg, M.J.; Zimmermann, M.; Hustedt, N.; Rossi, S.E.; Adam, S.; Melo, H.; et al. A Genetic Map of the Response to DNA Damage in Human Cells. Cell 2020, 182, 481–496.e21. [Google Scholar] [CrossRef] [PubMed]
- Crabbé, L.; Thomas, A.; Pantesco, V.; de Vos, J.; Pasero, P.; Lengronne, A. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 2010, 17, 1391–1397. [Google Scholar] [CrossRef]
- Naiki, T.; Kondo, T.; Nakada, D.; Matsumoto, K.; Sugimoto, K. Chl12 (Ctf18) Forms a Novel Replication Factor C-Related Complex and Functions Redundantly with Rad24 in the DNA Replication Checkpoint Pathway. Mol. Cell. Biol. 2001, 21, 5838–5845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gellon, L.; Razidlo, D.F.; Gleeson, O.; Verra, L.; Schulz, D.; Lahue, R.S.; Freudenreich, C.H. New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion. PLoS Genet. 2011, 7, e1001298. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, H.; Ohuchi, T.; Ui, A.; Tada, S.; Enomoto, T.; Seki, M. Ctf18 is required for homologous recombination-mediated double-strand break repair. Nucleic Acids Res. 2007, 35, 4989–5000. [Google Scholar] [CrossRef]
- Ogi, T.; Limsirichaikul, S.; Overmeer, R.M.; Volker, M.; Takenaka, K.; Cloney, R.; Nakazawa, Y.; Niimi, A.; Miki, Y.; Jaspers, N.G.; et al. Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Mol. Cell 2010, 37, 714–727. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Hiraga, S.-I.; Yamada, K.; Lamond, A.; Donaldson, A.D. Quantitative Proteomic Analysis of Chromatin Reveals that Ctf18 Acts in the DNA Replication Checkpoint. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansbach, A.B.; Noguchi, C.; Klansek, I.W.; Heidlebaugh, M.; Nakamura, T.M.; Noguchi, E. RFCCtf18and the Swi1-Swi3 Complex Function in Separate and Redundant Pathways Required for the Stabilization of Replication Forks to Facilitate Sister Chromatid Cohesion in Schizosaccharomyces pombe. Mol. Biol. Cell 2008, 19, 595–607. [Google Scholar] [CrossRef] [Green Version]
- Villa, F.; Simon, A.C.; Bazan, M.A.O.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [Green Version]
- Guan, C.; Li, J.; Sun, D.; Liu, Y.; Liang, H. The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome. J. Biol. Chem. 2017, 292, 9627–9636. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Kawasumi, R.; Giannattasio, M.; Dusi, S.; Yoshimoto, Y.; Miyata, K.; Umemura, K.; Hirota, K.; Branzei, D. AND-1 fork protection function prevents fork resection and is essential for proliferation. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Sengupta, S.; van Deursen, F.; de Piccoli, G.; Labib, K. Dpb2 Integrates the Leading-Strand DNA Polymerase into the Eukaryotic Replisome. Curr. Biol. 2013, 23, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Bermudez, V.P.; Farina, A.; Tappin, I.; Hurwitz, J. Influence of the Human Cohesion Establishment Factor Ctf4/AND-1 on DNA Replication. J. Biol. Chem. 2010, 285, 9493–9505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa-Sugata, N.; Masai, H. Roles of Human AND-1 in Chromosome Transactions in S Phase. J. Biol. Chem. 2009, 284, 20718–20728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calì, F.; Bharti, S.K.; di Perna, R.; Brosh, R.M., Jr.; Pisani, F.M. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016, 44, 705–717. [Google Scholar] [CrossRef]
- Yeeles, J.; Janska, A.; Early, A.; Diffley, J.F. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2016, 65, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Shyian, M.; Albert, B.; Zupan, A.M.; Ivanitsa, V.; Charbonnet, G.; Dilg, D.; Shore, D. Fork pausing complex engages topoisomerases at the replisome. Genes Dev. 2019, 34, 87–98. [Google Scholar] [CrossRef]
- Westhorpe, R.; Keszthelyi, A.; Minchell, N.E.; Jones, D.; Baxter, J. Separable functions of Tof1/Timeless in intra-S-checkpoint signalling, replisome stability and DNA topological stress. Nucleic Acids Res. 2020, 48, 12169–12187. [Google Scholar] [CrossRef]
- Hizume, K.; Endo, S.; Muramatsu, S.; Kobayashi, T.; Araki, H. DNA polymerase ε-dependent modulation of the pausing property of the CMG helicase at the barrier. Genes Dev. 2018, 32, 1315–1320. [Google Scholar] [CrossRef] [Green Version]
- Lerner, L.K.; Holzer, S.; Kilkenny, M.L.; Šviković, S.; Murat, P.; Schiavone, D.; Eldridge, C.B.; Bittleston, A.; Maman, J.D.; Branzei, D.; et al. Timeless couples G-quadruplex detection with processing by DDX 11 helicase during DNA replication. EMBO J. 2020, 39. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Mortusewicz, O.; Ma, H.T.; Herr, P.; Poon, R.Y.; Helleday, T.; Qian, C. Timeless Interacts with PARP-1 to Promote Homologous Recombination Repair. Mol. Cell 2015, 60, 163–176. [Google Scholar] [CrossRef] [Green Version]
- van der Lelij, P.; Chrzanowska, K.H.; Godthelp, B.C.; Rooimans, M.A.; Oostra, A.B.; Stumm, M.; Zdzienicka, M.Z.; Joenje, H.; de Winter, J.P. Warsaw Breakage Syndrome, a Cohesinopathy Associated with Mutations in the XPD Helicase Family Member DDX11/ChlR1. Am. J. Hum. Genet. 2010, 86, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Pisani, F.M. Spotlight on Warsaw Breakage Syndrome. Appl. Clin. Genet. 2019, 12, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, T.; Ooka, M.; Kawasumi, R.; Miyata, K.; Takata, M.; Hirota, K.; Branzei, D. Warsaw breakage syndrome DDX11 helicase acts jointly with RAD17 in the repair of bulky lesions and replication through abasic sites. Proc. Natl. Acad. Sci. USA 2018, 115, 8412–8417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jegadesan, N.K.; Branzei, D. DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc. Natl. Acad. Sci. USA 2021, 118, e2024258118. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Roig, M.B.; Nishino, T.; Kurze, A.; Uluocak, P.; Mishra, A.; Beckouet, F.; Underwood, P.; Metson, J.; Imre, R.; et al. Building Sister Chromatid Cohesion: Smc3 Acetylation Counteracts an Antiestablishment Activity. Mol. Cell 2009, 33, 763–774. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, X.; Li, Y.; Kim, B.-J.; Jia, J.; Huang, Z.; Yang, T.; Fu, X.; Jung, S.Y.; Wang, Y.; et al. Acetylation of Smc3 by Eco1 Is Required for S Phase Sister Chromatid Cohesion in Both Human and Yeast. Mol. Cell 2008, 31, 143–151. [Google Scholar] [CrossRef]
- Ben-Shahar, T.R.; Heeger, S.; Lehane, C.; East, P.; Flynn, H.; Skehel, M.; Uhlmann, F. Eco1-Dependent Cohesin Acetylation During Establishment of Sister Chromatid Cohesion. Science 2008, 321, 563–566. [Google Scholar] [CrossRef] [Green Version]
- Unnal, E.; Heidinger-Pauli, J.M.; Kim, W.; Guacci, V.; Onn, I.; Gygi, S.P.; Koshland, D.E. A Molecular Determinant for the Establishment of Sister Chromatid Cohesion. Science 2008, 321, 566–569. [Google Scholar] [CrossRef]
- Sutani, T.; Kawaguchi, T.; Kanno, R.; Itoh, T.; Shirahige, K. Budding Yeast Wpl1(Rad61)-Pds5 Complex Counteracts Sister Chromatid Cohesion-Establishing Reaction. Curr. Biol. 2009, 19, 492–497. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.-L.; Roig, M.B.; Hu, B.; Beckouet, F.; Metson, J.; Nasmyth, K. Cohesin’s DNA Exit Gate Is Distinct from Its Entrance Gate and Is Regulated by Acetylation. Cell 2012, 150, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Guacci, V.; Koshland, D. Cohesin-independent segregation of sister chromatids in budding yeast. Mol. Biol. Cell 2012, 23, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Guacci, V.; Stricklin, J.; Bloom, M.; Guō, X.; Bhatter, M.; Koshland, U. A novel mechanism for the establishment of sister chromatid cohesion by the ECO1 acetyltransferase. Mol. Biol. Cell 2015, 26, 117–133. [Google Scholar] [CrossRef] [Green Version]
- Hou, F.; Zou, H. Two Human Orthologues of Eco1/Ctf7 Acetyltransferases Are Both Required for Proper Sister-Chromatid Cohesion. Mol. Biol. Cell 2005, 16, 3908–3918. [Google Scholar] [CrossRef] [Green Version]
- Alomer, R.M.; da Silva, E.M.L.; Chen, J.; Piekarz, K.M.; McDonald, K.; Sansam, C.; Sansam, C.; Rankin, S. Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression. Proc. Natl. Acad. Sci. USA 2017, 114, 9906–9911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamino, M.; Ishibashi, M.; Nakato, R.; Akiyama, K.; Tanaka, H.; Kato, Y.; Negishi, L.; Hirota, T.; Sutani, T.; Bando, M.; et al. Esco1 Acetylates Cohesin via a Mechanism Different from That of Esco2. Curr. Biol. 2015, 25, 1694–1706. [Google Scholar] [CrossRef] [Green Version]
- Kawasumi, R.; Abe, T.; Arakawa, H.; Garre, M.; Hirota, K.; Branzei, D. ESCO1/2’s roles in chromosome structure and interphase chromatin organization. Genes Dev. 2017, 31, 2136–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bender, D.; da Silva, E.M.L.; Chen, J.; Poss, A.M.; Gawey, L.; Rulon, Z.; Rankin, S. Multivalent interaction of ESCO2 with the replication machinery is required for sister chromatid cohesion in vertebrates. Proc. Natl. Acad. Sci. USA 2019, 117, 1081–1089. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA Controls Establishment of Sister Chromatid Cohesion during S Phase. Mol. Cell 2006, 23, 723–732. [Google Scholar] [CrossRef]
- Lyons, N.A.; Fonslow, B.R.; Diedrich, J.K.; Yates, J.R.; Morgan, D.O. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nat. Struct. Mol. Biol. 2013, 20, 194–201. [Google Scholar] [CrossRef] [Green Version]
- Lyons, N.; Morgan, D.O. Cdk1-Dependent Destruction of Eco1 Prevents Cohesion Establishment after S Phase. Mol. Cell 2011, 42, 378–389. [Google Scholar] [CrossRef] [Green Version]
- Seoane, A.I.; Morgan, D.O. Firing of Replication Origins Frees Dbf4-Cdc7 to Target Eco1 for Destruction. Curr. Biol. 2017, 27, 2849–2855.e2. [Google Scholar] [CrossRef] [Green Version]
- Heidinger-Pauli, J.M.; Ünal, E.; Koshland, D. Distinct Targets of the Eco1 Acetyltransferase Modulate Cohesion in S Phase and in Response to DNA Damage. Mol. Cell 2009, 34, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Jones, M.J.K.; Jallepalli, P.V. Cohesin recruits the Esco1 acetyltransferase genome wide to repress transcription and promote cohesion in somatic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 11270–11275. [Google Scholar] [CrossRef] [Green Version]
- Wutz, G.; Ladurner, R.; St Hilaire, B.G.; Stocsits, R.R.; Nagasaka, K.; Pignard, B.; Sanborn, A.; Tang, W.; Várnai, C.; Ivanov, M.P.; et al. ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 2020, 9, e52091. [Google Scholar] [CrossRef]
- Yoshimura, A.; Sutani, T.; Shirahige, K. Functional control of Eco1 through the MCM complex in sister chromatid cohesion. Gene 2021, 784, 145584. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.L.; Ikeda, M.; Tanaka, H.; Nakagawa, T.; Bando, M.; Shirahige, K.; Kubota, Y.; Takisawa, H.; Masukata, H.; Takahashi, T. The Prereplication Complex Recruits XEco2 to Chromatin to Promote Cohesin Acetylation in Xenopus Egg Extracts. Curr. Biol. 2012, 22, 977–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuilkoski, C.M.; Skibbens, R.V. PCNA promotes context-specific sister chromatid cohesion establishment separate from that of chromatin condensation. Cell Cycle 2020, 19, 2436–2450. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, J.; Xin, S.; Jiang, M.; Zhang, J.; Li, Z.; Cao, Q.; Lou, H. Cul4-Ddb1 ubiquitin ligases facilitate DNA replication-coupled sister chromatid cohesion through regulation of cohesin acetyltransferase Esco2. PLoS Genet. 2019, 15, e1007685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Zhang, H.; Zhang, H.; Wang, Z.; Zhou, H.; Zhang, Z. A Cul4 E3 Ubiquitin Ligase Regulates Histone Hand-Off during Nucleosome Assembly. Cell 2013, 155, 817–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luciano, P.; Dehe, P.-M.; Audebert, S.; Géli, V.; Corda, Y. Replisome Function during Replicative Stress Is Modulated by Histone H3 Lysine 56 Acetylation Through Ctf4. Genetics 2015, 199, 1047–1063. [Google Scholar] [CrossRef]
- Ladurner, R.; Kreidl, E.; Ivanov, M.P.; Ekker, H.; Idarraga-Amado, M.H.; Busslinger, G.; Wutz, G.; Cisneros, D.; Peters, J. Sororin actively maintains sister chromatid cohesion. EMBO J. 2016, 35, 635–653. [Google Scholar] [CrossRef]
- Lafont, A.L.; Song, J.; Rankin, S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc. Natl. Acad. Sci. USA 2010, 107, 20364–20369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rankin, S.; Ayad, N.G.; Kirschner, M.W. Sororin, a Substrate of the Anaphase- Promoting Complex, Is Required for Sister Chromatid Cohesion in Vertebrates. Mol. Cell 2005, 18, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Zheng, G.; Tomchick, D.; Luo, X.; Yu, H. Structural Basis and IP6 Requirement for Pds5-Dependent Cohesin Dynamics. Mol. Cell 2016, 62, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Mitter, M.; Gasser, C.; Takacs, Z.; Langer, C.C.H.; Tang, W.; Jessberger, G.; Beales, C.T.; Neuner, E.; Ameres, S.L.; Peters, J.-M.; et al. Conformation of sister chromatids in the replicated human genome. Nature 2020, 586, 139–144. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sykora, M.M.; Veld, P.J.H.I.; Mechtler, K.; Peters, J.-M. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc. Natl. Acad. Sci. USA 2013, 110, 13404–13409. [Google Scholar] [CrossRef] [Green Version]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Tye, S.; Ronson, G.E.; Morris, J.R. A fork in the road: Where homologous recombination and stalled replication fork protection part ways. Semin. Cell Dev. Biol. 2020, 113, 14–26. [Google Scholar] [CrossRef]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakata, R.; Niwa, K.; La Torre, D.U.; Gu, C.; Tahara, E.; Takada, S.; Nishiyama, T. Opening of cohesin’s SMC ring is essential for timely DNA replication and DNA loop formation. Cell Rep. 2021, 35, 108999. [Google Scholar] [CrossRef]
- Carvajal-Maldonado, D.; Byrum, A.; Jackson, J.; Wessel, S.; Lemaçon, D.; Guitton-Sert, L.; Quinet, A.; Tirman, S.; Graziano, S.; Masson, J.-Y.; et al. Perturbing cohesin dynamics drives MRE11 nuclease-dependent replication fork slowing. Nucleic Acids Res. 2018, 47, 1294–1310. [Google Scholar] [CrossRef]
- Morales, C.; Ruiz-Torres, M.; Rodríguez-Acebes, S.; Lafarga, V.; Rodríguez-Corsino, M.; Megias, D.; Cisneros, D.A.; Peters, J.-M.; Méndez, J.; Losada, A. PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection. J. Biol. Chem. 2020, 295, 146–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peripolli, S.; Singh, T.; Patel, H.; Meneguello, L.; Kiso, K.; Thorpe, P.; Bertoli, C.; de Bruin, R.A.M. Oncogenic c-Myc induces replication stress by increasing cohesins chromatin occupancy. bioRxiv 2021. [Google Scholar] [CrossRef]
- Minchell, N.E.; Keszthelyi, A.; Baxter, J. Cohesin Causes Replicative DNA Damage by Trapping DNA Topological Stress. Mol. Cell 2020, 78, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Su, X.A.; Ma, D.; Parsons, J.V.; Replogle, J.M.; Amatruda, J.F.; Whittaker, C.A.; Stegmaier, K.; Amon, A. RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress. Genes Dev. 2021, 35, 556–572. [Google Scholar] [CrossRef]
- Brough, R.; Bajrami, I.; Vatcheva, R.; Natrajan, R.; Reis-Filho, J.S.; Lord, C.J.; Ashworth, A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012, 31, 1160–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couturier, A.M.; Fleury, H.; Patenaude, A.-M.; Bentley, V.L.; Rodrigue, A.; Coulombe, Y.; Niraj, J.; Pauty, J.; Berman, J.N.; Dellaire, G.; et al. Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res. 2016, 44, 10879–10897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frattini, C.; Hernandez, S.V.; Pellicanò, G.; Jossen, R.; Katou, Y.; Shirahige, K.; Bermejo, R. Cohesin Ubiquitylation and Mobilization Facilitate Stalled Replication Fork Dynamics. Mol. Cell 2017, 68, 758–772.e4. [Google Scholar] [CrossRef] [Green Version]
- Benedict, B.; van Schie, J.J.; Oostra, A.B.; Balk, J.A.; Wolthuis, R.M.; Riele, H.T.; de Lange, J. WAPL-Dependent Repair of Damaged DNA Replication Forks Underlies Oncogene-Induced Loss of Sister Chromatid Cohesion. Dev. Cell 2020, 52, 683–698.e7. [Google Scholar] [CrossRef] [Green Version]
- McAleenan, A.; Blanco, A.C.; Cordon-Preciado, V.; Sen, N.; Esteras, M.; Jarmuz, A.; Aragón, L. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature 2012, 493, 250–254. [Google Scholar] [CrossRef]
- Hellmuth, S.; Gutiérrez-Caballero, C.; Llano, E.; Pendás, A.M.; Stemmann, O. Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation. EMBO J. 2018, 37, e99184. [Google Scholar] [CrossRef]
- Nagao, K.; Adachi, Y.; Yanagida, M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 2004, 430, 1044–1048. [Google Scholar] [CrossRef]
- Nakamura, K.; Kustatscher, G.; Alabert, C.; Hödl, M.; Forne, I.; Völker-Albert, M.; Satpathy, S.; Beyer, T.E.; Mailand, N.; Choudhary, C.; et al. Proteome dynamics at broken replication forks reveal a distinct ATM-directed repair response suppressing DNA double-strand break ubiquitination. Mol. Cell 2021, 81, 1084–1099.e6. [Google Scholar] [CrossRef] [PubMed]
- Kukolj, E.; Kaufmann, T.; Dick, A.E.; Zeillinger, R.; Gerlich, D.W.; Slade, D. PARP inhibition causes premature loss of cohesion in cancer cells. Oncotarget 2017, 8, 103931–103951. [Google Scholar] [CrossRef]
- Masamsetti, V.P.; Low, R.R.J.; Mak, K.S.; O’Connor, A.; Riffkin, C.D.; Lamm, N.; Crabbe, L.; Karlseder, J.; Huang, D.C.S.; Hayashi, M.T.; et al. Replication stress induces mitotic death through parallel pathways regulated by WAPL and telomere deprotection. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Manning, A.L.; Yazinski, S.A.; Nicolay, B.; Bryll, A.; Zou, L.; Dyson, N.J. Suppression of Genome Instability in pRB-Deficient Cells by Enhancement of Chromosome Cohesion. Mol. Cell 2014, 53, 993–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lange, J.; Faramarz, A.; Oostra, A.B.; de Menezes, R.X.; van der Meulen, I.H.; Rooimans, M.A.; Rockx, D.A.; Brakenhoff, R.H.; van Beusechem, V.W.; King, R.W.; et al. Defective sister chromatid cohesion is synthetically lethal with impaired APC/C function. Nat. Commun. 2015, 6, 8399. [Google Scholar] [CrossRef] [Green Version]
- Stoepker, C.; Ameziane, N.; van der Lelij, P.; Kooi, I.E.; Oostra, A.B.; Rooimans, M.A.; van Mil, S.E.; Brink, A.; Dietrich, R.; Balk, J.A.; et al. Defects in the Fanconi Anemia Pathway and Chromatid Cohesion in Head and Neck Cancer. Cancer Res. 2015, 75, 3543–3553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Harn, T.; Foijer, F.; van Vugt, M.; Banerjee, R.; Yang, F.; Oostra, A.; Joenje, H.; Riele, H.T. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010, 24, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Schuck, P.L.; Ball, L.E.; Stewart, J.A. The DNA-binding protein CST associates with the cohesin complex and promotes chromosome cohesion. J. Biol. Chem. 2021, 297. [Google Scholar] [CrossRef] [PubMed]

| Homo sapiens | S. pombe | S. cerevisiae | Function | |
|---|---|---|---|---|
| SMC1A | Psm1 | Smc1 | Core cohesin subunits | |
| SMC3 | Psm3 | Smc3 | ||
| RAD21 | Rad21 | Mcd1/Scc1 | ||
| STAG1, STAG2 | Psc3 | Irr1/Scc3 | Cohesin associated factors | |
| PDS5A, PDS5B | Pds5 | Pds5 | ||
| NIPBL | Mis4 | Scc2 | Cohesin loader complex | |
| MAU2 | Ssl3 | Scc4 | ||
| WAPL | Wpl1 | Rad61/Wpl1 | Cohesin removal from chromatin | |
| ESPL1/SEPARASE | Cut1 | Esp1 | ||
| ESCO1, ESCO2 | Eso1 | Eco1/Ctf7 | SMC3 acetylation | |
| SORORIN | - | - | WAPL antagonist | |
| CDC6 | Cdc18 | Cdc6 | Pre-RC formation | |
| CDT1 | Cdt1 | Tah11/Cdt1 | ||
| CDC45 | Cdc45 | Cdc45/Sld4 | CMG (replicative helicase) | |
| MCM2-7 | Mcm2-7 | Mcm2-7 | ||
| GINS1-4 | Sld5-Psf1-Psf2-Psf3 (GINS) | Sld5-Psf1-Psf2-Psf3 (GINS) | ||
| PCNA | Pcn1 | Pol30/Pcna | Polymerase sliding clamp | |
| POLA1-POLA2-PRIM1-PRIM2 | Pol1-Spb70-Spp1-Spp2 | Pol1-Pol12-Pri1-Pri2 | Polymerase α-Primase complex | |
| POLD1-4 | Cdc6-Cdc1-Cdc27-Cdm1 | Pol3-Pol31-Pol32 | Polymerase δ complex | |
| POLE-POLE2-POLE3-POLE4 | Cdc20-Dpb2-Dpb3-Dpb4 | Pol2-Dpb2-Dpb3-Dpb4 | Polymerase ε complex | |
| FEN1 DNA2 | Rad2 Dna2 | Rad27 Dna2 | Okazaki fragment processing | |
| LIG1 | Cdc17 | Cdc9 | ||
| Epistasis group 1 | AND-1/WHDH1 | Mcl1 | Ctf4 | Mediating interactions at the replication fork |
| DDX11 | Chl1 | Chl1 | DNA helicase | |
| TIMELESS | Swi1 | Tof1 | Fork Protection Complex (FPC) | |
| TIPIN | Swi3 | Csm3 | ||
| Epistasis group 2 | CLASPIN | Mrc1 | Mrc1 | |
| CHTF18 | Ctf18 | Ctf18 | PCNA loader on leading strand (with Rfc2-5) | |
| CHTF8 | Ctf8 | Ctf8 | ||
| DSCC1 | Dcc1 | Dcc1 | ||
| RFC1 | Rfc1 | Rfc1/Cdc44 | PCNA loader on lagging strand (with Rfc2-5) | |
| ATAD5 | Elg1 | Elg1 | PCNA unloader (with Rfc2-5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Schie, J.J.M.; de Lange, J. The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks. Cells 2021, 10, 3455. https://doi.org/10.3390/cells10123455
van Schie JJM, de Lange J. The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks. Cells. 2021; 10(12):3455. https://doi.org/10.3390/cells10123455
Chicago/Turabian Stylevan Schie, Janne J. M., and Job de Lange. 2021. "The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks" Cells 10, no. 12: 3455. https://doi.org/10.3390/cells10123455






