New Insights into Epigenetic Regulation of T Cell Differentiation
Abstract
:1. Introduction
1.1. Generation of Differentiated T Cells
1.2. Major Epigenetic Processes and Regulators
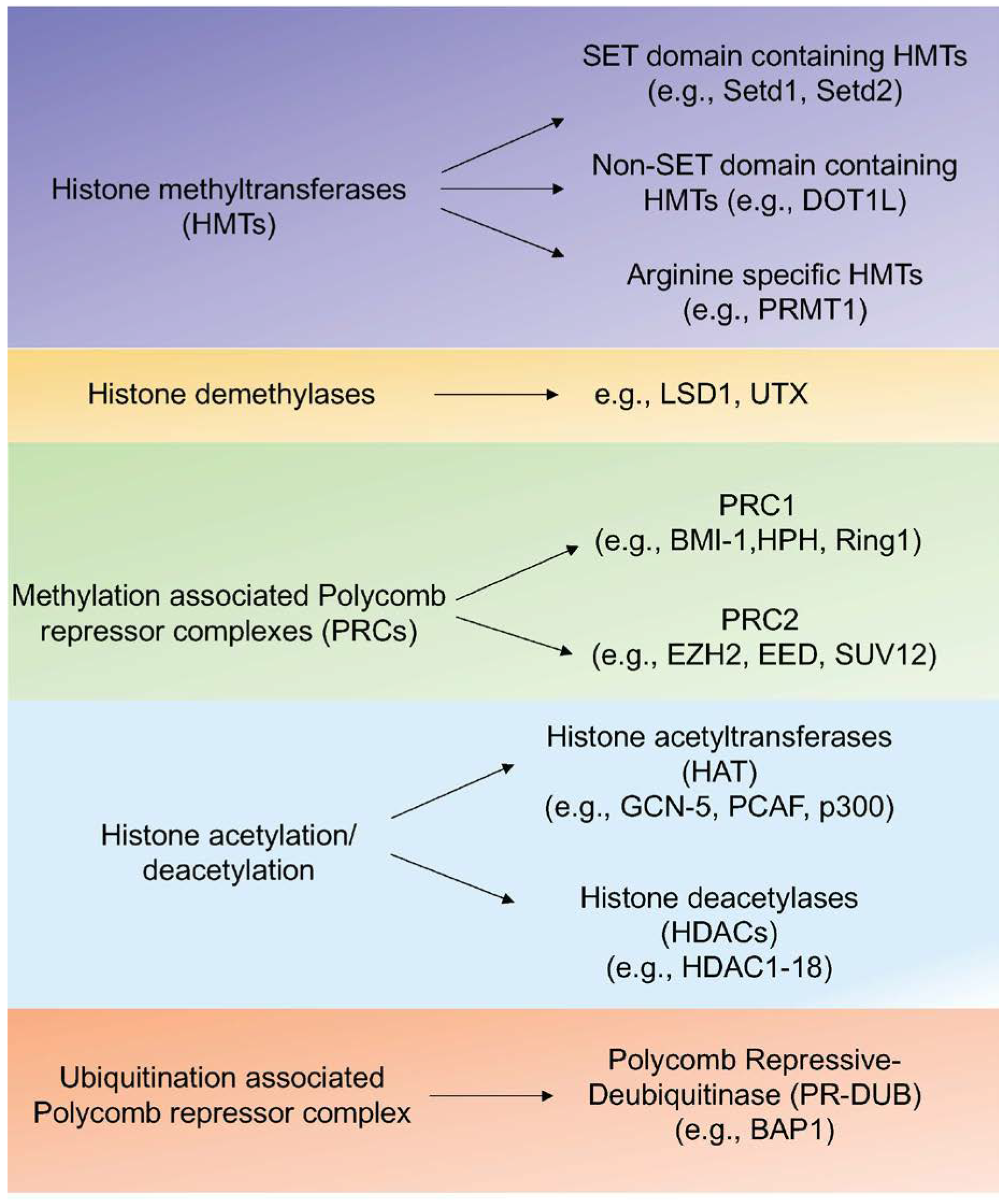
1.2.1. Histone Modifications
1.2.2. DNA Methylation
2. Epigenetic Changes during T Cell Differentiation
2.1. Epigenetic Control of CD4+ T Cell Differentiation
2.2. Epigenetics Associated with Treg Development and Maintenance
2.3. Epigenetic Regulation of CD8+ T Cell Differentiation
2.4. Epigenetic Directives of Memory T Cell Generation and Maintenance
2.5. Epigenetic Modulation of Natural Killer T Cells
3. Epigenetic Modification and Cancer
4. Perspective and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothenberg, E.V.; Moore, J.E.; Yui, M.A. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008, 8, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.; Adoro, S.; Park, J.H. Lineage fate and intense debate: Myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 2008, 8, 788–801. [Google Scholar] [CrossRef]
- Dutta, A.; Zhao, B.; Love, P.E. New insights into TCR β-selection. Trends Immunol. 2021, 42, 735–750. [Google Scholar] [CrossRef]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, J.; D’Angelo, W.; Badylak, S.F. Common Challenges in Tissue Regeneration; Elsevier: Amsterdam, The Netherlands, 2019; pp. 217–229. [Google Scholar]
- Van den Broek, T.; Borghans, J.A.M.; van Wijk, F. The full spectrum of human naive T cells. Nat. Rev. Immunol. 2018, 18, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.L.; Sousa, A.E. Establishment and Maintenance of the Human Naïve CD4+ T-Cell Compartment. Front. Pediatrics 2016, 4, 119. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [Green Version]
- Kondĕlková, K.; Vokurková, D.; Krejsek, J.; Borská, L.; Fiala, Z.; Ctirad, A. Regulatory T cells (TREG) and their roles in immune system with respect to immunopathological disorders. Acta Med. 2010, 53, 73–77. [Google Scholar] [CrossRef] [Green Version]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Sakaguchi, S. Epigenetic control of thymic Treg-cell development. Eur. J. Immunol. 2015, 45, 11–16. [Google Scholar] [CrossRef]
- Ono, M.; Yaguchi, H.; Ohkura, N.; Kitabayashi, I.; Nagamura, Y.; Nomura, T.; Miyachi, Y.; Tsukada, T.; Sakaguchi, S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 2007, 446, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.; deRoos, P.; Chaudhry, A.; Niec, R.E.; Arvey, A.; Samstein, R.M.; Leslie, C.; Shaffer, S.A.; Goodlett, D.R.; Rudensky, A.Y. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012, 13, 1010–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iizuka-Koga, M.; Nakatsukasa, H.; Ito, M.; Akanuma, T.; Lu, Q.; Yoshimura, A. Induction and maintenance of regulatory T cells by transcription factors and epigenetic modifications. J. Autoimmun. 2017, 83, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Benoist, C.; Mathis, D. Treg cells, life history, and diversity. Cold Spring Harb. Perspect. Biol. 2012, 4, a007021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tough, D.F.; Rioja, I.; Modis, L.K.; Prinjha, R.K. Epigenetic Regulation of T Cell Memory: Recalling Therapeutic Implications. Trends Immunol. 2020, 41, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Alfei, F.; Kanev, K.; Hofmann, M.; Wu, M.; Ghoneim, H.E.; Roelli, P.; Utzschneider, D.T.; Von Hoesslin, M.; Cullen, J.G.; Fan, Y.; et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 2019, 571, 265–269. [Google Scholar] [CrossRef]
- Kumar, A.; Suryadevara, N.; Hill, T.M.; Bezbradica, J.S.; Van Kaer, L.; Joyce, S. Natural Killer T Cells: An Ecological Evolutionary Developmental Biology Perspective. Front. Immunol. 2017, 8, 1858. [Google Scholar] [CrossRef] [Green Version]
- Tsagaratou, A. TET mediated epigenetic regulation of iNKT cell lineage fate choice and function. Mol. Immunol. 2018, 101, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Saito, H.; Liang, G.; Friedman, J.M. Epigenetic alterations and microRNA misexpresion in cancer and autoimmune diseases: A critical review. Clin. Rev. Allergy Immunol. 2014, 47, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jambhekar, A.; Dhall, A.; Shi, Y. Author Correction: Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2020, 21, 59. [Google Scholar] [CrossRef] [Green Version]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Whetstine, J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 2007, 25, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Foglizzo, M.; Middleton, A.J.; Burgess, A.E.; Crowther, J.M.; Dobson, R.C.J.; Murphy, J.M.; Day, C.L.; Mace, P.D. A bidentate Polycomb Repressive-Deubiquitinase complex is required for efficient activity on nucleosomes. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Schmidl, C.; Delacher, M.; Huehn, J.; Feuerer, M. Epigenetic mechanisms regulating T-cell responses. J. Allergy Clin. Immunol. 2018, 142, 728–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leoni, C.; Vincenzetti, L.; Emming, S.; Monticelli, S. Epigenetics of T lymphocytes in health and disease. Swiss Med. Wkly. 2015, 145, w14191. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, L.; Lu, L. Thymocyte selection: From signaling to epigenetic regulation. Adv. Immunol. 2019, 144, 1–22. [Google Scholar]
- Yoon, B.H.; Kim, M.; Kim, M.H.; Kim, H.J.; Kim, J.H.; Kim, J.H.; Kim, J.; Kim, Y.S.; Lee, D.; Kang, S.J.; et al. Dynamic Transcriptome, DNA Methylome, and DNA Hydroxymethylome Networks during T-Cell Lineage Commitment. Mol. Cells 2018, 41, 953–963. [Google Scholar]
- Scheer, S.; Runting, J.; Bramhall, M.; Russ, B.; Zaini, A.; Ellemor, J.; Rodrigues, G.; Ng, J.; Zaph, C. The Methyltransferase DOT1L Controls Activation and Lineage Integrity in CD4+ T Cells during Infection and Inflammation. Cell Rep. 2020, 33, 108505. [Google Scholar] [CrossRef]
- Kwesi-Maliepaard, E.M.; Aslam, M.A.; Alemdehy, M.F.; Van Den Brand, T.; McLean, C.; Vlaming, H.; Van Welsem, T.; Korthout, T.; Lancini, C.; Hendriks, S.; et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. USA 2020, 117, 20706–20716. [Google Scholar] [CrossRef]
- Onodera, A.; Kiuchi, M.; Kokubo, K.; Kato, M.; Ogino, T.; Horiuchi, S.; Kanai, U.; Hirahara, K.; Nakayama, T. Menin Controls the Memory Th2 Cell Function by Maintaining the Epigenetic Integrity of Th2 Cells. J. Immunol. 2017, 199, 1153–1162. [Google Scholar] [CrossRef] [Green Version]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef]
- Matkar, S.; Thiel, A.; Hua, X. Menin: A scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 2013, 38, 394–402. [Google Scholar] [CrossRef] [Green Version]
- Onodera, A.; Yamashita, M.; Endo, Y.; Kuwahara, M.; Tofukuji, S.; Hosokawa, H.; Kanai, A.; Suzuki, Y.; Nakayama, T. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J. Exp. Med. 2010, 207, 2493–2506. [Google Scholar] [CrossRef]
- Nakata, Y.; Brignier, A.C.; Jin, S.; Shen, Y.; Rudnick, S.I.; Sugita, M.; Gewirtz, A.M. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood 2010, 116, 1280–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiuchi, M.; Onodera, A.; Kokubo, K.; Ichikawa, T.; Morimoto, Y.; Kawakami, E.; Takayama, N.; Eto, K.; Koseki, H.; Hirahara, K.; et al. The Cxxc1 subunit of the Trithorax complex directs epigenetic licensing of CD4+ T cell differentiation. J. Exp. Med. 2021, 218, e20201690. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Meng, X.; Guo, Y.; Cao, W.; Liu, W.; Xia, Q.; Hui, Z.; Chen, J.; Hong, S.; Zhang, X.; et al. Epigenetic initiation of the TH17 differentiation program is promoted by Cxxc finger protein 1. Sci. Adv. 2019, 5, eaax1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Laurence, A.; Elias, K.M.; O’Shea, J.J. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 2007, 282, 34605–34610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camporeale, A.; Poli, V. IL-6, IL-17 and STAT3: A holy trinity in auto-immunity? Front. Biosci. (Landmark Ed.) 2012, 17, 2306–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timms, R.T.; Tchasovnikarova, I.A.; Antrobus, R.; Dougan, G.; Lehner, P.J. ATF7IP-Mediated Stabilization of the Histone Methyltransferase SETDB1 Is Essential for Heterochromatin Formation by the HUSH Complex. Cell Rep. 2016, 17, 653–659. [Google Scholar] [CrossRef] [Green Version]
- Sin, J.H.; Zuckerman, C.; Cortez, J.T.; Eckalbar, W.L.; Erle, D.J.; Anderson, M.S.; Waterfield, M.R. The epigenetic regulator ATF7ip inhibits Il2 expression, regulating Th17 responses. J. Exp. Med. 2019, 216, 2024–2037. [Google Scholar] [CrossRef] [Green Version]
- Cribbs, A.P.; Terlecki-Zaniewicz, S.; Philpott, M.; Baardman, J.; Ahern, D.; Lindow, M.; Obad, S.; Oerum, H.; Sampey, B.; Mander, P.K.; et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proc. Natl. Acad. Sci. USA 2020, 117, 6056–6066. [Google Scholar] [CrossRef] [Green Version]
- Dobenecker, M.-W.; Kim, J.K.; Marcello, J.; Fang, T.C.; Prinjha, R.; Bosselut, R.; Tarakhovsky, A. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J. Exp. Med. 2015, 212, 297–306. [Google Scholar] [CrossRef]
- Beyaz, S.; Kim, J.H.; Pinello, L.; Xifaras, M.E.; Hu, Y.; Huang, J.; Kerenyi, M.A.; Das, P.P.; Barnitz, R.A.; Herault, A.; et al. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat. Immunol. 2017, 18, 184–195. [Google Scholar] [CrossRef] [Green Version]
- Meng, M.; Liu, H.; Chen, S.; Zhao, H.; Gao, X.; Zhang, J.; Chen, D. Methylation of H3K27 and H3K4 in key gene promoter regions of thymus in RA mice is involved in the abnormal development and differentiation of iNKT cells. Immunogenetics 2019, 71, 489–499. [Google Scholar] [CrossRef]
- Hsu, F.-C.; Belmonte, P.J.; Constans, M.M.; Chen, M.W.; McWilliams, D.C.; Hiebert, S.W.; Shapiro, V.S. Histone Deacetylase 3 Is Required for T Cell Maturation. J. Immunol. 2015, 195, 1578–1590. [Google Scholar] [CrossRef]
- Thapa, P.; Das, J.; McWilliams, D.; Shapiro, M.; Sundsbak, R.; Nelson-Holte, M.; Tangen, S.; Anderson, J.; Desiderio, S.; Hiebert, S.; et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat. Commun. 2013, 4, 1582. [Google Scholar] [CrossRef] [PubMed]
- Tay, R.E.; Olawoyin, O.; Cejas, P.; Xie, Y.; Meyer, C.A.; Ito, Y.; Weng, Q.Y.; Fisher, D.E.; Long, H.W.; Brown, M.; et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J. Exp. Med. 2020, 217, e20191453. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Li, F.; Zeng, Z.; Zhao, Y.; Yu, S.; Shan, Q.; Li, Y.; Phillips, F.C.; Maina, P.K.; Qi, H.H.; et al. Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nat. Immunol. 2016, 17, 695–703. [Google Scholar] [CrossRef]
- Gao, B.; Kong, Q.; Zhang, Y.; Yun, C.; Dent, S.Y.R.; Song, J.; Zhang, D.D.; Wang, Y.; Li, X.; Fang, D. The Histone Acetyltransferase Gcn5 Positively Regulates T Cell Activation. J. Immunol. 2017, 198, 3927–3938. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yun, C.; Gao, B.; Xu, Y.; Zhang, Y.; Wang, Y.; Kong, Q.; Zhao, F.; Wang, C.-R.; Dent, S.Y.R.; et al. The Lysine Acetyltransferase GCN5 Is Required for iNKT Cell Development through EGR2 Acetylation. Cell Rep. 2017, 20, 600–612. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Wang, L.; Di Giorgio, E.; Akimova, T.; Beier, U.H.; Han, R.; Trevisanut, M.; Kalin, J.H.; Cole, P.A.; Hancock, W.W. Inhibiting the coregulator CoREST impairs Foxp3+ Treg function and promotes antitumor immunity. J. Clin. Investig. 2020, 130, 1830–1842. [Google Scholar] [CrossRef] [PubMed]
- Kalin, J.H.; Wu, M.; Gomez, A.V.; Song, Y.; Das, J.; Hayward, D.; Adejola, N.; Wu, M.; Panova, I.; Chung, H.J.; et al. Targeting the CoREST complex with dual histone deacetylase and demethylase inhibitors. Nat. Commun. 2018, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Gray, S.M.; Amezquita, R.A.; Guan, T.; Kleinstein, S.H.; Kaech, S.M. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity 2017, 46, 596–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Vasilatos, S.N.; Chen, L.; Wu, H.; Cao, Z.; Fu, Y.; Huang, M.; Vlad, A.M.; Lu, B.; Oesterreich, S.; et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 2019, 38, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.J.; McCuaig, R.D.; Tan, A.H.Y.; Hardy, K.; Seddiki, N.; Ali, S.; Dahlstrom, J.E.; Bean, E.G.; Dunn, J.; Forwood, J.; et al. Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch. Front. Immunol. 2020, 11, 1228. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Lafleur, M.W.; Nguyen, T.H.; Chen, S.; Chakravarthy, A.; Conway, J.R.; Li, Y.; Chen, H.; Yang, H.; Hsu, P.-H.; et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018, 174, 549–563.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, O.; Giles, J.R.; McDonald, S.; Manne, S.; Ngiow, S.F.; Patel, K.P.; Werner, M.T.; Huang, A.C.; Alexander, K.A.; Wu, J.E.; et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 2019, 571, 211–218. [Google Scholar] [CrossRef]
- Seo, H.; Chen, J.; González-Avalos, E.; Samaniego-Castruita, D.; Das, A.; Wang, Y.H.; López-Moyado, I.F.; Georges, R.O.; Zhang, W.; Onodera, A.; et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA 2019, 116, 12410–12415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, A.C.; Dündar, F.; Zumbo, P.; Chandran, S.S.; Klebanoff, C.A.; Shakiba, M.; Trivedi, P.; Menocal, L.; Appleby, H.; Camara, S.; et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 2019, 571, 270–274. [Google Scholar] [CrossRef]
- Berkley, A.M.; Hendricks, D.W.; Simmons, K.B.; Fink, P.J. Recent thymic emigrants and mature naive T cells exhibit differential DNA methylation at key cytokine loci. J. Immunol. 2013, 190, 6180–6186. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Kuroha, T.; Moriguchi, T.; Cummings, D.; Maillard, I.; Lim, K.C.; Engel, J.D. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009, 206, 2987–3000. [Google Scholar] [CrossRef]
- Wan, Y.Y. GATA3: A master of many trades in immune regulation. Trends Immunol. 2014, 35, 233–242. [Google Scholar] [CrossRef]
- Yagi, R.; Zhu, J.; Paul, W.E. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int. Immunol. 2011, 23, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Schewitz-Bowers, L.P.; Lait, P.J.P.; Copland, D.A.; Stimpson, M.L.; Li, J.J.; Liu, Y.; Dick, A.D.; Lee, R.W.J.; Wei, L. The Bromodomain and Extra-Terminal Protein Inhibitor OTX015 Suppresses T Helper Cell Proliferation and Differentiation. Curr. Mol. Med. 2019, 18, 594–601. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhao, Y.; Zou, L.; Zhang, D.; Aki, D.; Liu, Y.-C. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J. Exp. Med. 2019, 216, 1664–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, M.; Iizuka-Koga, M.; Ando, M.; Yoshimura, A. Development and Functional Modulation of Regulatory T Cells by Transcription Factors and Epigenetics. Cornea 2018, 37 (Suppl. S1), S42–S49. [Google Scholar] [CrossRef]
- Bellanti, J.A.; Li, D. Treg Cells and Epigenetic Regulation; Springer: Singapore, 2021; pp. 95–114. [Google Scholar]
- Ono, M. Control of regulatory T-cell differentiation and function by T-cell receptor signalling and Foxp3 transcription factor complexes. Immunology 2020, 160, 24–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkura, N.; Sakaguchi, S. Transcriptional and epigenetic basis of Treg cell development and function: Its genetic anomalies or variations in autoimmune diseases. Cell Res. 2020, 30, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Vanhanen, R.; Leskinen, K.; Mattila, I.P.; Saavalainen, P.; Arstila, T.P. Epigenetic and transcriptional analysis supports human regulatory T cell commitment at the CD4+CD8+ thymocyte stage. Cell. Immunol. 2020, 347, 104026. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Wu, C.-J.; Chan, C.-C.; Nguyen, D.T.; Lin, K.-R.; Lin, S.-J.; Chen, L.-C.; Yen, J.J.-Y.; Kuo, M.-L. DNA Methyltransferase Inhibitor Promotes Human CD4+CD25hFOXP3+ Regulatory T Lymphocyte Induction under Suboptimal TCR Stimulation. Front. Immunol. 2016, 7, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmin, K.A.; Morales-Nebreda, L.; Acosta, M.A.T.; Anekalla, K.R.; Chen, S.-Y.; Abdala-Valencia, H.; Politanska, Y.; Cheresh, P.; Akbarpour, M.; Steinert, E.M.; et al. Maintenance DNA methylation is essential for regulatory T cell development and stability of suppressive function. J. Clin. Investig. 2020, 130, 6571–6587. [Google Scholar] [CrossRef]
- Feng, Y.; Arvey, A.; Chinen, T.; van der Veeken, J.; Gasteiger, G.; Rudensky, A.Y. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 2014, 158, 749–763. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liang, Y.; LeBlanc, M.; Benner, C.; Zheng, Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 2014, 158, 734–748. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, J.; Martino, P.; Nguyen, L.; Li, X. Cutting Edge: CRISPR-Based Transcriptional Regulators Reveal Transcription-Dependent Establishment of Epigenetic Memory of Foxp3 in Regulatory T Cells. J. Immunol. 2020, 205, 2953–2958. [Google Scholar] [CrossRef] [PubMed]
- Chorro, L.; Suzuki, M.; Chin, S.S.; Williams, T.M.; Snapp, E.L.; Odagiu, L.; Labrecque, N.; Lauvau, G. Interleukin 2 modulates thymic-derived regulatory T cell epigenetic landscape. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Kidani, Y.; Vandenbon, A.; Hirota, K.; Kawakami, R.; Yasuda, K.; Motooka, D.; Nakamura, S.; Kondo, M.; et al. Addendum: Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017, 18, 1270. [Google Scholar] [CrossRef] [Green Version]
- Dobenecker, M.-W.; Park, J.S.; Marcello, J.; McCabe, M.T.; Gregory, R.; Knight, S.D.; Rioja, I.; Bassil, A.K.; Prinjha, R.K.; Tarakhovsky, A. Signaling function of PRC2 is essential for TCR-driven T cell responses. J. Exp. Med. 2018, 215, 1101–1113. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, M.M.; Bamidele, A.O.; Svingen, P.A.; Sagstetter, M.R.; Smyrk, T.C.; Gaballa, J.M.; Hamdan, F.H.; Kosinsky, R.L.; Gibbons, H.R.; Sun, Z.; et al. BMI1 maintains the Treg epigenomic landscape to prevent inflammatory bowel disease. J. Clin. Investig. 2021, 131, e140755. [Google Scholar] [CrossRef]
- Xu, T.; Stewart, K.M.; Wang, X.; Liu, K.; Xie, M.; Ryu, J.K.; Li, K.; Ma, T.; Wang, H.; Ni, L.; et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 2017, 548, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Zhang, K.; Milner, J.J.; Toma, C.; Chen, R.; Scott-Browne, J.P.; Pereira, R.M.; Crotty, S.; Chang, J.T.; Pipkin, M.E.; et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 2017, 18, 573–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jadhav, R.R.; Gustafson, C.E.; Le Saux, S.; Ye, Z.; Li, X.; Tian, L.; Weyand, C.M.; Goronzy, J.J. Distinct Age-Related Epigenetic Signatures in CD4 and CD8 T Cells. Front. Immunol. 2020, 11, 585168. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Christofides, A.; Bardhan, K.; Li, L.; Boussiotis, V.A. Corrigendum: Regulation of T Cell Differentiation and Function by EZH2. Front. Immunol. 2016, 7, 346. [Google Scholar] [CrossRef] [Green Version]
- Stairiker, C.J.; Thomas, G.D.; Salek-Ardakani, S. EZH2 as a Regulator of CD8+ T Cell Fate and Function. Front. Immunol. 2020, 11, 593203. [Google Scholar] [CrossRef] [PubMed]
- Grewal, I.S.; Foellmer, H.G.; Grewal, K.D.; Xu, J.; Hardardottir, F.; Baron, J.L.; Janeway, C.A., Jr.; Flavell, R.A. Requirement for CD40 ligand in costimulation induction, T cell activation, and experimental allergic encephalomyelitis. Science 1996, 273, 1864–1867. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Naito, T.; Tenno, M.; Maruyama, M.; Koseki, H.; Taniuchi, I.; Naoe, Y. ThPOK represses CXXC5, which induces methylation of histone H3 lysine 9 in Cd40lg promoter by association with SUV39H1: Implications in repression of CD40L expression in CD8+ cytotoxic T cells. J. Leukoc. Biol. 2016, 100, 327–338. [Google Scholar] [CrossRef]
- Kallies, A.; Xin, A.; Belz, G.T.; Nutt, S.L. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 2009, 31, 283–295. [Google Scholar] [CrossRef] [Green Version]
- Piccirillo, A.R.; Cattley, R.T.; D’Cruz, L.M.; Hawse, W.F. Histone acetyltransferase CBP is critical for conventional effector and memory T-cell differentiation in mice. J. Biol. Chem. 2019, 294, 2397–2406. [Google Scholar] [CrossRef] [Green Version]
- Carty, S.A.; Gohil, M.; Banks, L.B.; Cotton, R.M.; Johnson, M.E.; Stelekati, E.; Wells, A.D.; Wherry, E.J.; Koretzky, G.A.; Jordan, M.S. The Loss of TET2 Promotes CD8+ T Cell Memory Differentiation. J. Immunol. 2018, 200, 82–91. [Google Scholar] [CrossRef] [Green Version]
- Youngblood, B.; Hale, J.S.; Kissick, H.T.; Ahn, E.; Xu, X.; Wieland, A.; Araki, K.; West, E.E.; Ghoneim, H.E.; Fan, Y.; et al. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 2017, 552, 404–409. [Google Scholar] [CrossRef]
- Ladle, B.H.; Li, K.-P.; Phillips, M.J.; Pucsek, A.B.; Haile, A.; Powell, J.D.; Jaffee, E.M.; Hildeman, D.A.; Gamper, C.J. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. USA 2016, 113, 10631–10636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durek, P.; Nordström, K.; Gasparoni, G.; Salhab, A.; Kressler, C.; de Almeida, M.; Bassler, K.; Ulas, T.; Schmidt, F.; Xiong, J.; et al. Epigenomic Profiling of Human CD4+ T Cells Supports a Linear Differentiation Model and Highlights Molecular Regulators of Memory Development. Immunity 2016, 45, 1148–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, L.; Goudot, C.; Zueva, E.; Gueguen, P.; Burgdorf, N.; Waterfall, J.J.; Quivy, J.-P.; Almouzni, G.; Amigorena, S. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 2018, 359, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Schauder, D.M.; Shen, J.; Chen, Y.; Kasmani, M.Y.; Kudek, M.R.; Burns, R.; Cui, W. E2A-regulated epigenetic landscape promotes memory CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2013452118. [Google Scholar] [CrossRef]
- Li, K.-P.; Ladle, B.H.; Kurtulus, S.; Sholl, A.; Shanmuganad, S.; Hildeman, D.A. T-cell receptor signal strength and epigenetic control of Bim predict memory CD8+ T-cell fate. Cell Death Differ. 2020, 27, 1214–1224. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, R.-B.; Cao, Q.; Fan, K.-Q.; Huang, L.-J.; Yu, J.-S.; Gao, Z.-J.; Huang, T.; Zhong, J.-Y.; Mao, X.-T.; et al. USP16-mediated deubiquitination of calcineurin A controls peripheral T cell maintenance. J. Clin. Investig. 2019, 129, 2856–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Tang, K.; Ma, J.; Zhou, L.; Liu, J.; Zeng, L.; Zhu, L.; Xu, P.; Chen, J.; Wei, K.; et al. Ketogenesis-generated β-hydroxybutyrate is an epigenetic regulator of CD8+ T-cell memory development. Nat. Cell Biol. 2020, 22, 18–25. [Google Scholar] [CrossRef]
- Mak, T.W.; Saunders, M.E. Bridging Innate and Adaptive Immunity: NK, γδ T, and NKT Cells; Elsevier: Amsterdam, The Netherlands, 2006; pp. 517–552. [Google Scholar]
- Tumes, D.; Hirahara, K.; Papadopoulos, M.; Shinoda, K.; Onodera, A.; Kumagai, J.; Yip, K.H.; Pant, H.; Kokubo, K.; Kiuchi, M.; et al. Ezh2 controls development of natural killer T cells, which cause spontaneous asthma-like pathology. J. Allergy Clin. Immunol. 2019, 144, 549–560.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Deng, C.; Hu, X.; Patel, B.; Fu, X.; Qiu, Y.; Brand, M.; Zhao, K.; Huang, S. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene 2012, 31, 5007–5018. [Google Scholar] [CrossRef] [Green Version]
- Hayami, S.; Kelly, J.D.; Cho, H.S.; Yoshimatsu, M.; Unoki, M.; Tsunoda, T.; Field, H.I.; Neal, D.E.; Yamaue, H.; Ponder, B.A.; et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 2011, 128, 574–586. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, P.; Yu, B. Advances toward LSD1 inhibitors for cancer therapy. Future Med. Chem. 2017, 9, 1227–1242. [Google Scholar] [CrossRef] [Green Version]
- Muthuswamy, R.; Berk, E.; Junecko, B.F.; Zeh, H.J.; Zureikat, A.H.; Normolle, D.; Luong, T.M.; Reinhart, T.A.; Bartlett, D.L.; Kalinski, P. NF-κB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res. 2012, 72, 3735–3743. [Google Scholar] [CrossRef] [Green Version]
- Loo Yau, H.; Bell, E.; Ettayebi, I.; De Almeida, F.C.; Boukhaled, G.M.; Shen, S.Y.; Allard, D.; Morancho, B.; Marhon, S.A.; Ishak, C.A.; et al. DNA hypomethylating agents increase activation and cytolytic activity of CD8+ T cells. Mol. Cell 2021, 81, 1469–1483.e8. [Google Scholar] [CrossRef]
- Henning, A.N.; Roychoudhuri, R.; Restifo, N.P. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 2018, 18, 340–356. [Google Scholar] [CrossRef]
- Philip, M.; Fairchild, L.; Sun, L.; Horste, E.L.; Camara, S.; Shakiba, M.; Scott, A.C.; Viale, A.; Lauer, P.; Merghoub, T.; et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017, 545, 452–456. [Google Scholar] [CrossRef]
- Peirs, S.; Van der Meulen, J.; Van de Walle, I.; Taghon, T.; Speleman, F.; Poppe, B.; Van Vlierberghe, P. Epigenetics in T-cell acute lymphoblastic leukemia. Immunol. Rev. 2015, 263, 50–67. [Google Scholar] [CrossRef]

| Regulator | Cell Types | Modification/Function | References |
|---|---|---|---|
| Dot1L | Th-2, CD8+ | H3K79me2 | [36,37] |
| Menin | Th-2 | Menin/TrxG complex promotes H3K4me3 | [38,39,40,41,42] |
| Cxxc1 | Th-1, Th-2, Th-17 | Cxxc1/TrxG complex inhibits/promotes H3K4me3 in gene specific manner | [43,44,45,46] |
| ATF7ip | Th-17 | ATF7ip/SETDB1 complex promotes H3K9me3 | [47,48] |
| Utx | Th-17, iNKT | Histone demethylase | [49,50,51,52] |
| Hdac3 | CD4+, CD8+, iNKT | Histone deacetylation | [53,54,55] |
| Tcf1/ Lef1 | CD4+, CD8+ | Histone deacetylation | [56] |
| Gcn5 | Th-1, Th-17, iNKT | Histone acetylation | [57,58] |
| Rcor1 | Treg | Rcor1 is a part of CoREST complex which mediates histone deacetylation | [59,60] |
| Ezh2 | CD8+, iNKT | H3K27me3 | [37,50,61] |
| Lsd1 | CD8+ CTLs | H3K4 and H3K79 demethylase | [62,63,64] |
| Tox | Exhausted CD8+ | Binds to epigenetic regulators to change gene expression | [65,66,67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutta, A.; Venkataganesh, H.; Love, P.E. New Insights into Epigenetic Regulation of T Cell Differentiation. Cells 2021, 10, 3459. https://doi.org/10.3390/cells10123459
Dutta A, Venkataganesh H, Love PE. New Insights into Epigenetic Regulation of T Cell Differentiation. Cells. 2021; 10(12):3459. https://doi.org/10.3390/cells10123459
Chicago/Turabian StyleDutta, Avik, Harini Venkataganesh, and Paul E. Love. 2021. "New Insights into Epigenetic Regulation of T Cell Differentiation" Cells 10, no. 12: 3459. https://doi.org/10.3390/cells10123459
APA StyleDutta, A., Venkataganesh, H., & Love, P. E. (2021). New Insights into Epigenetic Regulation of T Cell Differentiation. Cells, 10(12), 3459. https://doi.org/10.3390/cells10123459







