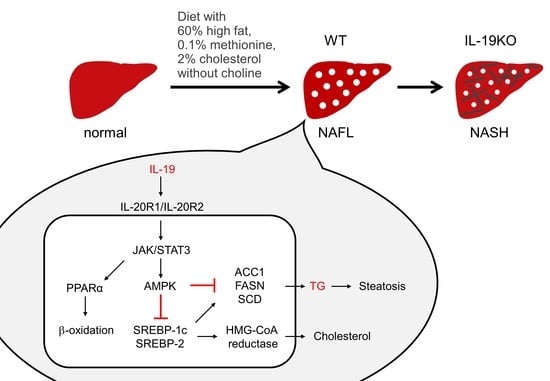
IL-19 Contributes to the Development of Nonalcoholic Steatohepatitis by Altering Lipid Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. NASH Induction
2.3. Kupffer Cells, Hepatocytes Isolation and Immunocytochemical Analysis
2.4. In Vitro Steatosis Assay in HepG2 Cells
2.5. Reporter Cells
2.6. Aminotransferase and Lactose Dehydrogenase Levels
2.7. Liver Histology and Immunohistochemical Analysis
2.8. RNA Isolation and Quantitative Real-Time PCR (QPCR)
2.9. Western Blot
2.10. Statistical Analysis
3. Results
3.1. IL-19 Expression in the Kupffer Cells
3.2. Body and Liver Weights
3.3. ALT and AST
3.4. Liver Histology
3.5. IL-19 Expression and Factors Involved in NASH Progression
3.6. Effect of IL-19 on In Vitro Steatosis Model in HepG2 Cells
3.7. Effect of IL-19 on the Response in Hepatocyte
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | acetyl-CoA carboxylase |
| ACLY | ATP citrate lyase |
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| CDAHFD | 60% high fat, 0.1% methionine, and 2% cholesterol without choline |
| FASN | fatty acid synthase |
| H&E | hematoxylin and eosin |
| IL | interleukin |
| KO | gene-deficient |
| NAFLD | nonalcoholic fatty liver disease |
| NAFL | nonalcoholic fatty liver |
| NASH | nonalcoholic steatohepatitis |
| PPAR | peroxisome proliferator-activated receptor |
| PPRE | PPAR-response element |
| QPCR | quantitative Real-Time PCR |
| SCD | stearoyl-CoA desaturase |
| SD | standard diet |
| SIE | sis-inducible element |
| SREBP | sterol regulatory element-binding protein |
| TG | triglyceride |
| WT | wild-type |
References
- Azuma, Y.T.; Nakajima, H.; Takeuchi, T. IL-19 as a potential therapeutic in autoimmune and inflammatory diseases. Curr. Pharm. Des. 2011, 17, 3776–3780. [Google Scholar] [CrossRef]
- Matsuo, Y.; Azuma, Y.T.; Kuwamura, M.; Kuramoto, N.; Nishiyama, K.; Yoshida, N.; Ikeda, Y.; Fujimoto, Y.; Nakajima, H.; Takeuchi, T. Interleukin 19 reduces inflammation in chemically induced experimental colitis. Int. Immunopharmacol. 2015, 29, 468–475. [Google Scholar] [CrossRef]
- Azuma, Y.T.; Matsuo, Y.; Kuwamura, M.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.J.; Nakajima, H.; Karow, M.; Takeuchi, T. Interleukin-19 protects mice from innate-mediated colonic inflammation. Inflamm. Bowel Dis. 2010, 16, 1017–1028. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Azuma, Y.T.; Matsuo, Y.; Kuwamura, M.; Kuramoto, N.; Miki, M.; Azuma, N.; Teramoto, M.; Nishiyama, K.; Izawa, T.; et al. Interleukin-19 contributes as a protective factor in experimental Th2-mediated colitis. Naunyn Schmiedebergs Arch. Pharmacol. 2017, 390, 261–268. [Google Scholar] [CrossRef][Green Version]
- Fujimoto, Y.; Fujita, T.; Kuramoto, N.; Kuwamura, M.; Izawa, T.; Nishiyama, K.; Yoshida, N.; Nakajima, H.; Takeuchi, T.; Azuma, Y.T. The role of interleukin-19 in contact hypersensitivity. Biol. Pharm. Bull. 2018, 41, 182–189. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Aono, K.; Azuma, Y.T. The clarified role of interleukin-19 in the inflammatory bowel disease and hypersensitivity: Insights from animal models and humans. J. Vet. Med. Sci. 2019, 81, 1067–1073. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Kuramoto, N.; Yoneyama, M.; Azuma, Y.T. Interleukin-19 as an immunoregulatory cytokine. Curr. Mol. Pharmacol. 2021, 14, 191–199. [Google Scholar] [CrossRef]
- Wegermann, K.; Suzuki, A.; Mavis, A.M.; Abdelmalek, M.F.; Diehl, A.M.; Moylan, C.A. Tackling Nonalcoholic Fatty Liver Disease: Three Targeted Populations. Hepatology 2021, 73, 1199–1206. [Google Scholar] [CrossRef]
- Balakrishnan, M.; Loomba, R. The Role of Noninvasive Tests for Differentiating NASH From NAFL and Diagnosing Advanced Fibrosis Among Patients With NAFLD. J. Clin. Gastroenterol. 2020, 54, 107–113. [Google Scholar] [CrossRef]
- Fujii, H.; Kawada, N.; Japan Study Group of Nafld Jsg-Nafld. The Role of Insulin Resistance and Diabetes in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2020, 21, 3863. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Tabas, I.; Pajvani, U.B. Mechanisms of Fibrosis Development in Nonalcoholic Steatohepatitis. Gastroenterology 2020, 158, 1913–1928. [Google Scholar] [CrossRef] [PubMed]
- Caparrós, E.; Francés, R. The Interleukin-20 Cytokine Family in Liver Disease. Front. Immunol. 2018, 9, 1155. [Google Scholar] [CrossRef]
- Kirsch, R.; Clarkson, V.; Shephard, E.G.; Marais, D.A.; Jaffer, M.A.; Woodburne, V.E.; Kirsch, R.E.; de Hall, P.L. Rodent nutritional model of non-alcoholic steatohepatitis: Species, strain and sex difference studies. J. Gastroenterol. Hepatol. 2003, 18, 1272–1282. [Google Scholar] [CrossRef]
- Shiri-Sverdlov, R.; Wouters, K.; van Gorp, P.J.; Gijbels, M.J.; Noel, B.; Buffat, L.; Staels, B.; Maeda, N.; van Bilsen, M.; Hofker, M.H. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. J. Hepatol. 2006, 44, 732–741. [Google Scholar] [CrossRef]
- Kita, Y.; Takamura, T.; Misu, H.; Ota, T.; Kurita, S.; Takeshita, Y.; Uno, M.; Matsuzawa-Nagata, N.; Kato, K.; Ando, H.; et al. Metformin prevents and reverses inflammation in a non-diabetic mouse model of nonalcoholic steatohepatitis. PLoS ONE 2012, 7, e43056. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Hada, N.; Sakamaki, Y.; Uno, A.; Shiga, T.; Tanaka, C.; Ito, T.; Katsume, A.; Sudoh, M. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int. J. Exp. Pathol. 2013, 94, 93–103. [Google Scholar] [CrossRef]
- Ioannou, G.N. The role of cholesterol in the pathogenesis of NASH. Trends Endocrinol. Metab. 2016, 27, 84–95. [Google Scholar] [CrossRef]
- Joshi-Barve, S.; Barve, S.S.; Amancherla, K.; Gobejishvili, L.; Hill, D.; Cave, M.; Hote, P.; McClain, C.J. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 2007, 46, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Kuwamura, M.; Azuma, Y.T. Deficiency of interleukin-19 exacerbates lipopolysaccharide/D-galactosamine-induced acute liver failure. J. Vet. Med. Sci. 2020, 82, 1450–1455. [Google Scholar] [CrossRef]
- Azuma, Y.T.; Nishiyama, K.; Matsuo, Y.; Kuwamura, M.; Morioka, A.; Nakajima, H.; Takeuchi, T. PPARα contributes to colonic protection in mice with DSS-induced colitis. Int. Immunopharmacol. 2010, 10, 1261–1267. [Google Scholar] [CrossRef]
- Azuma, Y.T.; Hagi, K.; Shintani, N.; Kuwamura, M.; Nakajima, H.; Hashimoto, H.; Baba, A.; Takeuchi, T. PACAP provides colonic protection against dextran sodium sulfate induced colitis. J. Cell. Physiol. 2008, 216, 111–119. [Google Scholar] [CrossRef]
- Azuma, Y.T.; Nishiyama, K.; Kita, S.; Komuro, I.; Nakajima, H.; Iwamoto, T.; Takeuchi, T. Na+/Ca2+ exchanger 2-heterozygote knockout mice display decreased acetylcholine release and altered colonic motility in vivo. Neurogastroenterol. Motil. 2012, 24, e600–e610. [Google Scholar] [CrossRef]
- Nishiyama, K.; Aono, K.; Fujimoto, Y.; Kuwamura, M.; Okada, T.; Tokumoto, H.; Izawa, T.; Okano, R.; Nakajima, H.; Takeuchi, T.; et al. Chronic kidney disease after 5/6 nephrectomy disturbs the intestinal microbiota and alters intestinal motility. J. Cell Physiol. 2019, 234, 6667–6678. [Google Scholar] [CrossRef]
- Yoneda, Y.; Azuma, Y.; Inoue, K.; Ogita, K.; Mitani, A.; Zhang, L.; Masuda, S.; Higashihara, M.; Kataoka, K. Positive correlation between prolonged potentiation of binding of double-stranded oligonucleotide probe for the transcription factor AP1 and resistance to transient forebrain ischemia in gerbil hippocampus. Neuroscience 1997, 79, 1023–1037. [Google Scholar] [CrossRef]
- Horiuchi, H.; Parajuli, B.; Wang, Y.; Azuma, Y.T.; Mizuno, T.; Takeuchi, H.; Suzumura, A. Interleukin-19 acts as a negative autocrine regulator of activated microglia. PLoS ONE 2015, 10, e0118640. [Google Scholar] [CrossRef]
- Cai, D.; Li, Y.; Zhang, K.; Zhou, B.; Guo, F.; Holm, L.; Liu, H.Y. Co-option of PPARα in the regulation of lipogenesis and fatty acid oxidation in CLA-induced hepatic steatosis. J. Cell Physiol. 2021, 236, 4387–4402. [Google Scholar] [CrossRef]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Q.; Fu, J.; Ren, R. Polysaccharides derived from natural sources regulate triglyceride and cholesterol metabolism: A review of the mechanisms. Food Funct. 2019, 10, 2330–2339. [Google Scholar] [CrossRef]
- Igal, R.A.; Sinner, D.I. Stearoyl-CoA desaturase 5 (SCD5), a delta-9 fatty acyl desaturase in search of a function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158840. [Google Scholar] [CrossRef]
- Ouyang, S.; Mo, Z.; Sun, S.; Yin, K.; Lv, Y. Emerging role of Insig-1 in lipid metabolism and lipid disorders. Clin. Chim. Acta 2020, 508, 206–212. [Google Scholar] [CrossRef]
- Wang, M.; Ma, L.J.; Yang, Y.; Xiao, Z.; Wan, J.B. n-3 Polyunsaturated fatty acids for the management of alcoholic liver disease: A critical review. Crit. Rev. Food Sci. Nutr. 2019, 59, S116–S129. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARalpha. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, J.; Shi, Y.; Ye, J.M. Discovery of Novel Anti-Diabetic Drugs by Targeting Lipid Metabolism. Curr. Drug Targets 2015, 16, 1372–1380. [Google Scholar] [CrossRef]
- Horn, C.L.; Morales, A.L.; Savard, C.; Farrell, G.C.; Ioannou, G.N. Role of Cholesterol-Associated Steatohepatitis in the Development of NASH. Hepatol. Commun. 2021, in press. [Google Scholar] [CrossRef]
- Yata, Y.; Gotwals, P.; Koteliansky, V.; Rockey, D.C. Dose-dependent inhibition of hepatic fibrosis in mice by a TGF-beta soluble receptor: Implications for antifibrotic therapy. Hepatology 2002, 35, 1022–1030. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef]
- Zhong, X.; Huang, M.; Kim, H.G.; Zhang, Y.; Chowdhury, K.; Cai, W.; Saxena, R.; Schwabe, R.F.; Liangpunsakul, S.; Dong, X.C. SIRT6 protects against liver fibrosis by deacetylation and suppression of SMAD3 in hepatic stellate cells. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 341–364. [Google Scholar] [CrossRef]
- Roehlen, N.; Crouchet, E.; Baumert, T.F. Liver fibrosis: Mechanistic concepts and therapeutic perspectives. Cells 2020, 9, 875. [Google Scholar] [CrossRef]
- El-Kehdy, H.; Najar, M.; De Kock, J.; Agha, D.M.; Rogiers, V.; Merimi, M.; Lagneaux, L.; Sokal, E.M.; Najimi, M. Inflammation differentially modulates the biological features of adult derived human liver stem/progenitor cells. Cells 2020, 9, 1640. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azuma, Y.-T.; Fujita, T.; Izawa, T.; Hirota, K.; Nishiyama, K.; Ikegami, A.; Aoyama, T.; Ike, M.; Ushikai, Y.; Kuwamura, M.; et al. IL-19 Contributes to the Development of Nonalcoholic Steatohepatitis by Altering Lipid Metabolism. Cells 2021, 10, 3513. https://doi.org/10.3390/cells10123513
Azuma Y-T, Fujita T, Izawa T, Hirota K, Nishiyama K, Ikegami A, Aoyama T, Ike M, Ushikai Y, Kuwamura M, et al. IL-19 Contributes to the Development of Nonalcoholic Steatohepatitis by Altering Lipid Metabolism. Cells. 2021; 10(12):3513. https://doi.org/10.3390/cells10123513
Chicago/Turabian StyleAzuma, Yasu-Taka, Takashi Fujita, Takeshi Izawa, Kana Hirota, Kazuhiro Nishiyama, Airi Ikegami, Tomoko Aoyama, Mikihito Ike, Yumi Ushikai, Mitsuru Kuwamura, and et al. 2021. "IL-19 Contributes to the Development of Nonalcoholic Steatohepatitis by Altering Lipid Metabolism" Cells 10, no. 12: 3513. https://doi.org/10.3390/cells10123513
APA StyleAzuma, Y.-T., Fujita, T., Izawa, T., Hirota, K., Nishiyama, K., Ikegami, A., Aoyama, T., Ike, M., Ushikai, Y., Kuwamura, M., Fujii, H., & Tsuneyama, K. (2021). IL-19 Contributes to the Development of Nonalcoholic Steatohepatitis by Altering Lipid Metabolism. Cells, 10(12), 3513. https://doi.org/10.3390/cells10123513