Adhesion GPCR GPR56 Expression Profiling in Human Tissues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
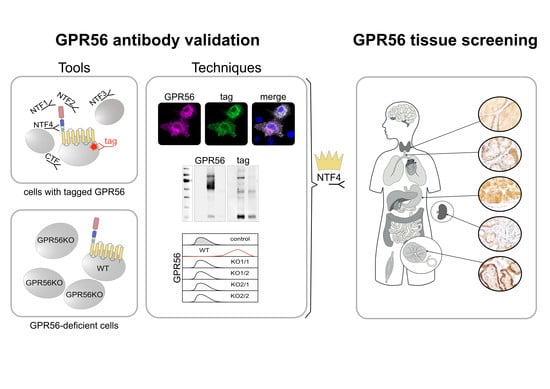
2.2. Antibodies (Abs), Plasmids, and Cell Lines
2.3. Generation of GPR56KO Clones
2.4. Immunolabeling of Cells and Tissues
2.4.1. Attached Cells and Cryosections
2.4.2. Paraffin-Embedded Cells and Tissues
2.5. Flow Cytometry
2.6. Western Blot Analysis
2.7. qRT-PCR and RNA-Sequencing
3. Results
3.1. Validating GPR56 Abs with Cells Expressing Tagged GPR56
3.2. Validating GPR56 Abs Using ADGRG1-Deficient Cells
3.3. GPR56 Is Mainly Present in Non-Squamous Secreting Epithelia
3.4. Microglia Express GPR56 in Adult Brains
3.5. Cellular GPR56 Localization Varies Depending on Cell Type and Context
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| adhesion GPCR | adhesion G-protein-coupled receptor |
| Ab | antibody |
| CNS | central nervous system |
| CTF | C-terminal fragment |
| ECD | extracellular domain |
| GPCR | G-protein-coupled receptor |
| GAIN | GPCR autoproteolysis-inducing |
| GPS | GPCR proteolysis site |
| ICD | intracellular domain |
| KO | knock-out |
| NTF | N-terminal fragment |
| WT | wild type |
References
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Yang, L.; Begum, S.; Xu, L. GPR56 is essential for testis development and male fertility in mice. Dev. Dyn. 2010, 239, 3358–3367. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.-M.; van de Garde, M.D.B.; Cheng, K.-F.; Baars, P.A.; Remmerswaal, E.B.M.; van Lier, R.A.W.; Mackay, C.R.; Lin, H.-H.; Hamann, J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011, 90, 735–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.P.; Wrann, C.D.; Rao, R.R.; Nair, S.K.; Jedrychowski, M.P.; You, J.-S.; Martínez-Redondo, V.; Gygi, S.P.; Ruas, J.L.; Hornberger, T.A.; et al. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc. Natl. Acad. Sci. USA 2014, 111, 15756–15761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piao, X.; Hill, R.S.; Bodell, A.; Chang, B.S.; Basel-Vanagaite, L.; Straussberg, R.; Dobyns, W.B.; Qasrawi, B.; Winter, R.M.; Innes, A.M.; et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 2004, 303, 2033–2036. [Google Scholar] [CrossRef] [Green Version]
- Bae, B.-I.; Tietjen, I.; Atabay, K.D.; Evrony, G.D.; Johnson, M.B.; Asare, E.; Wang, P.P.; Murayama, A.Y.; Im, K.; Lisgo, S.N.; et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 2014, 343, 764–768. [Google Scholar] [CrossRef] [Green Version]
- Giera, S.; Deng, Y.; Luo, R.; Ackerman, S.D.; Mogha, A.; Monk, K.R.; Ying, Y.; Jeong, S.-J.; Makinodan, M.; Bialas, A.R.; et al. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 2015, 6, 6121. [Google Scholar] [CrossRef] [Green Version]
- Ackerman, S.D.; Luo, R.; Poitelon, Y.; Mogha, A.; Harty, B.L.; D’Rozario, M.; Sanchez, N.E.; Lakkaraju, A.K.K.; Gamble, P.; Li, J.; et al. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J. Exp. Med. 2018, 215, 941–961. [Google Scholar] [CrossRef]
- Li, T.; Chiou, B.; Gilman, C.K.; Luo, R.; Koshi, T.; Yu, D.; Oak, H.C.; Giera, S.; Johnson-Venkatesh, E.; Muthukumar, A.K.; et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020, 39, e104136. [Google Scholar] [CrossRef] [PubMed]
- Belzeaux, R.; Gorgievski, V.; Fiori, L.M.; Lopez, J.P.; Grenier, J.; Lin, R.; Nagy, C.; Ibrahim, E.C.; Gascon, E.; Courtet, P.; et al. GPR56/ADGRG1 is associated with response to antidepressant treatment. Nat. Commun. 2020, 11, 1635. [Google Scholar] [CrossRef]
- Hamann, J.; Aust, G.; Arac, D.; Engel, F.B.; Formstone, C.; Frederiksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCI. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef] [PubMed]
- Salzman, G.S.; Ackerman, S.D.; Ding, C.; Koide, A.; Leon, K.; Luo, R.; Stoveken, H.M.; Fernandez, C.G.; Tall, G.G.; Piao, X.; et al. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron 2016, 91, 1292–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.E.; Han, J.M.; Park, C.R.; Shin, K.J.; Ahn, C.; Seong, J.Y.; Hwang, J.I. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J. Cancer Res. Clin. Oncol. 2010, 138, 47–53. [Google Scholar] [CrossRef]
- Zendman, A.J.; Cornelissen, I.M.; Weidle, U.H.; Ruiter, D.J.; van Muijen, G.N. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett. 1999, 446, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Parker, R.M.; Darby, K.; Eyre, H.J.; Copeland, N.G.; Crawford, J.; Gilbert, D.J.; Sutherland, G.R.; Jenkins, N.A.; Herzog, H. GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics 1999, 55, 296–305. [Google Scholar] [CrossRef]
- Chiang, N.Y.; Hsiao, C.C.; Huang, Y.S.; Chen, H.Y.; Hsieh, I.J.; Chang, G.W.; Lin, H.H. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J. Biol. Chem. 2011, 286, 14215–14225. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.W.; Hsiao, C.C.; Peng, Y.M.; Vieira Braga, F.A.; Kragten, N.A.; Remmerswaal, E.B.; van de Garde, M.D.; Straussberg, R.; Konig, G.M.; Kostenis, E.; et al. The Adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 2016, 15, 1757–1770. [Google Scholar] [CrossRef] [Green Version]
- Pabst, C.; Bergeron, A.; Lavallee, V.P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016, 127, 2018–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shashidhar, S.; Lorente, G.; Nagavarapu, U.; Nelson, A.; Kuo, J.; Cummins, J.; Nikolich, K.; Urfer, R.; Foehr, E.D. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene 2005, 24, 1673–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Arendt, T.; Morawski, M.; Gärtner, U.; Fröhlich, N.; Schulze, F.; Wohmann, N.; Jäger, C.; Eisenlöffel, C.; Gertz, H.-J.; Mueller, W.; et al. Inhomogeneous distribution of Alzheimer pathology along the isocortical relief. Are cortical convolutions an Achilles heel of evolution? Brain Pathol. 2017, 27, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, I.; Reimann, K.; Jankuhn, S.; Kirilina, E.; Stieler, J.; Sonntag, M.; Meijer, J.; Weiskopf, N.; Reinert, T.; Arendt, T.; et al. Cell specific quantitative iron mapping on brain slices by immuno-µPIXE in healthy elderly and Parkinson’s disease. Acta Neuropathol. Commun. 2021, 9, 47. [Google Scholar] [CrossRef]
- Takaki, T. An epithelial cell line (KNS-62) derived from a brain metastasis of bronchial squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 1980, 96, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veninga, H.; Becker, S.; Hoek, R.M.; Wobus, M.; Wandel, E.; van der Kaa, J.; van der Valk, M.; de Vos, A.F.; Haase, H.; Owens, B.; et al. Analysis of CD97 expression and manipulation: Antibody treatment but not gene targeting curtails granulocyte migration. J. Immunol. 2008, 181, 6574–6583. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Fortney, K.; Dobriban, E.; Garagnani, P.; Pirazzini, C.; Monti, D.; Mari, D.; Atzmon, G.; Barzilai, N.; Franceschi, C.; Owen, A.B.; et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet. 2015, 11, e1005728. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Olaniru, O.E.; Pingitore, A.; Giera, S.; Piao, X.; Castañera González, R.; Jones, P.M.; Persaud, S.J. The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve β-cell function. Cell. Mol. Life Sci. 2018, 75, 4007–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knierim, A.B.; Rothe, J.; Cakir, M.V.; Lede, V.; Wilde, C.; Liebscher, I.; Thor, D.; Schoneberg, T. Genetic basis of functional variability in adhesion G protein-coupled receptors. Sci. Rep. 2019, 9, 11036. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jin, Z.; Koirala, S.; Bu, L.; Xu, L.; Hynes, R.O.; Walsh, C.A.; Corfas, G.; Piao, X. GPR56 regulates pial basement membrane integrity and cortical lamination. J. Neurosci. 2008, 28, 5817–5826. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Tietjen, I.; Bu, L.; Liu-Yesucevitz, L.; Gaur, S.K.; Walsh, C.A.; Piao, X. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum. Mol. Genet. 2007, 16, 1972–1985. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Jeong, S.J.; Jin, Z.; Strokes, N.; Li, S.; Piao, X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 2011, 108, 12925–12930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, N.-Y.; Chang, G.-W.; Huang, Y.-S.; Peng, Y.-M.; Hsiao, C.-C.; Kuo, M.-L.; Lin, H.-H. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 2016, 129, 2156–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, G.; Sakitani, K.; Wang, H.; Jin, Y.; Dubeykovskiy, A.; Worthley, D.L.; Tailor, Y.; Wang, T.C. The G-protein coupled receptor 56, expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget 2017, 8, 40606–40619. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Hsiao, C.-C.; Sankowski, R.; Prinz, M.; Smolders, J.; Huitinga, I.; Hamann, J. GPCRomics of homeostatic and disease-associated human microglia. Front. Immunol. 2021, 12, 674189. [Google Scholar] [CrossRef]
- Ehrlich, A.T.; Maroteaux, G.; Robe, A.; Venteo, L.; Nasseef, M.T.; van Kempen, L.C.; Mechawar, N.; Turecki, G.; Darcq, E.; Kieffer, B.L. Expression map of 78 brain-expressed mouse orphan GPCRs provides a translational resource for neuropsychiatric research. Commun. Biol. 2018, 1, 102. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- van der Poel, M.; Ulas, T.; Mizee, M.R.; Hsiao, C.-C.; Miedema, S.S.M.; Adelia; Schuurman, K.G.; Helder, B.; Tas, S.W.; Schultze, J.L.; et al. Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 2019, 10, 1139. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaiser, F.; Morawski, M.; Krohn, K.; Rayes, N.; Hsiao, C.-C.; Quaas, M.; Aust, G. Adhesion GPCR GPR56 Expression Profiling in Human Tissues. Cells 2021, 10, 3557. https://doi.org/10.3390/cells10123557
Kaiser F, Morawski M, Krohn K, Rayes N, Hsiao C-C, Quaas M, Aust G. Adhesion GPCR GPR56 Expression Profiling in Human Tissues. Cells. 2021; 10(12):3557. https://doi.org/10.3390/cells10123557
Chicago/Turabian StyleKaiser, Fyn, Markus Morawski, Knut Krohn, Nada Rayes, Cheng-Chih Hsiao, Marianne Quaas, and Gabriela Aust. 2021. "Adhesion GPCR GPR56 Expression Profiling in Human Tissues" Cells 10, no. 12: 3557. https://doi.org/10.3390/cells10123557
APA StyleKaiser, F., Morawski, M., Krohn, K., Rayes, N., Hsiao, C.-C., Quaas, M., & Aust, G. (2021). Adhesion GPCR GPR56 Expression Profiling in Human Tissues. Cells, 10(12), 3557. https://doi.org/10.3390/cells10123557