MiR-200c-3p Contrasts PD-L1 Induction by Combinatorial Therapies and Slows Proliferation of Epithelial Ovarian Cancer through Downregulation of β-Catenin and c-Myc
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biopsies and Cell Lines
2.2. RNA Extraction from EOC Biopsies and Cell Lines
2.3. Real-Time qPCR
2.4. Transfections
2.5. Cell Treatments
2.6. Colony Formation Assay
2.7. Immunoblotting
2.8. Nuclear and Cytoplasmic Protein Extraction
2.9. MiR-200c-3p and PD-L1, β-catenin, c-Myc Expression Correlation Studies in Cancer Cell Line Encyclopedia (CCLE)
2.10. Immunohistochemistry of EOC Paraffin Sections
2.11. Statistical and Bioinformatics Analyses
3. Results
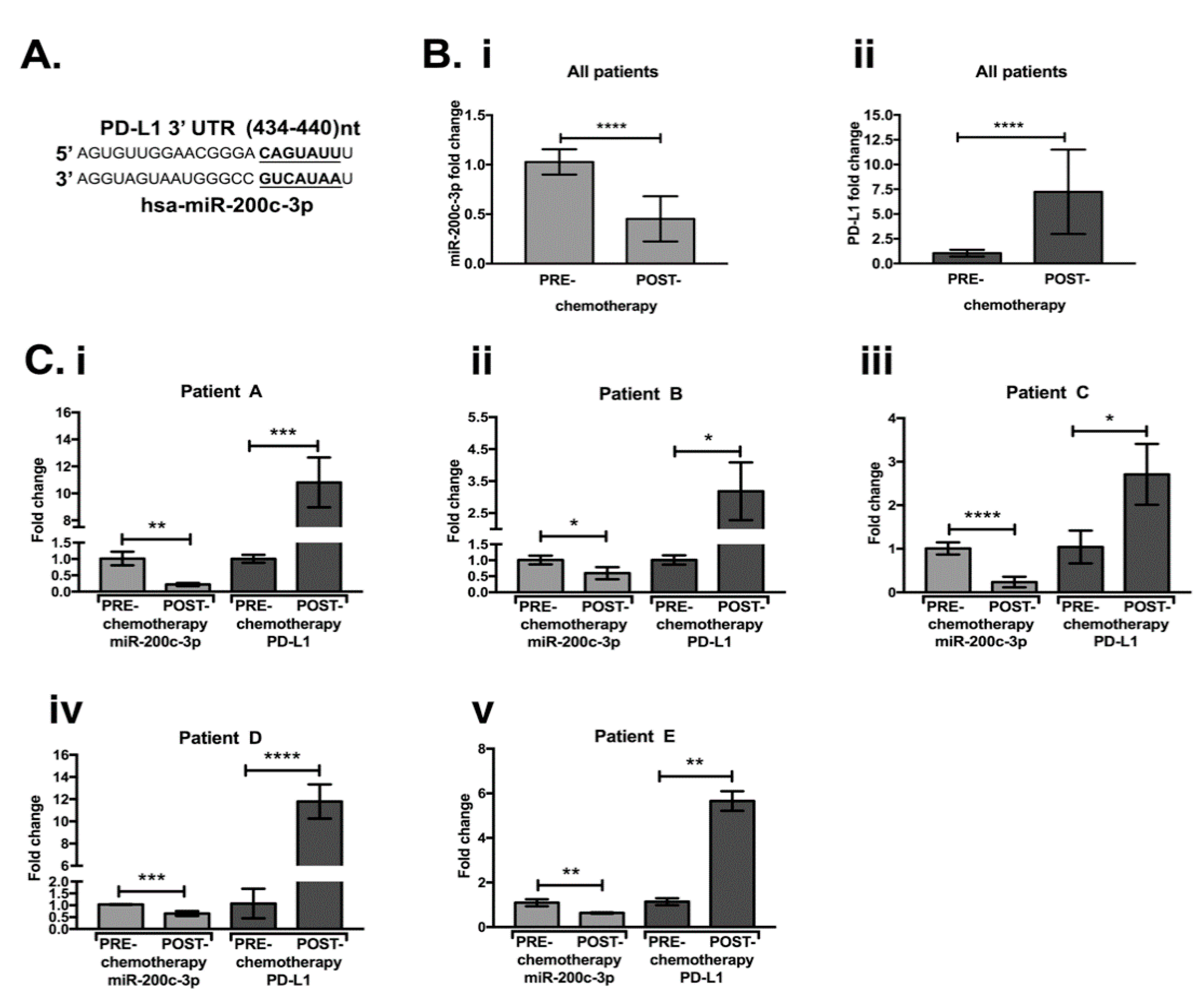
3.1. MiR-200c-3p and PDL1 Expression in EOC Biopsies before and after Chemotherapy
3.2. PD-L1 Is Targeted by miR-200c-3p in SKOV3 Cell Line
3.3. PD-L1, β-Catenin (CTNNB1) and c-Myc Expression and Its Correlation with miR-200c-3p in OC Cell Lines through CCLE Data Portal
3.4. MiR-200c-3p Inhibits Its Target Genes in Olaparib/Ionizing Radiation (IR) Treated and Untreated SKOV3 Cells
3.5. Olaparib and IR Effect on miR-200c-3p and PD-L1 mRNA in SKOV3 Cells
3.6. MiR-200c Reduces Colony Formation Ability of SKOV3 Cells
3.7. Subcellular Localization of β-Catenin and c-Myc Is Altered by miR-200c
3.8. Differential Expression of PD-L1, c-Myc, β-Catenin and CD3+ T-Cell Infiltration in Clinical Samples Obtained before and after Chemotherapy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vang, R.; Shih Ie, M.; Kurman, R.J. Ovarian low-grade and high-grade serous carcinoma: Pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv. Anat. Pathol. 2009, 16, 267–282. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian Cancers: Genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Prat, J. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication. J. Gynecol. Oncol. 2015, 26, 87–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Kaufman, M.; Cruz, A.; Thompson, J.; Reddy, S.; Bansal, N.; Cohen, J.G.; Wu, Y.; Vadgama, J.; Farias-Eisner, R. A review of the effects of healthcare disparities on the experience and survival of ovarian cancer patients of different racial and ethnic backgrounds. J. Cancer Metastasis Treat. 2019, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccarelli, S.; Megiorni, F.; Bellavia, D.; Marchese, C.; Screpanti, I.; Checquolo, S. Notch3 Targeting: A novel weapon against ovarian cancer stem cells. Stem Cells Int. 2019, 2019, 6264931. [Google Scholar] [CrossRef]
- Yan, Y.; Kumar, A.B.; Finnes, H.; Markovic, S.N.; Park, S.; Dronca, R.S.; Dong, H. Combining immune checkpoint inhibitors with conventional cancer therapy. Front. Immunol. 2018, 9, 1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA approval summary: Olaparib monotherapy in patients with deleterious germline brca-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [Green Version]
- Faraoni, I.; Graziani, G. Role of BRCA mutations in cancer treatment with Poly(ADP-ribose) Polymerase (PARP) inhibitors. Cancers 2018, 10, 487. [Google Scholar] [CrossRef] [Green Version]
- Coleman, R.L.; Fleming, G.F.; Brady, M.F.; Swisher, E.M.; Steffensen, K.D.; Friedlander, M.; Okamoto, A.; Moore, K.N.; Efrat Ben-Baruch, N.; Werner, T.L.; et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N. Engl. J. Med. 2019, 381, 2403–2415. [Google Scholar] [CrossRef]
- Bi, Y.; Verginadis, I.I.; Dey, S.; Lin, L.; Guo, L.; Zheng, Y.; Koumenis, C. Radiosensitization by the PARP inhibitor olaparib in BRCA1-proficient and deficient high-grade serous ovarian carcinomas. Gynecol. Oncol. 2018, 150, 534–544. [Google Scholar] [CrossRef]
- Boussios, S.; Karihtala, P.; Moschetta, M.; Karathanasi, A.; Sadauskaite, A.; Rassy, E.; Pavlidis, N. Combined strategies with Poly (ADP-Ribose) Polymerase (PARP) inhibitors for the treatment of ovarian cancer: A Literature Review. Diagnostics 2019, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Césaire, M.; Thariat, J.; Candéias, S.M.; Stefan, D.; Saintigny, Y.; Chevalier, F. Combining PARP inhibition, radiation, and immunotherapy: A possible strategy to improve the treatment of cancer? Int. J. Mol. Sci. 2018, 19, 3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L. Prognostic effect of programmed death-ligand 1 (PD-L1) in ovarian cancer: A systematic review, meta-analysis and bioinformatics study. J. Ovarian Res. 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and antitumor activity of Anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Waggoner, S.; Vidal, G.A.; Mita, M.; Moroney, J.W.; Holloway, R.; Van Le, L.; Sachdev, J.C.; Chapman-Davis, E.; Colon-Otero, G.; et al. Single-Arm Phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. 2019, 5, 1141–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Zhang, Z. The prognostic value of the miR-200 family in ovarian cancer: A meta-analysis. Acta Obstet. Gynecol. Scand. 2016, 95, 505–512. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Cloonan, N. Re-thinking miRNA-mRNA interactions: Intertwining issues confound target discovery. Bioessays 2015, 37, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, E.; Faggioni, A.; Trivedi, P.; Slack, F.J. The Nefarious Nexus of Noncoding RNAs in Cancer. Int. J. Mol. Sci. 2018, 19, 2072. [Google Scholar] [CrossRef] [Green Version]
- Forterre, A.; Komuro, H.; Aminova, S.; Harada, M. A Comprehensive Review of Cancer MicroRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [Google Scholar] [CrossRef]
- Hu, X.; Macdonald, D.M.; Huettner, P.C.; Feng, Z.; El Naqa, I.M.; Schwarz, J.K.; Mutch, D.G.; Grigsby, P.W.; Powell, S.N.; Wang, X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol. Oncol. 2009, 114, 457–464. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Bendoraite, A.; Knouf, E.C.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; O’Briant, K.C.; Ventura, A.P.; Godwin, A.K.; Karlan, B.Y.; Drescher, C.W.; et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: Evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol. 2010, 116, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Jiao, A.; Sui, M.; Zhang, L.; Sun, P.; Geng, D.; Zhang, W.; Wang, X.; Li, J. MicroRNA-200c inhibits the metastasis of non-small cell lung cancer cells by targeting ZEB2, an epithelial-mesenchymal transition regulator. Mol. Med. Rep. 2016, 13, 3349–3355. [Google Scholar] [CrossRef] [PubMed]
- Rebustini, I.T.; Hayashi, T.; Reynolds, A.D.; Dillard, M.L.; Carpenter, E.M.; Hoffman, M.P. miR-200c regulates FGFR-dependent epithelial proliferation via Vldlr during submandibular gland branching morphogenesis. Development 2012, 139, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amici, S.; Ceccarelli, S.; Vescarelli, E.; Romano, F.; Frati, L.; Marchese, C.; Angeloni, A. TNFalpha modulates Fibroblast Growth Factor Receptor 2 gene expression through the pRB/E2F1 pathway: Identification of a non-canonical E2F binding motif. PLoS ONE 2013, 8, e61491. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Zhang, X.; Feng, X.; Fan, X.; Jin, Z. The crosstalk between microRNAs and the Wnt/β-catenin signaling pathway in cancer. Oncotarget 2017, 8, 14089–14106. [Google Scholar] [CrossRef] [Green Version]
- Grzywa, T.M.; Klicka, K.; Włodarski, P.K. Regulators at Every Step-How microRNAs Drive Tumor Cell Invasiveness and Metastasis. Cancers 2020, 12, 3709. [Google Scholar] [CrossRef]
- Bodnar, L.; Stanczak, A.; Cierniak, S.; Smoter, M.; Cichowicz, M.; Kozlowski, W.; Szczylik, C.; Wieczorek, M.; Lamparska-Przybysz, M. Wnt/beta-catenin pathway as a potential prognostic and predictive marker in patients with advanced ovarian cancer. J. Ovarian Res. 2014, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Pyzer, A.R.; Stroopinsky, D.; Rosenblatt, J.; Anastasiadou, E.; Rajabi, H.; Washington, A.; Tagde, A.; Chu, J.H.; Coll, M.; Jiao, A.L.; et al. MUC1 inhibition leads to decrease in PD-L1 levels via upregulation of miRNAs. Leukemia 2017, 31, 2780–2790. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, E.; Stroopinsky, D.; Alimperti, S.; Jiao, A.L.; Pyzer, A.R.; Cippitelli, C.; Pepe, G.; Severa, M.; Rosenblatt, J.; Etna, M.P.; et al. Epstein-Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia 2019, 33, 132–147. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.; Slack, F.J.; Anastasiadou, E. Epstein-Barr virus: From kisses to cancer, an ingenious immune evader. Oncotarget 2018, 9, 36411–36412. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mundo, L.; Ambrosio, M.R.; Raimondi, F.; Del Porro, L.; Guazzo, R.; Mancini, V.; Granai, M.; Jim Rocca, B.; Lopez, C.; Bens, S.; et al. Molecular switch from MYC to MYCN expression in MYC protein negative Burkitt lymphoma cases. Blood Cancer J. 2019, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef]
- Lengyel, E.; Burdette, J.E.; Kenny, H.A.; Matei, D.; Pilrose, J.; Haluska, P.; Nephew, K.P.; Hales, D.B.; Stack, M.S. Epithelial ovarian cancer experimental models. Oncogene 2014, 33, 3619–3633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, E.K.; Schmidt, H.; Anastasiadou, E.; Altucci, L.; Angelini, M.; Badimon, L.; Balligand, J.L.; Benincasa, G.; Capasso, G.; Conte, F.; et al. Molecular networks in Network Medicine: Development and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1489. [Google Scholar] [CrossRef] [PubMed]
- DelloRusso, C.; Welcsh, P.L.; Wang, W.; Garcia, R.L.; King, M.C.; Swisher, E.M. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 2007, 5, 35–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vescarelli, E.; Gerini, G.; Megiorni, F.; Anastasiadou, E.; Pontecorvi, P.; Solito, L.; De Vitis, C.; Camero, S.; Marchetti, C.; Mancini, R.; et al. MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. J. Exp. Clin. Cancer Res. 2020, 39, 1–15. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef]
- Bali, A.; O’Brien, P.M.; Edwards, L.S.; Sutherland, R.L.; Hacker, N.F.; Henshall, S.M. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin. Cancer Res. 2004, 10, 5168–5177. [Google Scholar] [CrossRef] [Green Version]
- Dang, C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, G.L.; Price, M.J.; Cheadle, E.J.; Hawkins, R.E.; Gilham, D.E.; Edmondson, R.J. Ex vivo expanded tumour-infiltrating lymphocytes from ovarian cancer patients release anti-tumour cytokines in response to autologous primary ovarian cancer cells. Cancer Immunol. Immunother. 2018, 67, 1519–1531. [Google Scholar] [CrossRef] [Green Version]
- Heredia-Soto, V.; López-Guerrero, J.A.; Redondo, A.; Mendiola, M. The hallmarks of ovarian cancer: Focus on angiogenesis and micro-environment and new models for their characterisation. EJC Suppl. 2020, 15, 49–55. [Google Scholar] [CrossRef]
- Cao, Q.; Lu, K.; Dai, S.; Hu, Y.; Fan, W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 2392–2401. [Google Scholar]
- Philippou, Y.; Sjoberg, H.T.; Murphy, E.; Alyacoubi, S.; Jones, K.I.; Gordon-Weeks, A.N.; Phyu, S.; Parkes, E.E.; Gillies McKenna, W.; Lamb, A.D.; et al. Impacts of combining anti-PD-L1 immunotherapy and radiotherapy on the tumour immune microenvironment in a murine prostate cancer model. Br. J. Cancer 2020, 123, 1089–1100. [Google Scholar] [CrossRef]
- Sato, H.; Okonogi, N.; Nakano, T. Rationale of combination of anti-PD-1/PD-L1 antibody therapy and radiotherapy for cancer treatment. Int. J. Clin. Oncol. 2020, 25, 801–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lampert, E.J.; Zimmer, A.; Padget, M.; Cimino-Mathews, A.; Nair, J.R.; Liu, Y.; Swisher, E.M.; Hodge, J.W.; Nixon, A.B.; Nichols, E.; et al. Combination of PARP Inhibitor Olaparib, and PD-L1 Inhibitor Durvalumab, in Recurrent Ovarian Cancer: A Proof-of-Concept Phase II Study. Clin. Cancer Res. 2020, 26, 4268. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Xia, W.; Yamaguchi, H.; Wei, Y.; Chen, M.K.; Hsu, J.M.; Hsu, J.L.; Yu, W.H.; Du, Y.; Lee, H.H.; et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin. Cancer Res. 2017, 23, 3711–3720. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.M.; McMellen, A.; Watson, Z.L.; Aguilera, J.; Ferguson, R.; Nurmemmedov, E.; Thakar, T.; Moldovan, G.L.; Kim, H.; Cittelly, D.M.; et al. Activation of Wnt signaling promotes olaparib resistant ovarian cancer. Mol. Carcinog. 2019, 58, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Evan, G.I.; Wyllie, A.H.; Gilbert, C.S.; Littlewood, T.D.; Land, H.; Brooks, M.; Waters, C.M.; Penn, L.Z.; Hancock, D.C. Induction of apoptosis in fibroblasts by c-myc protein. Cell 1992, 69, 119–128. [Google Scholar] [CrossRef]
- Bai, J.X.; Yan, B.; Zhao, Z.N.; Xiao, X.; Qin, W.W.; Zhang, R.; Jia, L.T.; Meng, Y.L.; Jin, B.Q.; Fan, D.M.; et al. Tamoxifen represses miR-200 microRNAs and promotes epithelial-to-mesenchymal transition by up-regulating c-Myc in endometrial carcinoma cell lines. Endocrinology 2013, 154, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Hua, Y.; Qiu, H.; Hao, J.; Zou, K.; Li, Z.; Hu, S.; Guo, P.; Chen, M.; Sui, S.; et al. PD-L1 promotes tumor growth and progression by activating WIP and β-catenin signaling pathways and predicts poor prognosis in lung cancer. Cell Death Dis. 2020, 11, 506. [Google Scholar] [CrossRef]
- Nguyen, V.H.L.; Hough, R.; Bernaudo, S.; Peng, C. Wnt/β-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019, 12, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCluggage, W.G.; Lyness, R.W.; Atkinson, R.J.; Dobbs, S.P.; Harley, I.; McClelland, H.R.; Price, J.H. Morphological effects of chemotherapy on ovarian carcinoma. J. Clin. Pathol. 2002, 55, 27–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, H.; Li, G.; Wen, X.; Lv, Y.; Zhang, M.; Wei, Y.; Xie, W.; Zhong, P.; Pang, L. Prognostic significance of beta-catenin expression in patients with ovarian cancer: A meta-analysis. Gene 2018, 678, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Reyes-González, J.M.; Armaiz-Peña, G.N.; Mangala, L.S.; Valiyeva, F.; Ivan, C.; Pradeep, S.; Echevarría-Vargas, I.M.; Rivera-Reyes, A.; Sood, A.K.; Vivas-Mejía, P.E. Targeting c-MYC in Platinum-Resistant Ovarian Cancer. Mol. Cancer Ther. 2015, 14, 2260–2269. [Google Scholar] [CrossRef] [Green Version]
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009, 114, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.T.; Adams, S.F.; Tahirovic, E.; Hagemann, I.S.; Coukos, G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2012, 124, 192–198. [Google Scholar] [CrossRef] [Green Version]







| Patient | Age | Grade | FIGO | Chemotherapy Regimen | BRCA1/2 Status |
|---|---|---|---|---|---|
| A | 56 | G3 | IV | Carboplatin auc 5 a + TXL 175 b + BEVA 15 c (3 cycles) | WT |
| B | 43 | G3 | IIIC | Carboplatin auc 5 + Caelyx 30 d (3 cycles) | mutBRCA1 class 5 |
| C | 46 | G3 | IIIC | Carboplatin auc 5 + TXL 80 (3 cycles) | WT |
| D | 69 | G3 | IIIC | Carboplatin auc 5 + TXL 175 (3 cycles) | WT |
| E | 55 | n.d. | IIIC | Carboplatin auc 3 + TXL 80 (cycles n.d.) | WT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anastasiadou, E.; Messina, E.; Sanavia, T.; Mundo, L.; Farinella, F.; Lazzi, S.; Megiorni, F.; Ceccarelli, S.; Pontecorvi, P.; Marampon, F.; et al. MiR-200c-3p Contrasts PD-L1 Induction by Combinatorial Therapies and Slows Proliferation of Epithelial Ovarian Cancer through Downregulation of β-Catenin and c-Myc. Cells 2021, 10, 519. https://doi.org/10.3390/cells10030519
Anastasiadou E, Messina E, Sanavia T, Mundo L, Farinella F, Lazzi S, Megiorni F, Ceccarelli S, Pontecorvi P, Marampon F, et al. MiR-200c-3p Contrasts PD-L1 Induction by Combinatorial Therapies and Slows Proliferation of Epithelial Ovarian Cancer through Downregulation of β-Catenin and c-Myc. Cells. 2021; 10(3):519. https://doi.org/10.3390/cells10030519
Chicago/Turabian StyleAnastasiadou, Eleni, Elena Messina, Tiziana Sanavia, Lucia Mundo, Federica Farinella, Stefano Lazzi, Francesca Megiorni, Simona Ceccarelli, Paola Pontecorvi, Francesco Marampon, and et al. 2021. "MiR-200c-3p Contrasts PD-L1 Induction by Combinatorial Therapies and Slows Proliferation of Epithelial Ovarian Cancer through Downregulation of β-Catenin and c-Myc" Cells 10, no. 3: 519. https://doi.org/10.3390/cells10030519
APA StyleAnastasiadou, E., Messina, E., Sanavia, T., Mundo, L., Farinella, F., Lazzi, S., Megiorni, F., Ceccarelli, S., Pontecorvi, P., Marampon, F., Di Gioia, C. R. T., Perniola, G., Panici, P. B., Leoncini, L., Trivedi, P., Lenzi, A., & Marchese, C. (2021). MiR-200c-3p Contrasts PD-L1 Induction by Combinatorial Therapies and Slows Proliferation of Epithelial Ovarian Cancer through Downregulation of β-Catenin and c-Myc. Cells, 10(3), 519. https://doi.org/10.3390/cells10030519














