Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress
Abstract
1. Introduction
2. Material and Methods
2.1. The Turkish Accessions
2.2. Metabolomic Analyses
2.3. Drought Experiment
3. Results
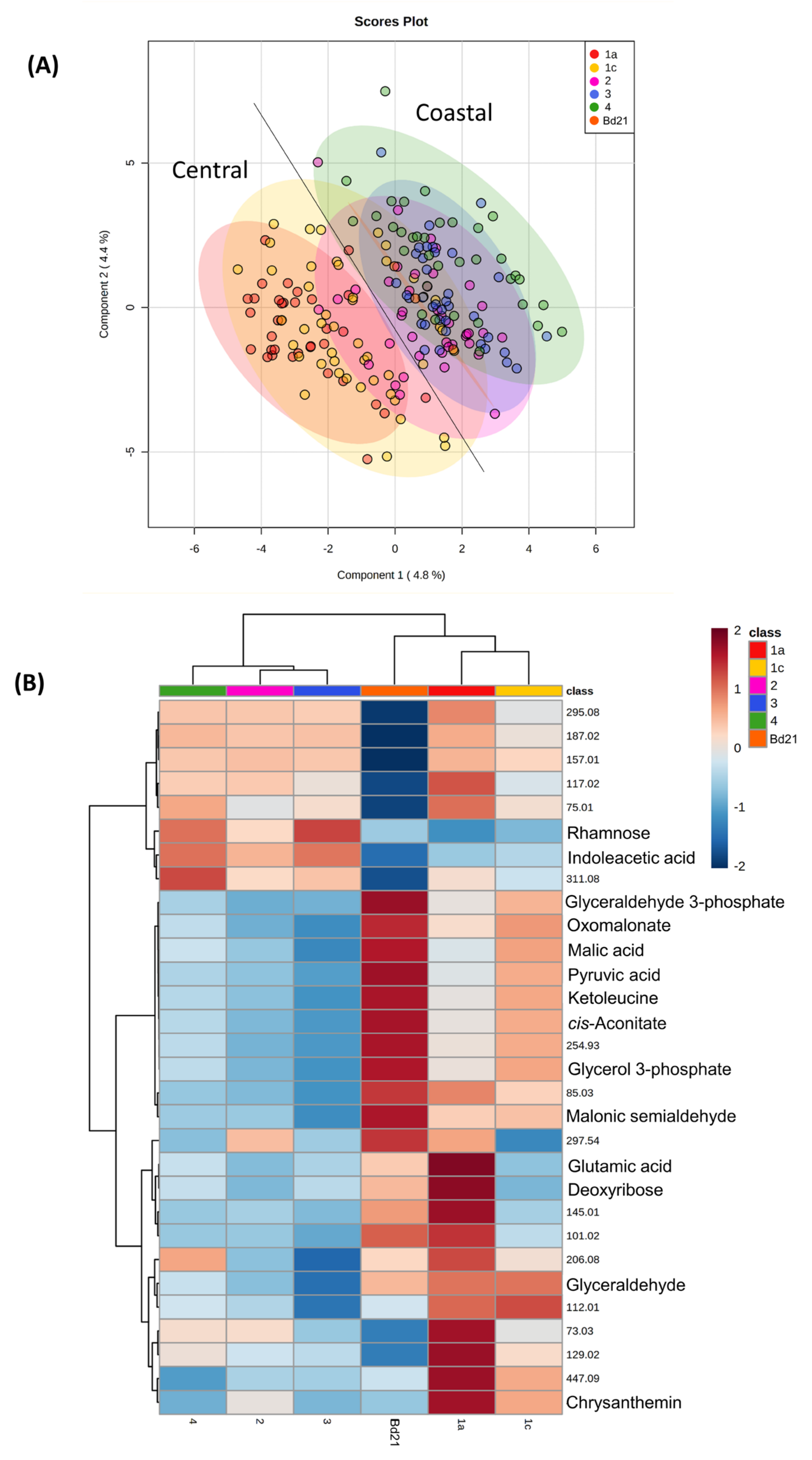
3.1. Metabolite Profiling of Brachypodium Distachyon
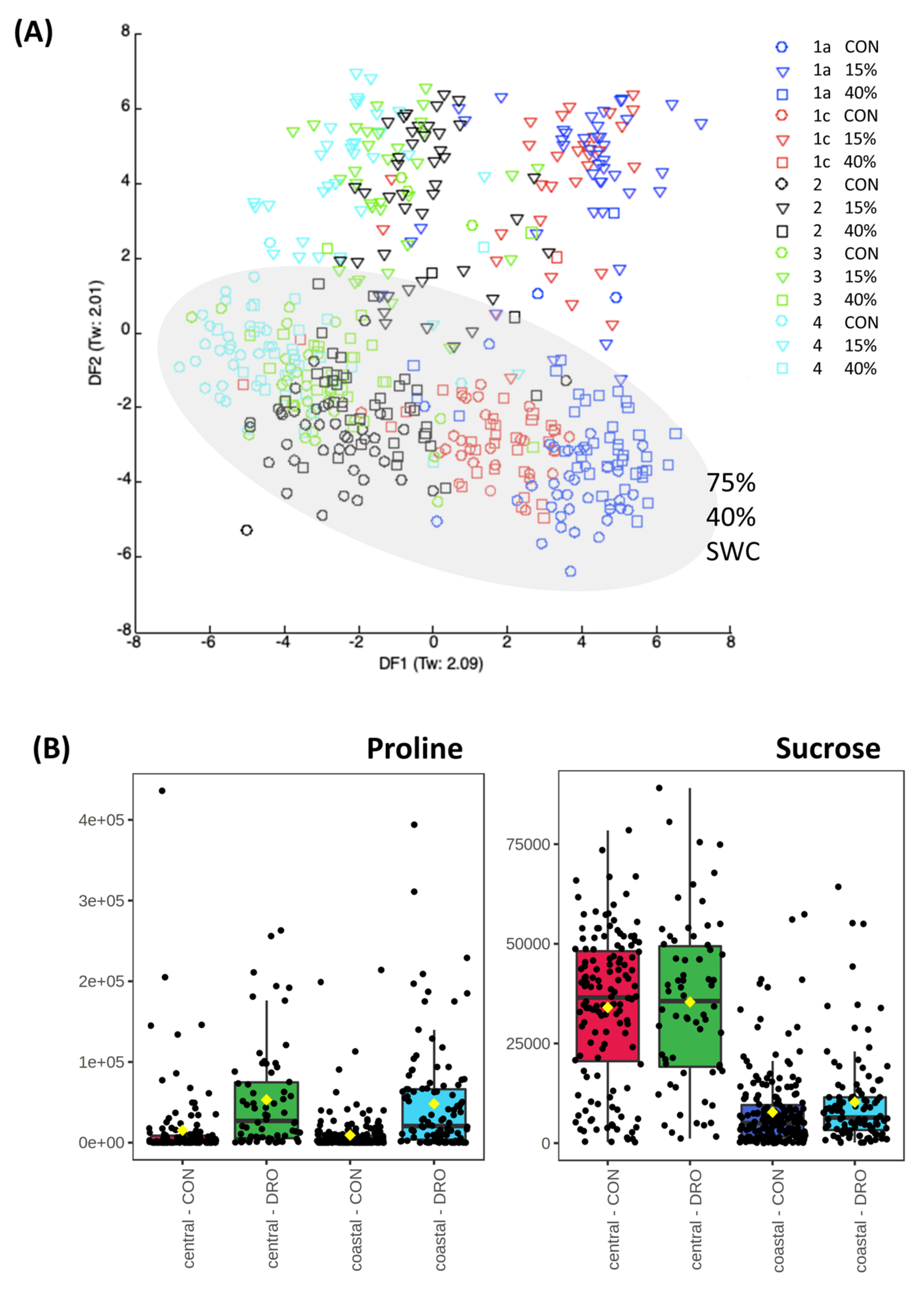
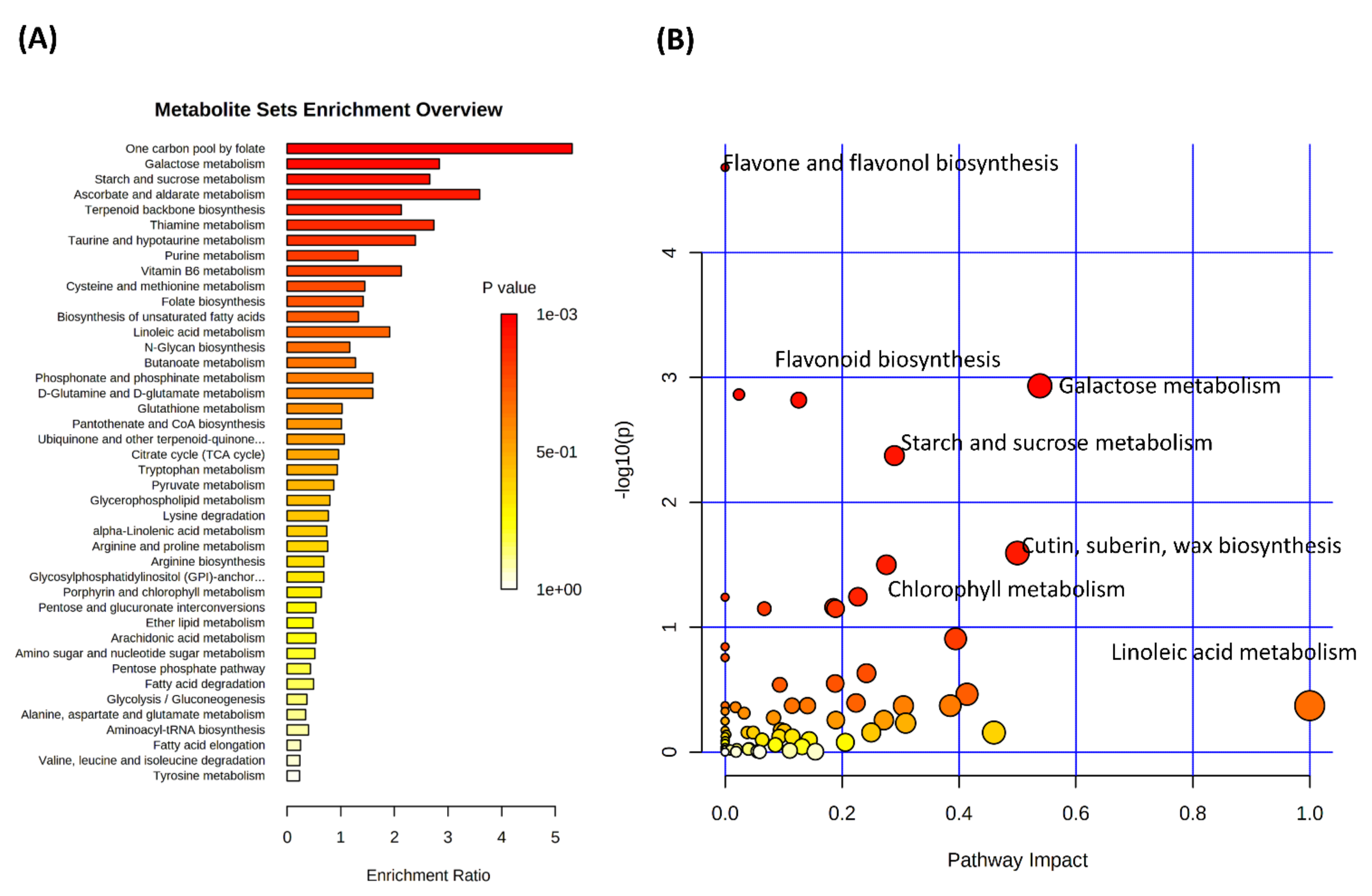
3.2. Drought Stress and Metabolome Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Draper, J.; Mur, L.A.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Allainguillaume, J.; Catalan, P.; Hasterok, R.; Jenkins, G.; Lesniewska, K.; Thomas, I.; Vogel, J. Exploiting the brachypodium tool box in cereal and grass research. New Phytol. 2011, 191, 334–347. [Google Scholar] [CrossRef]
- Brkljacic, J.; Grotewold, E.; Scholl, R.; Mockler, T.; Garvin, D.F.; Vain, P.; Brutnell, T.; Sibout, R.; Bevan, M.; Budak, H.; et al. Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 2011, 157, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.C.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 2184. [Google Scholar] [CrossRef]
- Wilson, P.B.; Streich, J.C.; Murray, K.D.; Eichten, S.R.; Cheng, R.; Aitken, N.C.; Spokas, K.; Warthmann, N.; Gordon, S.P.; Accession, C.; et al. Global diversity of the Brachypodium species complex as a resource for genome-wide association studies demonstrated for agronomic traits in response to climate. Genetics 2019, 211, 317–331. [Google Scholar] [CrossRef]
- Skalska, A.; Stritt, C.; Wyler, M.; Williams, H.W.; Vickers, M.; Han, J.; Tuna, M.; Savas Tuna, G.; Susek, K.; Swain, M.; et al. Genetic and methylome variation in turkish brachypodium distachyon accessions differentiate two geographically distinct subpopulations. Int. J. Mol. Sci. 2020, 21, 6700. [Google Scholar] [CrossRef]
- Herrera, C.M.; Bazaga, P. Genetic and epigenetic divergence between disturbed and undisturbed subpopulations of a Mediterranean shrub: A 20-year field experiment. Ecol. Evol. 2016, 6, 3832–3847. [Google Scholar] [CrossRef]
- Herrera, C.M.; Medrano, M.; Bazaga, P. Comparative spatial genetics and epigenetics of plant populations: Heuristic value and a proof of concept. Mol. Ecol. 2016, 25, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, H.; Kliebenstein, D.J.; West, M.A.; Kim, K.; van Poecke, R.; Katagiri, F.; Michelmore, R.W.; Doerge, R.W.; St Clair, D.A. Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 2007, 19, 2099–2110. [Google Scholar] [CrossRef] [PubMed]
- Clauw, P.; Coppens, F.; De Beuf, K.; Dhondt, S.; Van Daele, T.; Maleux, K.; Storme, V.; Clement, L.; Gonzalez, N.; Inze, D. Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 2015, 167, 800–816. [Google Scholar] [CrossRef]
- Kerwin, R.; Feusier, J.; Corwin, J.; Rubin, M.; Lin, C.; Muok, A.; Larson, B.; Li, B.; Joseph, B.; Francisco, M.; et al. Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. Elife 2015, 4, e05604. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Guy, C.; Kaplan, F.; Kopka, J.; Selbig, J.; Hincha, D.K. Metabolomics of temperature stress. Physiol. Plant 2008, 132, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Szarka, A.; Tomasskovics, B.; Bánhegyi, G. The ascorbate-glutathione-α-tocopherol triad in abiotic stress response. Int. J. Mol. Sci. 2012, 13, 4458–4483. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in plant stress physiology. Adv. Biochem. Eng. Biotechnol. 2018, 164, 187–236. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Innovation-Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 757. [Google Scholar] [CrossRef]
- Skirycz, A.; De Bodt, S.; Obata, T.; De Clercq, I.; Claeys, H.; De Rycke, R.; Andriankaja, M.; Van Aken, O.; Van Breusegem, F.; Fernie, A.R.; et al. Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 2010, 152, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Witt, S.; Galicia, L.; Lisec, J.; Cairns, J.; Tiessen, A.; Araus, J.L.; Palacios-Rojas, N.; Fernie, A.R. Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol. Plant 2012, 5, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Do, P.T.; Degenkolbe, T.; Erban, A.; Heyer, A.G.; Kopka, J.; Kohl, K.I.; Hincha, D.K.; Zuther, E. Dissecting rice polyamine metabolism under controlled long-term drought stress. PLoS ONE 2013, 8, e60325. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef]
- Korn, M.; Gartner, T.; Erban, A.; Kopka, J.; Selbig, J.; Hincha, D.K. Predicting arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol. Plant 2010, 3, 224–235. [Google Scholar] [CrossRef]
- Kim, J.K.; Bamba, T.; Harada, K.; Fukusaki, E.; Kobayashi, A. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J. Exp. Bot. 2007, 58, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Widodo; Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Lopez-Alvarez, D.; Zubair, H.; Beckmann, M.; Draper, J.; Catalan, P. Diversity and association of phenotypic and metabolomic traits in the close model grasses Brachypodium distachyon, B. stacei and B. hybridum. Ann. Bot. 2017, 119, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Wang, W.; Wietsma, T.W.; Winkler, T.; Lange, I.; Jansson, C.; Lange, B.M.; McDowell, N.G. Metabolic shifts associated with drought-induced senescence in Brachypodium. Plant Sci. 2019, 289, 110278. [Google Scholar] [CrossRef]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Ellis, D.I.; Heald, J.K.; Goodacre, R.; Mur, L.A. Metabolomic approaches reveal that phosphatidic and phosphatidyl glycerol phospholipids are major discriminatory non-polar metabolites in responses by Brachypodium distachyon to challenge by Magnaporthe grisea. Plant J. 2006, 46, 351–368. [Google Scholar] [CrossRef]
- Routledge, A.P.; Shelley, G.; Smith, J.V.; Talbot, N.J.; Draper, J.; Mur, L.A. Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 2004, 5, 253–265. [Google Scholar] [CrossRef]
- Fisher, L.H.; Han, J.; Corke, F.M.; Akinyemi, A.; Didion, T.; Nielsen, K.K.; Doonan, J.H.; Mur, L.A.; Bosch, M. Linking Dynamic Phenotyping with metabolite analysis to study natural variation in drought responses of Brachypodium distachyon. Front. Plant Sci. 2016, 7, 1751. [Google Scholar] [CrossRef]
- Lenk, I.; Fisher, L.H.C.; Vickers, M.; Akinyemi, A.; Didion, T.; Swain, M.; Jensen, C.S.; Mur, L.A.J.; Bosch, M. Transcriptional and metabolomic analyses indicate that cell wall properties are associated with drought tolerance in Brachypodium distachyon. Int. J. Mol. Sci. 2019, 20, 1758. [Google Scholar] [CrossRef]
- Brennan, L. Use of metabotyping for optimal nutrition. Curr. Opin. Biotechnol. 2017, 44, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Sawada, Y.; Kanaya, S.; Kuromori, T.; Kobayashi, M.; Klausnitzer, R.; Hanada, K.; Akiyama, K.; Sakurai, T.; Saito, K.; et al. Toward genome-wide metabolotyping and elucidation of metabolic system: Metabolic profiling of large-scale bioresources. J. Plant Res. 2010, 123, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Baptista, R.; Fazakerley, D.M.; Beckmann, M.; Baillie, L.; Mur, L.A.J. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Sci. Rep. 2018, 8, 5084. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Bino, R.J.; Hall, R.D.; Fiehn, O.; Kopka, J.; Saito, K.; Draper, J.; Nikolau, B.J.; Mendes, P.; Roessner-Tunali, U.; Beale, M.H.; et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004, 9, 418–425. [Google Scholar] [CrossRef]
- O’Donovan, C.B.; Walsh, M.C.; Woolhead, C.; Forster, H.; Celis-Morales, C.; Fallaize, R.; Macready, A.L.; Marsaux, C.F.M.; Navas-Carretero, S.; Rodrigo San-Cristobal, S.; et al. Metabotyping for the development of tailored dietary advice solutions in a European population: The Food4Me study. Br. J. Nutr. 2017, 118, 561–569. [Google Scholar] [CrossRef]
- Botelho, P.B.; Fioratti, C.O.; Abdalla, D.S.P.; Bertolami, M.C.; Castro, I.A. Classification of individuals with dyslipidaemia controlled by statins according to plasma biomarkers of oxidative stress using cluster analysis. Br. J. Nutr. 2010, 103, 256–265. [Google Scholar] [CrossRef]
- Wilcox, M.; Li, Q.; Sun, Y.; Stang, P.; Berlin, J.; Wang, D. Genome-wide association study for empirically derived metabolic phenotypes in the framingham heart study offspring cohort. BMC Proc. 2009, 3 (Suppl. 7), S53. [Google Scholar] [CrossRef]
- Yu, X.; Xiao, J.; Chen, S.; Yu, Y.; Ma, J.; Lin, Y.; Li, R.; Lin, J.; Fu, Z.; Zhou, Q.; et al. Metabolite signatures of diverse Camellia sinensis tea populations. Nat. Commun. 2020, 11, 5586. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Krahmer, A.; Herwig, N.; Schulz, H.; Hadian, J.; Meiners, T. Variation of secondary metabolite profile of zataria multiflora boiss. populations linked to geographic, climatic, and edaphic factors. Front. Plant Sci. 2020, 11, 969. [Google Scholar] [CrossRef]
- Saldanha, L.L.; Allard, P.M.; Afzan, A.; de Melo, F.; Marcourt, L.; Queiroz, E.F.; Vilegas, W.; Furlan, C.M.; Dokkedal, A.L.; Wolfender, J.L. Metabolomics of Myrcia bella Populations in Brazilian Savanna Reveals Strong Influence of Environmental Factors on Its Specialized Metabolism. Molecules 2020, 25, 2954. [Google Scholar] [CrossRef]
- Zeliou, K.; Koui, E.M.; Papaioannou, C.; Koulakiotis, N.S.; Iatrou, G.; Tsarbopoulos, A.; Papasotiropoulos, V.; Lamari, F.N. Metabolomic fingerprinting and genetic discrimination of four Hypericum taxa from Greece. Phytochemistry 2020, 174, 112290. [Google Scholar] [CrossRef]
- Karimi, A.; Krahmer, A.; Herwig, N.; Hadian, J.; Schulz, H.; Meiners, T. Metabolomics approaches for analyzing effects of geographic and environmental factors on the variation of root essential oils of Ferula assa-foetida L. J. Agric. Food Chem. 2020, 68, 9940–9952. [Google Scholar] [CrossRef]
- Fang, C.; Luo, J. Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism. Plant J. 2019, 97, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic profiling of soybeans (Glycine max l.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Kolmer, J.A.; Xu, W.W.; Garvin, D.F. Lr34-mediated leaf rust resistance in wheat: Transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Mol. Plant Microbe Interact. 2008, 21, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. The power of auxin in plants. Plant Physiol. 2010, 154, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Tanimoto, E. Regulation of root growth by plant hormones-Roles for auxin and gibberellin. Crit. Rev. Plant Sci. 2005, 24, 249–265. [Google Scholar] [CrossRef]
- Comas, L.H.; Becker, S.R.; Cruz, V.M.; Byrne, P.F.; Dierig, D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013, 4, 442. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.; Unal, B.T.; Garcia-Caparros, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2020. [Google Scholar] [CrossRef]
- Wiame, E.; Delpierre, G.; Collard, F.; Van Schaftingen, E. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J. Biol. Chem. 2002, 277, 42523–42529. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.L.; Jin, X.H.; Yang, Y.; Lin, Z.W.; Zhang, Y.L. The regulatory role of riboflavin in the drought tolerance of tobacco plants depends on ROS production. Plant Growth Regul. 2014, 72, 269–277. [Google Scholar] [CrossRef]
- He, M.; Min, J.W.; Kong, W.L.; He, X.H.; Li, J.X.; Peng, B.W. A review on the pharmacological effects of vitexin and isovitexin. Fitoterapia 2016, 115, 74–85. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Fellows, F.C. Biosynthesis and degradation of saccharopine, an intermediate of lysine metabolism. Biochem. J. 1973, 136, 321–327. [Google Scholar] [CrossRef]
- Qiu, X.M.; Sun, Y.Y.; Ye, X.Y.; Li, Z.G. Signaling role of glutamate in plants. Front. Plant Sci. 2019, 10, 1743. [Google Scholar] [CrossRef]
- Li, Z.G.; Ye, X.Y.; Qiu, X.M. Glutamate signaling enhances the heat tolerance of maize seedlings by plant glutamate receptor-like channels-mediated calcium signaling. Protoplasma 2019, 256, 1165–1169. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Yu, G.; Li, J.; Wang, Y.; Wang, X. Comprehensive multiomics analysis reveals key roles of NACs in plant growth and development and its environmental adaption mechanism by regulating metabolite pathways. Genomics 2020, 112, 4897–4911. [Google Scholar] [CrossRef]
- Sharma, S.; Sahu, R.; Navathe, S.; Mishra, V.K.; Chand, R.; Singh, P.K.; Joshi, A.K.; Pandey, S.P. Natural variation in elicitation of defense-signaling associates to field resistance against the spot blotch disease in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 636. [Google Scholar] [CrossRef] [PubMed]
- Allevato, D.M.; Kiyota, E.; Mazzafera, P.; Nixon, K.C. Ecometabolomic analysis of wild populations of Pilocarpus pennatifolius (Rutaceae) using unimodal analyses. Front. Plant Sci. 2019, 10, 258. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalska, A.; Beckmann, M.; Corke, F.; Savas Tuna, G.; Tuna, M.; Doonan, J.H.; Hasterok, R.; Mur, L.A.J. Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress. Cells 2021, 10, 683. https://doi.org/10.3390/cells10030683
Skalska A, Beckmann M, Corke F, Savas Tuna G, Tuna M, Doonan JH, Hasterok R, Mur LAJ. Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress. Cells. 2021; 10(3):683. https://doi.org/10.3390/cells10030683
Chicago/Turabian StyleSkalska, Aleksandra, Manfred Beckmann, Fiona Corke, Gulsemin Savas Tuna, Metin Tuna, John H. Doonan, Robert Hasterok, and Luis A. J. Mur. 2021. "Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress" Cells 10, no. 3: 683. https://doi.org/10.3390/cells10030683
APA StyleSkalska, A., Beckmann, M., Corke, F., Savas Tuna, G., Tuna, M., Doonan, J. H., Hasterok, R., & Mur, L. A. J. (2021). Metabolomic Variation Aligns with Two Geographically Distinct Subpopulations of Brachypodium Distachyon before and after Drought Stress. Cells, 10(3), 683. https://doi.org/10.3390/cells10030683







