Muscle Glycogen Phosphorylase and Its Functional Partners in Health and Disease
Abstract
:1. Introduction
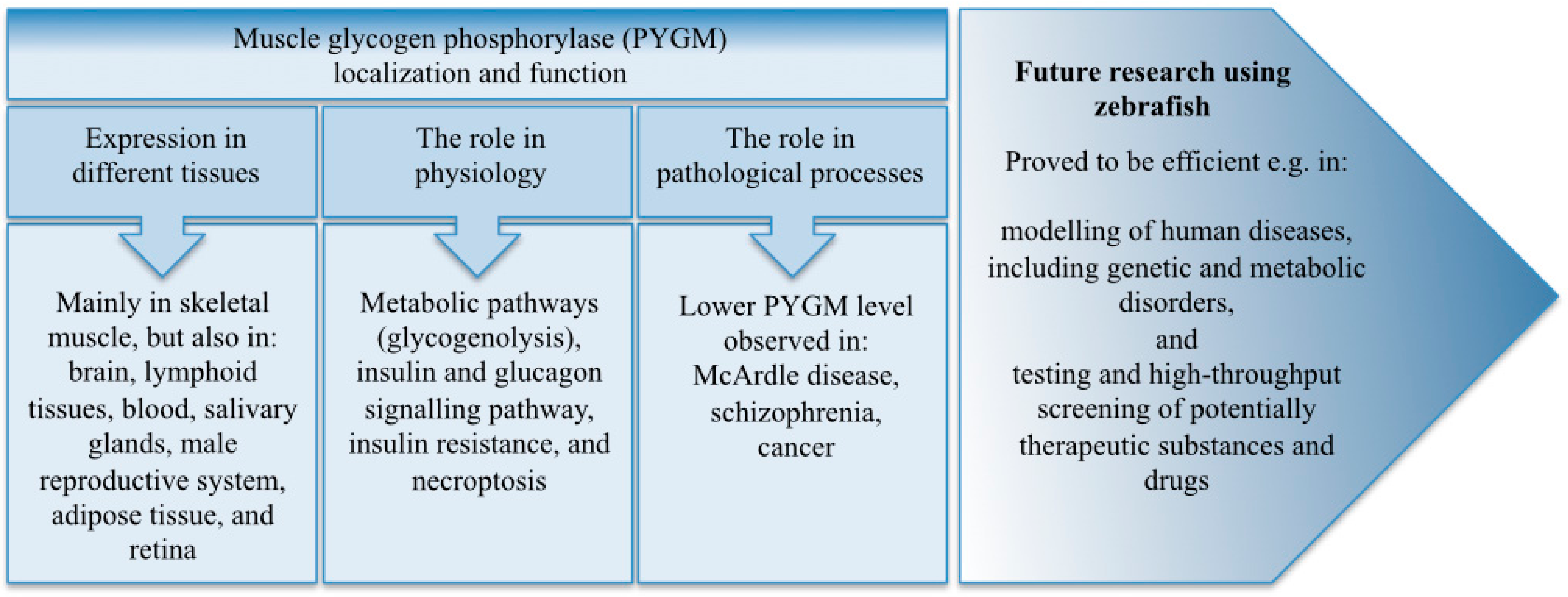
2. PYGM Expression in Different Tissues and Organs
3. The Biological Importance of PYGM
3.1. The Role of PYGM in Physiology
3.2. The Role of PYGM in Pathological Processes
3.2.1. Muscle Glycogen Phosphorylase Deficiency (McArdle Disease)
3.2.2. PYGM in Schizophrenia
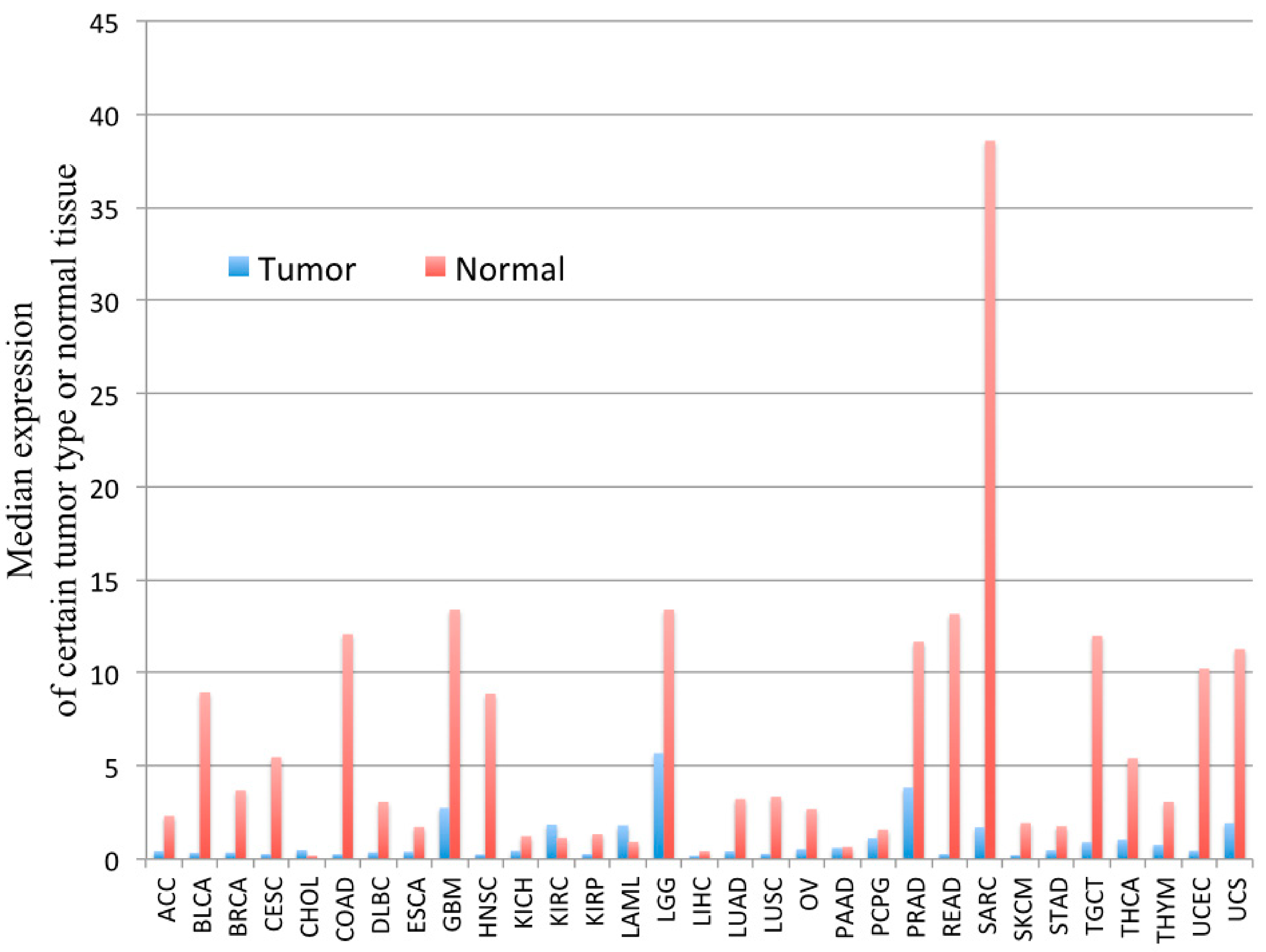
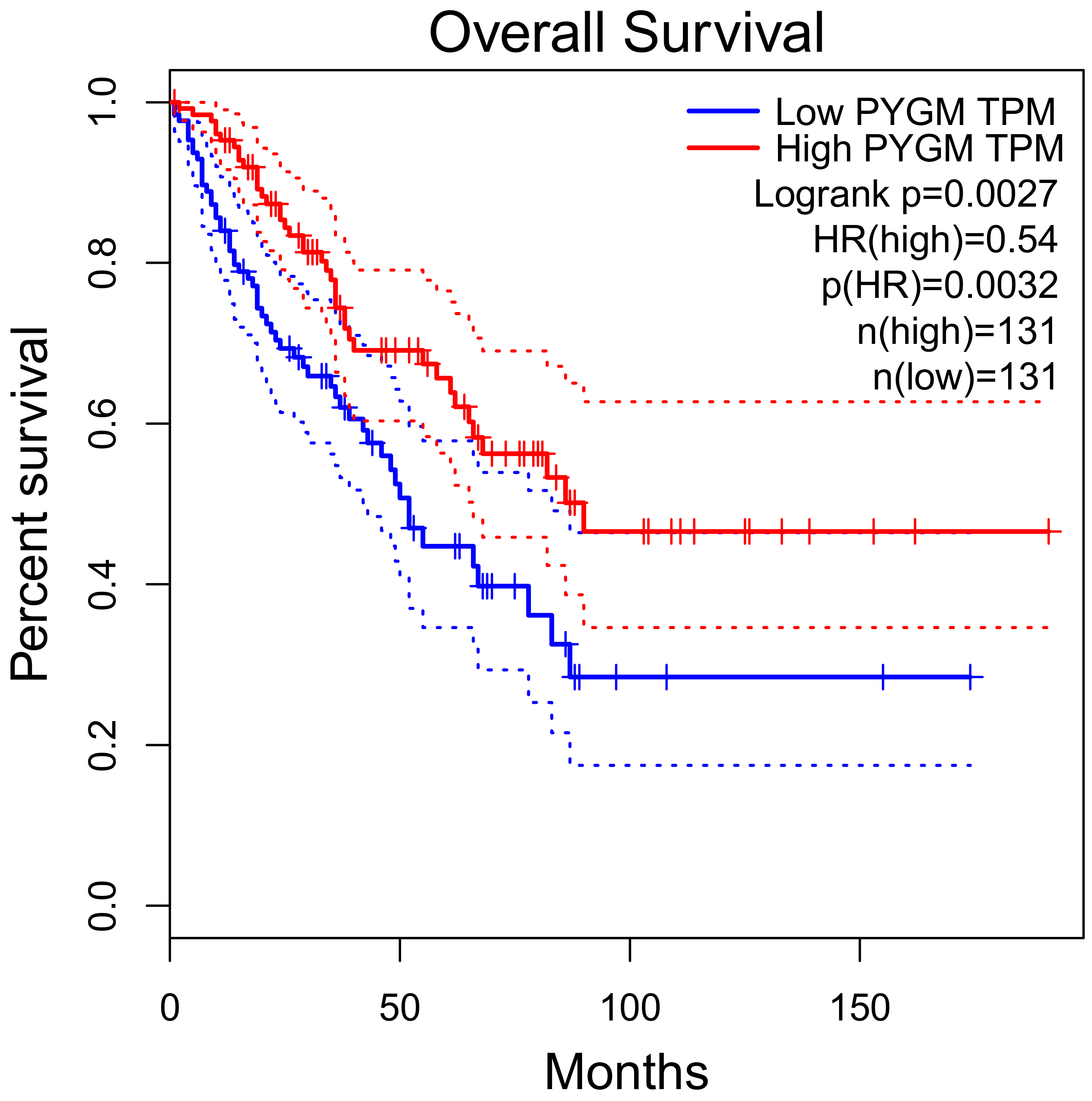
3.2.3. PYGM in Cancer
4. Why Use Zebrafish to Study PYGM?
5. Summary and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Mauro, S. Muscle glycogenoses: An overview. Acta Myol. 2007, 26, 35–41. [Google Scholar] [PubMed]
- Nogales-Gadea, G.; Santalla, A.; Brull, A.; De Luna, N.; Lucia, A.; Pinós, T. The pathogenomics of McArdle disease—Genes, enzymes, models, and therapeutic implications. J. Inherit. Metab. Dis. 2015, 38, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.; Bartlett, J.B.; Convey, G.; Hardern, I.; Teague, J.L.; Loxham, S.J.G.; Allen, J.M.; Poucher, S.M.; Charles, A.D. Sensitivity of glycogen phosphorylase isoforms to indole site inhibitors is markedly dependent on the activation state of the enzyme. Br. J. Pharmacol. 2006, 149, 775–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llavero, F.; Sastre, A.A.; Montoro, M.L.; Gálvez, P.; Lacerda, H.M.; Parada, L.A.; Zugaza, J.L. McArdle Disease: New Insights into Its Underlying Molecular Mechanisms. Int. J. Mol. Sci. 2019, 20, 5919. [Google Scholar] [CrossRef] [Green Version]
- Hudson, J.W.; Golding, G.; Crerar, M.M. Evolution of Allosteric Control in Glycogen Phosphorylase. J. Mol. Biol. 1993, 234, 700–721. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and Metabolic Syndrome in Circadian Clock Mutant Mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [Green Version]
- Stenvers, D.J.; Scheer, F.A.J.L.; Schrauwen, P.; La Fleur, S.E.; Kalsbeek, A. Circadian clocks and insulin resistance. Nat. Rev. Endocrinol. 2019, 15, 75–89. [Google Scholar] [CrossRef]
- Baek, M.; Virgilio, S.; Lamb, T.M.; Ibarra, O.; Andrade, J.M.; Gonçalves, R.D.; Dovzhenok, A.; Lim, S.; Bell-Pedersen, D.; Bertolini, M.C.; et al. Circadian clock regulation of the glycogen synthase (GSN) gene by WCC is critical for rhythmic glycogen metabolism inNeurospora crassa. Proc. Natl. Acad. Sci. USA 2019, 116, 10435–10440. [Google Scholar] [CrossRef] [Green Version]
- Scheer, F.A.J.L.; Hilton, M.F.; Mantzoros, C.S.; Shea, S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 2009, 106, 4453–4458. [Google Scholar] [CrossRef] [Green Version]
- Chasiotis, D. The regulation of glycogen phosphorylase and glycogen breakdown in human skeletal muscle. Acta Physiol. Scand. Suppl. 1983, 518, 1–68. [Google Scholar]
- Hue, L.; Bontemps, F.; Hers, H. The effects of glucose and of potassium ions on the interconversion of the two forms of glycogen phosphorylase and of glycogen synthetase in isolated rat liver preparations. Biochem. J. 1975, 152, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.-J.; Li, G.-Y.; Xu, C.-D.; Wu, Y.; Zhou, Z.-S.; Wang, S.-G.; Li, C. Regulatory Functions of Nilaparvata lugens GSK-3 in Energy and Chitin Metabolism. Front. Physiol. 2020, 11, 518876. [Google Scholar] [CrossRef] [PubMed]
- Madsen, N.B.; Avramovic-Zikic, O.; Honikel, K.O. Structure-Function Relationships in Glycogen Phosphorylase with Respect to Its Control Characteristics. Ann. N. Y. Acad. Sci. 1973, 210, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Lillpopp, L.; Tzikas, S.; Ojeda, F.; Zeller, T.; Baldus, S.; Bickel, C.; Sinning, C.R.; Wild, P.S.; Genth-Zotz, S.; Warnholtz, A.; et al. Prognostic Information of Glycogen Phosphorylase Isoenzyme BB in Patients with Suspected Acute Coronary Syndrome. Am. J. Cardiol. 2012, 110, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Pudil, R.; Vašatová, M.; Lenco, J.; Tichy, M.; Řeháček, V.; Fucikova, A.; Horacek, J.M.; Vojacek, J.; Pleskot, M.; Stulik, J.; et al. Plasma glycogen phosphorylase BB is associated with pulmonary artery wedge pressure and left ventricle mass index in patients with hypertrophic cardiomyopathy. Clin. Chem. Lab. Med. 2010, 48, 1193–1195. [Google Scholar] [CrossRef]
- Crerar, M.M.; Karlsson, O.; Fletterick, R.J.; Hwang, P.K. Chimeric Muscle and Brain Glycogen Phosphorylases Define Protein Domains Governing Isozyme-specific Responses to Allosteric Activation. J. Biol. Chem. 1995, 270, 13748–13756. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Schmid, H.; Pfeiffer-Guglielmi, B.; Dolderer, B.; Thiess, U.; Verleysdonk, S.; Hamprecht, B. Expression of the Brain and Muscle Isoforms of Glycogen Phosphorylase in Rat Heart. Neurochem. Res. 2009, 34, 581–586. [Google Scholar] [CrossRef]
- Abugessaisa, I.; Noguchi, S.; Hasegawa, A.; Harshbarger, J.; Kondo, A.; Lizio, M.; Severin, J.; Carninci, P.; Kawaji, H.; Kasukawa, T. FANTOM5 CAGE profiles of human and mouse reprocessed for GRCh38 and GRCm38 genome assemblies. Sci. Data 2017, 4, 170107. [Google Scholar] [CrossRef]
- Jakobsen, E.; Bak, L.K.; Walls, A.B.; Reuschlein, A.-K.; Schousboe, A.; Waagepetersen, H.S. Glycogen Shunt Activity and Glycolytic Supercompensation in Astrocytes May Be Distinctly Mediated via the Muscle form of Glycogen Phosphorylase. Neurochem. Res. 2017, 89, 537–2494. [Google Scholar] [CrossRef]
- Pfeiffer-Guglielmi, B.; Fleckenstein, B.; Jung, G.; Hamprecht, B. Immunocytochemical localization of glycogen phosphorylase isozymes in rat nervous tissues by using isozyme-specific antibodies. J. Neurochem. 2003, 85, 73–81. [Google Scholar] [CrossRef]
- Pinacho, R.; Vila, E.; Prades, R.; Tarragó, T.; Castro, E.; Ferrer, I.; Ramos, B. The glial phosphorylase of glycogen isoform is reduced in the dorsolateral prefrontal cortex in chronic schizophrenia. Schizophr. Res. 2016, 177, 37–43. [Google Scholar] [CrossRef]
- Arrizabalaga, O.; Lacerda, H.M.; Zubiaga, A.M.; Zugaza, J.L. Rac1 Protein Regulates Glycogen Phosphorylase Activation and Controls Interleukin (IL)-2-dependent T Cell Proliferation. J. Biol. Chem. 2012, 287, 11878–11890. [Google Scholar] [CrossRef] [Green Version]
- De Luna, N.; Brull, A.; Lucia, A.; Santalla, A.; Garatachea, N.; Martí, R.; Andreu, A.L.; Pinós, T. PYGM expression analysis in white blood cells: A complementary tool for diagnosing McArdle disease? Neuromuscul. Disord. 2014, 24, 1079–1086. [Google Scholar] [CrossRef]
- Llavero, F.; Urzelai, B.; Osinalde, N.; Gálvez, P.; Lacerda, H.M.; Parada, L.A.; Zugaza, J.L. Guanine Nucleotide Exchange Factor αPIX Leads to Activation of the Rac 1 GTPase/Glycogen Phosphorylase Pathway in Interleukin (IL)-2-stimulated T Cells. J. Biol. Chem. 2015, 290, 9171–9182. [Google Scholar] [CrossRef] [Green Version]
- Llavero, F.; Artaso, A.; Lacerda, H.M.; Parada, L.A.; Zugaza, J.L. Lck/PLCγ control migration and proliferation of interleukin (IL)-2-stimulated T cells via the Rac1 GTPase/glycogen phosphorylase pathway. Cell. Signal. 2016, 28, 1713–1724. [Google Scholar] [CrossRef] [Green Version]
- Schmid, H.; Dolderer, B.; Thiess, U.; Verleysdonk, S.; Hamprecht, B. Renal Expression of the Brain and Muscle Isoforms of Glycogen Phosphorylase in Different Cell Types. Neurochem. Res. 2008, 33, 2575–2582. [Google Scholar] [CrossRef]
- Hernández, C.; Garcia-Ramírez, M.; García-Rocha, M.; Saez-López, C.; Valverde, Á.M.; Guinovart, J.J.; Simó, R. Glycogen storage in the human retinal pigment epithelium: A comparative study of diabetic and non-diabetic donors. Acta Diabetol. 2014, 51, 543–552. [Google Scholar] [CrossRef]
- Vaclavik, V.; Naderi, F.; Schaller, A.; Escher, P. Longitudinal case study and phenotypic multimodal characterization of McArdle disease-linked retinopathy: Insight into pathomechanisms. Ophthalmic Genet. 2020, 41, 73–78. [Google Scholar] [CrossRef]
- Johnson, L.N. Glycogen phosphorylase: Control by phosphorylation and allosteric effectors. FASEB J. 1992, 6, 2274–2282. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Blandin, G.; Marchand, S.; Charton, K.; Danièle, N.; Gicquel, E.; Boucheteil, J.-B.; Bentaib, A.; Barrault, L.; Stockholm, D.; Bartoli, M.; et al. A human skeletal muscle interactome centered on proteins involved in muscular dystrophies: LGMD interactome. Skelet. Muscle 2013, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Adeva-Andany, M.M.; González-Lucán, M.; Donapetry-García, C.; Fernández-Fernández, C.; Ameneiros-Rodríguez, E. Glycogen metabolism in humans. BBA Clin. 2016, 5, 85–100. [Google Scholar] [CrossRef] [Green Version]
- Zang, J.; Neuhauss, S.C.F. The Binding Properties and Physiological Functions of Recoverin. Front. Mol. Neurosci. 2018, 11, 473. [Google Scholar] [CrossRef]
- Alsberge, J.B.; Chen, J.J.; Zaidi, A.A.; Fu, A.D. Retinal Dystrophy in a Patient with Mcardle Disease. Retin. Cases Brief. Rep. 2018. [Google Scholar] [CrossRef]
- Leonardy, N.J.; Harbin, R.L.; Sternberg, P. Pattern Dystrophy of the Retinal Pigment Epithelium in a Patient with McArdle’s Disease. Am. J. Ophthalmol. 1988, 106, 741–742. [Google Scholar] [CrossRef]
- Mahroo, O.A.; Khan, K.N.; Wright, G.; Ockrim, Z.; Scalco, R.S.; Robson, A.G.; Tufail, A.; Michaelides, M.; Quinlivan, R.; Webster, A.R. Retinopathy Associated with Biallelic Mutations in PYGM (McArdle Disease). Ophthalmology 2019, 126, 320–322. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, T.; Lei, T.; Zhang, D.; Du, S.; Girani, L.; Qi, D.; Lin, C.; Tong, R.; Wang, Y. RIP1/RIP3-regulated necroptosis as a target for multifaceted disease therapy (Review). Int. J. Mol. Med. 2019, 44, 771–786. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-W.; Shao, J.; Lin, J.; Zhang, N.; Lu, B.-J.; Lin, S.-C.; Dong, M.-Q.; Han, J. RIP3, an Energy Metabolism Regulator That Switches TNF-Induced Cell Death from Apoptosis to Necrosis. Science 2009, 325, 332–336. [Google Scholar] [CrossRef]
- Llavero, F.; Montoro, M.L.; Sastre, A.A.; Fernández-Moreno, D.; Lacerda, H.M.; Parada, L.A.; Lucia, A.; Zugaza, J.L. Epidermal growth factor receptor controls glycogen phosphorylase in T cells through small GTPases of the RAS family. J. Biol. Chem. 2019, 294, 4345–4358. [Google Scholar] [CrossRef] [PubMed]
- Tixier, V.; Bataillé, L.; Etard, C.; Jagla, T.; Weger, M.; Daponte, J.P.; Strähle, U.; Dickmeis, T.; Jagla, K. Glycolysis supports embryonic muscle growth by promoting myoblast fusion. Proc. Natl. Acad. Sci. USA 2013, 110, 18982–18987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migocka-Patrzałek, M.; Lewicka, A.; Elias, M.; Daczewska, M. The effect of muscle glycogen phosphorylase (Pygm) knockdown on zebrafish morphology. Int. J. Biochem. Cell Biol. 2020, 118, 105658. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almodóvar-Payá, A.; Villarreal-Salazar, M.; De Luna, N.; Nogales-Gadea, G.; Real-Martínez, A.; Andreu, A.L.; Martín, M.A.; Arenas, J.; Lucia, A.; Vissing, J.; et al. Preclinical Research in Glycogen Storage Diseases: A Comprehensive Review of Current Animal Models. Int. J. Mol. Sci. 2020, 21, 9621. [Google Scholar] [CrossRef]
- Nogales-Gadea, G.; Brull, A.; Santalla, A.; Andreu, A.L.; Arenas, J.; Martín, M.A.; Lucia, A.; De Luna, N.; Pinós, T. McArdle Disease: Update of Reported Mutations and Polymorphisms in thePYGMGene. Hum. Mutat. 2015, 36, 669–678. [Google Scholar] [CrossRef]
- Nogales-Gadea, G.; Pinós, T.; Lucia, A.; Arenas, J.; Cámara, Y.; Brull, A.; De Luna, N.; Martín, M.A.; Garcia-Arumí, E.; Marti, R.; et al. Knock-in mice for the R50X mutation in the PYGM gene present with McArdle disease. Brain 2012, 135, 2048–2057. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.A.S.; Christofolini, D.M.; Perez, M.M.; Alves, B.C.A.; Rodart, I.; Figueiredo, F.W.S.; Turke, K.C.; Feder, D.; Junior, M.C.F.; Nucci, A.M.; et al. PYGM mRNA expression in McArdle disease: Demographic, clinical, morphological and genetic features. PLoS ONE 2020, 15, e0236597. [Google Scholar] [CrossRef]
- Casalino, G.; Chan, W.; McAvoy, C.; Coppola, M.; Bandello, F.; Bird, A.C.; Chakravarthy, U. Multimodal imaging of posterior ocular involvement in McArdle’s disease. Clin. Exp. Optom. 2018, 101, 412–415. [Google Scholar] [CrossRef]
- Sears, S.M.; Hewett, S.J. Influence of glutamate and GABA transport on brain excitatory/inhibitory balance. Exp. Biol. Med. 2021. [Google Scholar] [CrossRef]
- Uno, Y.; Coyle, J.T. Glutamate hypothesis in schizophrenia. Psychiatry Clin. Neurosci. 2019, 73, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Briski, K.P.; Ibrahim, M.M.H.; Mahmood, A.S.M.H.; Alshamrani, A.A. Norepinephrine Regulation of Ventromedial Hypothalamic Nucleus Astrocyte Glycogen Metabolism. Int. J. Mol. Sci. 2021, 22, 759. [Google Scholar] [CrossRef]
- Moghaddam, B.; Javitt, D.C. From Revolution to Evolution: The Glutamate Hypothesis of Schizophrenia and its Implication for Treatment. Neuropsychopharmacology 2011, 37, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Scalco, R.S.; EUROMAC Consortium; Lucia, A.; Santalla, A.; Martinuzzi, A.; Vavla, M.; Reni, G.; Toscano, A.; Musumeci, O.; Voermans, N.C.; et al. Data from the European registry for patients with McArdle disease and other muscle glycogenoses (EUROMAC). Orphanet. J. Rare Dis. 2020, 15, 1–8. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Yang, Y. Bioinformatics-based discovery of PYGM and TNNC2 as potential biomarkers of head and neck squamous cell carcinoma. Biosci. Rep. 2019, 39, 20191612. [Google Scholar] [CrossRef] [Green Version]
- Nogales-Gadea, G.; Consuegra-García, I.; Rubio, J.C.; Arenas, J.; Cuadros, M.; Camara, Y.; Torres-Torronteras, J.; Fiuza-Luces, C.; Lucia, A.; Martín, M.A.; et al. A Transcriptomic Approach to Search for Novel Phenotypic Regulators in McArdle Disease. PLoS ONE 2012, 7, e31718. [Google Scholar] [CrossRef] [Green Version]
- Dieci, M.V.; Smutná, V.; Scott, V.; Yin, G.; Xu, R.; Vielh, P.; Mathieu, M.-C.; Vicier, C.; Laporte, M.; Drusch, F.; et al. Whole exome sequencing of rare aggressive breast cancer histologies. Breast Cancer Res. Treat. 2016, 156, 21–32. [Google Scholar] [CrossRef]
- Al-Salameh, A.; Baudry, C.; Cohen, R. Update on multiple endocrine neoplasia Type 1 and 2. La Presse Médicale 2018, 47, 722–731. [Google Scholar] [CrossRef]
- Kedra, D.; Seroussi, E.; Fransson, I.; Trifunovic, J.; Clark, M.; Lagercrantz, J.; Blennow, E.; Mehlin, H.; Dumanski, J. The germinal center kinase gene and a novel CDC25-like gene are located in the vicinity of the PYGM gene on 11q13. Qual. Life Res. 1997, 100, 611–619. [Google Scholar] [CrossRef]
- Lemmes, I.; Van de Ven, W.J.M.; Kas, K.; Zhang, C.X.; Giraud, S.; Wautot, V.; Buisson, N.; Pugeat, M.; Peix, J.L.; Caldener, A.; et al. The search for the MEN1 gene. The European Consortium on MEN-1. Intern. Med. 1998, 243, 441–446. [Google Scholar] [CrossRef]
- Asteria, C.; Anagni, M.; Persani, L.; Beck-Peccoz, P. Loss of heterozygosity of the MEN1 gene in a large series of TSH-secreting pituitary adenomas. J. Endocrinol. Investig. 2001, 24, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Bièche, I.; Lidereau, R. Genetic alterations in breast cancer. Genes Chromosom. Cancer 1995, 14, 227–251. [Google Scholar] [CrossRef] [PubMed]
- Debelenko, L.V.; Brambilla, E.; Agarwal, S.K.; Swalwell, J.I.; Kester, M.B.; Lubensky, I.A.; Zhuang, Z.; Guru, S.C.; Manickam, P.; Olufemi, S.-E.; et al. Identification of MEN1 gene mutations in sporadic carcinoid tumors of the lung. Hum. Mol. Genet. 1997, 6, 2285–2290. [Google Scholar] [CrossRef] [PubMed]
- Petzmann, S.; Ullmann, R.; Klemen, H.; Renner, H.; Popper, H.H. Loss of heterozygosity on chromosome arm 11q in lung carcinoids. Hum. Pathol. 2001, 32, 333–338. [Google Scholar] [CrossRef]
- Zhuang, Z.; Merino, M.J.; Chuaqui, R.; Liotta, L.; Emmert-Buck, M.R. Identical allelic loss on chromosome 11q13 in microdissected in situ and invasive human breast cancer. Cancer Res. 1995, 55, 467–471. [Google Scholar]
- Vandenberghe, E.; Peeters, C.D.W.; Wlodarska, I.; Stul, M.; Louwagie, A.; Verhoef, G.; Thomas, J.; Criel, A.; Cassiman, J.J.; Mecucci, C.; et al. Chromosome 11q rearrangements in B non Hodgkin’s lymphoma. Br. J. Haematol. 1992, 81, 212–217. [Google Scholar] [CrossRef]
- Pastor, M.; Nogal, A.; Molina-Pinelo, S.; Melendez, R.; Salinas, A.; De La Peña, M.G.; Martín-Juan, J.; Corral, J.; Garcia-Carbonero, R.; Carnero, A.; et al. Identification of proteomic signatures associated with lung cancer and COPD. J. Proteom. 2013, 89, 227–237. [Google Scholar] [CrossRef]
- Tashima, S.; Shimada, S.; Yamaguchi, K.; Tsuruta, J.; Ogawa, M. Expression of brain-type glycogen phosphorylase is a potentially novel early biomarker in the carcinogenesis of human colorectal carcinomas. Am. J. Gastroenterol. 2000, 95, 255–263. [Google Scholar] [CrossRef]
- Cui, G.; Wang, H.; Liu, W.; Xing, J.; Song, W.; Zeng, Z.; Liu, L.; Wang, H.; Wang, X.; Luo, H.; et al. Glycogen Phosphorylase B Is Regulated by miR101-3p and Promotes Hepatocellular Carcinoma Tumorigenesis. Front. Cell Dev. Biol. 2020, 8, 566494. [Google Scholar] [CrossRef]
- Wang, Z.; Han, G.; Liu, Q.; Zhang, W.; Wang, J. Silencing of PYGB suppresses growth and promotes the apoptosis of prostate cancer cells via the NF-κB/Nrf2 signaling pathway. Mol. Med. Rep. 2018, 18, 3800–3808. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.K.; Kim, J.H.; Lee, C.H.; Kim, J.M.; Kang, C.D.; Kim, Y.D.; Choi, K.U.; Kim, H.W.; Kim, J.Y.; Park, D.Y.; et al. Clinicopathological significance of BGP expression in non-small-cell lung carcinoma: Relationship with histological type, microvessel density and patients’ survival. Pathology 2006, 38, 555–560. [Google Scholar] [CrossRef]
- Zhou, Y.; Jin, Z.; Wang, C. Glycogen phosphorylase B promotes ovarian cancer progression via Wnt/β-catenin signaling and is regulated by miR-133a-3p. Biomed. Pharmacother. 2019, 120, 109449. [Google Scholar] [CrossRef]
- Davis, A.; Gao, R.; Navin, N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1867, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Real-Martinez, A.; Brull, A.; Huerta, J.; Tarrasó, G.; Lucia, A.; Martin, M.A.; Arenas, J.; Andreu, A.L.; Nogales-Gadea, G.; Vissing, J.; et al. Low survival rate and muscle fiber-dependent aging effects in the McArdle disease mouse model. Sci. Rep. 2019, 9, 5116. [Google Scholar] [CrossRef]
- Krag, T.O.; Pinós, T.; Nielsen, T.L.; Duran, J.; Garcia-Rocha, M.; Andreu, A.L.; Vissing, J. Differential glucose metabolism in mice and humans affected by McArdle disease. Am. J. Physiol. Integr. Comp. Physiol. 2016, 311, R307–R314. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, T.L.; Pinós, T.; Brull, A.; Vissing, J.; Krag, T.O. Exercising with blocked muscle glycogenolysis: Adaptation in the McArdle mouse. Mol. Genet. Metab. 2018, 123, 21–27. [Google Scholar] [CrossRef]
- Dubińska-Magiera, M.; Daczewska, M.; Lewicka, A.; Migocka-Patrzałek, M.; Niedbalska-Tarnowska, J.; Jagla, K. Zebrafish: A Model for the Study of Toxicants Affecting Muscle Development and Function. Int. J. Mol. Sci. 2016, 17, 1941. [Google Scholar] [CrossRef]
- Plantié, E.; Migocka-Patrzałek, M.; Daczewska, M.; Jagla, K. Model Organisms in the Fight against Muscular Dystrophy: Lessons from Drosophila and Zebrafish. Molecules 2015, 20, 6237–6253. [Google Scholar] [CrossRef] [Green Version]
- Benchoula, K.; Khatib, A.; Jaffar, A.; Ahmed, Q.U.; Sulaiman, W.M.A.W.; Wahab, R.A.; El-Seedi, H.R. The promise of zebrafish as a model of metabolic syndrome. Exp. Anim. 2019, 68, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Bradford, Y.M.; Toro, S.; Ramachandran, S.; Ruzicka, L.; Howe, D.G.; Eagle, A.; Kalita, P.; Martin, R.; Moxon, S.A.T.; Schaper, K.; et al. Zebrafish Models of Human Disease: Gaining Insight into Human Disease at ZFIN. ILAR J. 2017, 58, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Seth, A.; Stemple, D.L.; Barroso, I. The emerging use of zebrafish to model metabolic disease. Dis. Model. Mech. 2013, 6, 1080–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.A. Zebrafish: A model system to examine the neurodevelopmental basis of schizophrenia. Prog. Brain Res. 2009, 179, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hason, M.; Bartůněk, P. Zebrafish Models of Cancer—New Insights on Modeling Human Cancer in a Non-Mammalian Vertebrate. Genes 2019, 10, 935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassar, S.; Adatto, I.; Freeman, J.L.; Gamse, J.T.; Iturria, I.; Lawrence, C.; Muriana, A.; Peterson, R.T.; Van Cruchten, S.; Zon, L.I. Use of Zebrafish in Drug Discovery Toxicology. Chem. Res. Toxicol. 2020, 33, 95–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishnani, P.S.; on behalf of the ACMG Work Group on Diagnosis and Management of Glycogen Storage Diseases Type VI and IX; Goldstein, J.; Austin, S.L.; Arn, P.; Bachrach, B.; Bali, D.S.; Chung, W.K.; El-Gharbawy, A.; Brown, L.M.; et al. Diagnosis and management of glycogen storage diseases type VI and IX: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2019, 21, 772–789. [Google Scholar] [CrossRef] [Green Version]
- Camus, S.; Quevedo, C.; Menéndez, S.; Paramonov, I.; Stouten, P.F.W.; Janssen, R.A.J.; Rueb, S.; He, S.; Snaar-Jagalska, B.; Laricchia-Robbio, L.; et al. Identification of phosphorylase kinase as a novel therapeutic target through high-throughput screening for anti-angiogenesis compounds in zebrafish. Oncogene 2011, 31, 4333–4342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 4 February 2021).
- UniProt. Available online: https://www.uniprot.org/uniprot (accessed on 4 February 2021).
- ZFIN. Available online: http://zfin.org/ (accessed on 4 February 2021).
- Quach, H.N.B.; Tao, S.; Vrljicak, P.; Joshi, A.; Ruan, H.; Sukumaran, R.; Varshney, G.K.; LaFave, M.C.; Burgess, S.M.; Winkler, C.; et al. A Multifunctional Mutagenesis System for Analysis of Gene Function in Zebrafish. G3 Genes Genomes Genet. 2015, 5, 1283–1299. [Google Scholar] [CrossRef] [Green Version]
- Dagli, A.; Sentner, C.P.; Weinstein, D.A. Glycogen Storage Disease Type III. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Henn, K.; Braunbeck, T. Dechorionation as a tool to improve the fish embryo toxicity test (FET) with the zebrafish (Danio rerio). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 153, 91–98. [Google Scholar] [CrossRef]
- Grech, A.; Tebby, C.; Brochot, C.; Bois, F.Y.; Bado-Nilles, A.; Dorne, J.-L.; Quignot, N.; Beaudouin, R. Generic physiologically-based toxicokinetic modelling for fish: Integration of environmental factors and species variability. Sci. Total Environ. 2019, 651, 516–531. [Google Scholar] [CrossRef]
- De Abreu, M.S.; Kalueff, A.V. Of mice and zebrafish: The impact of the experimenter identity on animal behavior. Lab. Anim. 2021, 50, 7. [Google Scholar] [CrossRef]
- Stygar, D.; Andrare, D.; Bażanów, B.; Chełmecka, E.; Sawczyn, T.; Skrzep-Poloczek, B.; Olszańska, E.; Karcz, K.W.; Jochem, J. The Impact of DJOS Surgery, a High Fat Diet and a Control Diet on the Enzymes of Glucose Metabolism in the Liver and Muscles of Sprague-Dawley Rats. Front. Physiol. 2019, 10, 571. [Google Scholar] [CrossRef]
- Pelletier, J.; Bellot, G.; Gounon, P.; Lacas-Gervais, S.; Pouysségur, J.; Mazure, N.M. Glycogen Synthesis is Induced in Hypoxia by the Hypoxia-Inducible Factor and Promotes Cancer Cell Survival. Front. Oncol. 2012, 2, 18. [Google Scholar] [CrossRef] [Green Version]

| Zebrafish (Danio rerio) Orthologs of Some Human (Homo sapiens) Protein Partners of Muscle Glycogen Phosphorylase (PYGM) | |||
|---|---|---|---|
| Human | Zebrafish | ||
| Protein | Systematic Name | Protein | Systematic Name |
| AGL | P35573 | agla | A0A0R4IA63 |
| UGP2 | Q16851 | ugp2a and ugp2b | B8JMZ1 and Q6NWJ8 |
| PHK (PHKG1) | Q16816 | phkg1b | Q503G9 |
| PPP1R3A | Q16821 | ppp1r3ab and ppp1r3aa | E7EZR5 and E7F487 |
| PPP1R3B | Q86XI6 | ppp1r3b | Q803M0 |
| ALDH18A1 | P54886 | aldh18a1 | A4IGC8 |
| FLRT1 | Q9NZU1 | flrt1a | A8BBF0 |
| CALM1 | P0DP23 | calm1a and calm1b | Q6PI52 and Q6PI52 |
| CALM2 | P0DP24 | calm2a and calm2b | Q6PI52 and Q6PI52 |
| CALM3 | P0DP25 | calm3a and calm3b | Q6PI52 and Q6PI52 |
| RCVRN | P35243 | rcvrn2 | Q6PC38 |
| RIPK1 | Q13546 | ripk1l | A8DZG7 |
| RAC1 | P63000 | rac1a and rac1b | Q7ZSZ9 and Q29RC5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Migocka-Patrzałek, M.; Elias, M. Muscle Glycogen Phosphorylase and Its Functional Partners in Health and Disease. Cells 2021, 10, 883. https://doi.org/10.3390/cells10040883
Migocka-Patrzałek M, Elias M. Muscle Glycogen Phosphorylase and Its Functional Partners in Health and Disease. Cells. 2021; 10(4):883. https://doi.org/10.3390/cells10040883
Chicago/Turabian StyleMigocka-Patrzałek, Marta, and Magdalena Elias. 2021. "Muscle Glycogen Phosphorylase and Its Functional Partners in Health and Disease" Cells 10, no. 4: 883. https://doi.org/10.3390/cells10040883
APA StyleMigocka-Patrzałek, M., & Elias, M. (2021). Muscle Glycogen Phosphorylase and Its Functional Partners in Health and Disease. Cells, 10(4), 883. https://doi.org/10.3390/cells10040883






