Biomechanical Properties of Cancer Cells
Abstract
:1. Introduction
2. Interplay between Biochemical and Biomechanical Features of Tumor Cells in Their Microenvironment
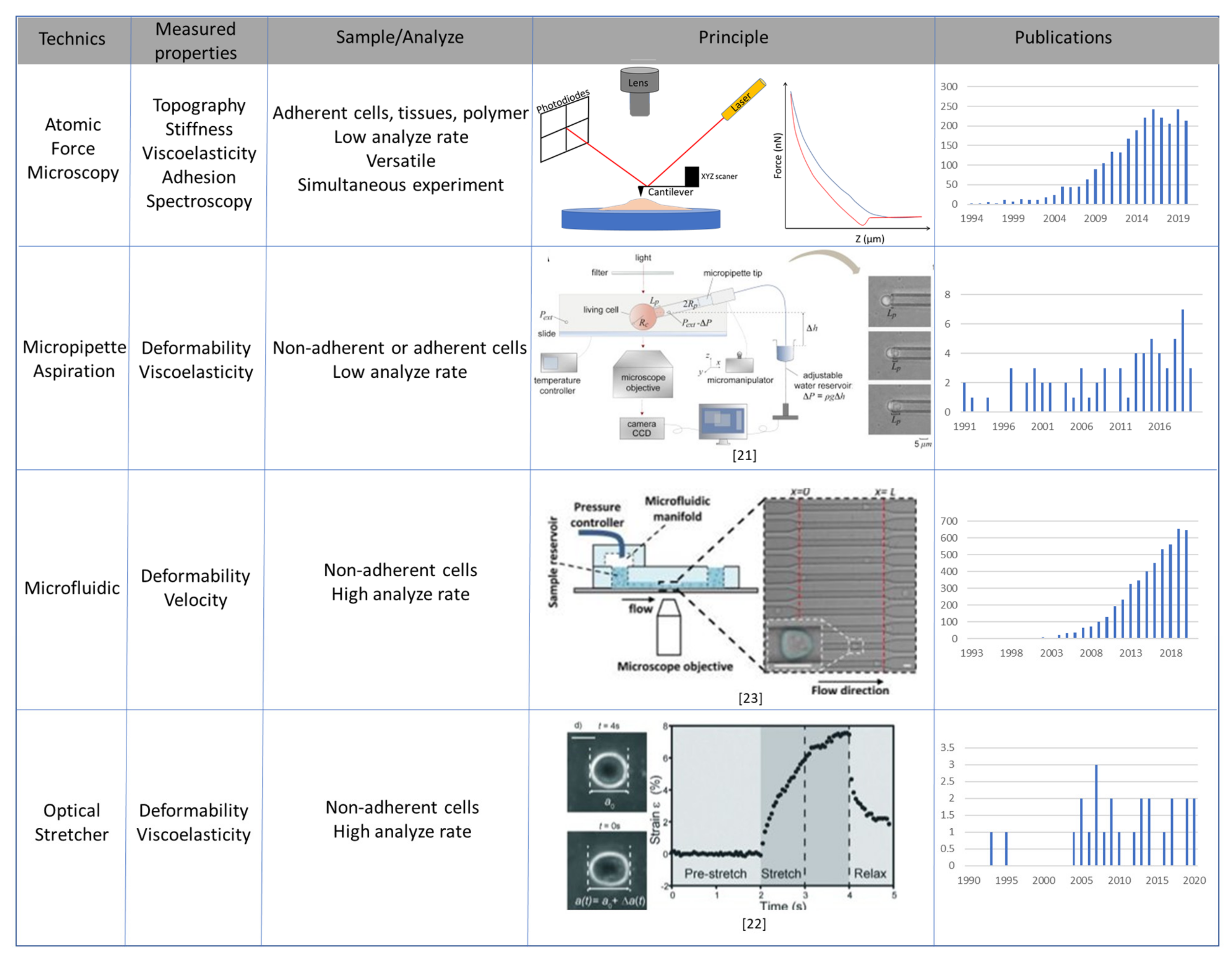
3. Main Methods to Study Mechanical Properties of Cancer Cells
3.1. Atomic Force Microscopy
3.2. Micropipette Aspiration, Optical Stretcher and Microfluidic Systems
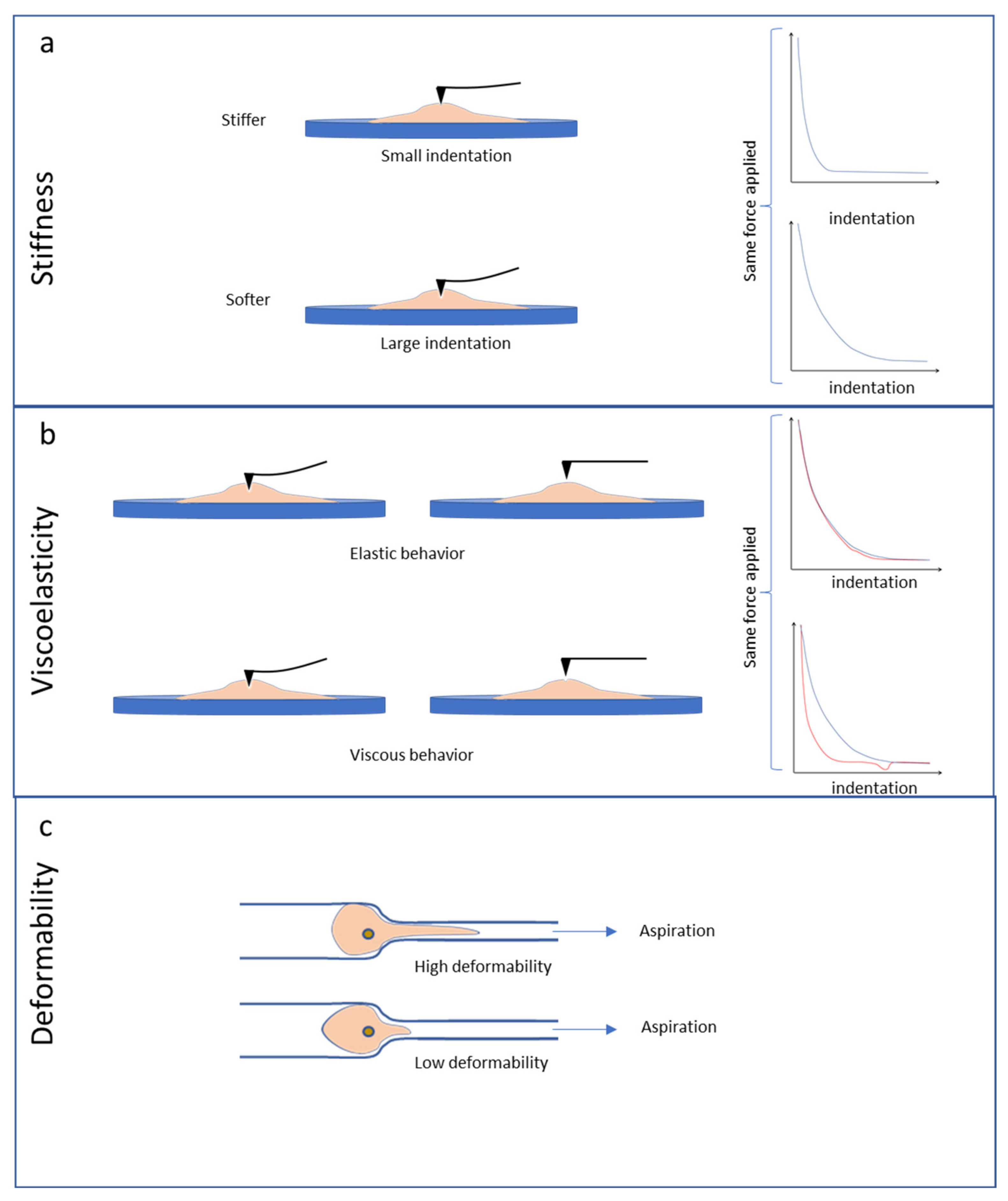
4. Stiffness, Viscoelasticity and Deformability of Cultured Cancer Cells
4.1. Pioneering Experiments on the Biomechanics in Cancer Cells
4.2. Stiffness and Viscoelasticity of Cancer Cells
4.3. Deformability Properties of Cancer Cells
4.4. Ex-Vivo Cancer Cell Analyses
5. Cancer Cell Cytoskeleton, Cell Morphology and Biomechanical Properties
5.1. Cytoskeleton Disruption by Drugs and Biomechanics in Cancer Cells
5.2. Link between Cytoskeleton, Morphology and Nanomechanical Properties of Cancer Cells
6. Cell-Environment Mechanical Interaction in Cancer
6.1. Mechanoreciprocity between Tumor Cells and the Surrounding ECM
6.2. Mechanoreciprocity during Local Invasion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Poudineh, M.; Sargent, E.H.; Pantel, K.; Kelley, S.O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2018, 2, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Gizem Sonugür, F.; Akbulut, H. The role of tumor microenvironment in genomic instability of malignant tumors. Front. Genet. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Broders-Bondon, F.; Ho-Bouldoires, T.H.N.; Fernandez-Sanchez, M.E.; Farge, E. Mechanotransduction in tumor progression: The dark side of the force. J. Cell Biol. 2018, 217, 1571–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riching, K.M.; Cox, B.L.; Salick, M.R.; Pehlke, C.; Riching, A.S.; Ponik, S.M.; Bass, B.R.; Crone, W.C.; Jiang, Y.; Weaver, A.M.; et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 2015, 107, 2546–2558. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Friedrichs, J.; Song, Y.H.; Werner, C.; Estroff, L.A.; Fischbach, C. Intrafibrillar, bone-mimetic collagen mineralization regulates breast cancer cell adhesion and migration. Biomaterials 2019, 198, 95–106. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, L.; Yang, K.; Iwasawa, K.; Kadekaro, A.L.; Takebe, T.; Andl, T.; Zhang, Y. The β-catenin/YAP signaling axis is a key regulator of melanoma-associated fibroblasts. Signal Transduct. Target. Ther. 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Binning, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [Green Version]
- Pyne, A.; Thompson, R.; Leung, C.; Roy, D.; Hoogenboom, B.W. Single-molecule reconstruction of oligonucleotide secondary structure by atomic force microscopy. Small 2014, 10, 3257–3261. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Dang, D.; Liu, L.; Xi, N.; Wang, Y. Atomic force microscopy in characterizing cell mechanics for biomedical applications: A review. IEEE Trans. Nanobiosci. 2017, 16, 523–540. [Google Scholar] [CrossRef]
- Almqvist, N.; Backman, L.; Fredriksson, S. Imaging human erythrocyte spectrin with atomic force microscopy. Micron 1994, 25, 227–232. [Google Scholar] [CrossRef]
- Putman, C.A.; van der Werf, K.O.; de Grooth, B.G.; van Hulst, N.F.; Greve, J. Viscoelasticity of living cells allows high resolution imaging by tapping mode atomic force microscopy. Biophys. J. 1994, 67, 1749–1753. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Dang, D.; Xi, N.; Wang, Y.; Liu, L. Nanoscale imaging and force probing of biomolecular systems using atomic force microscopy: From single molecules to living cells. Nanoscale 2017, 9, 17643–17666. [Google Scholar] [CrossRef] [PubMed]
- Cheong, L.Z.; Zhao, W.; Song, S.; Shen, C. Lab on a tip: Applications of functional atomic force microscopy for the study of electrical properties in biology. Acta Biomater. 2019, 99, 33–52. [Google Scholar] [CrossRef]
- Bouchonville, N.; Nicolas, A. Quantification of the Elastic Properties of Soft and Sticky Materials Using AFM. Methods Mol. Biol. 2019, 1886, 281–290. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Okajima, T.; Raman, A. Measuring viscoelasticity of soft biological samples using atomic force microscopy. Soft Matter 2019, 16, 64–81. [Google Scholar] [CrossRef]
- Evans, E.A. New Membrane Concept Applied to the Analysis of Fluid Shear- and Micropipette-Deformed Red Blood Cells. Biophys. J. 1973, 13, 941–954. [Google Scholar] [CrossRef] [Green Version]
- González-Bermúdez, B.; Guinea, G.V.; Plaza, G.R. Advances in Micropipette Aspiration: Applications in Cell Biomechanics, Models, and Extended Studies. Biophys. J. 2019, 116, 587–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faigle, C.; Lautenschläger, F.; Whyte, G.; Homewood, P.; Martín-Badosa, E.; Guck, J. A monolithic glass chip for active single-cell sorting based on mechanical phenotyping. Lab Chip 2015, 15, 1267–1275. [Google Scholar] [CrossRef] [Green Version]
- Ahmmed, S.M.; Bithi, S.S.; Pore, A.A.; Mubtasim, N.; Schuster, C.; Gollahon, L.S.; Vanapalli, S.A. Multi-sample deformability cytometry of cancer cells. APL Bioeng. 2018, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, W.H.; Schindl, M.; Cardozo, T.J.; Ezzell, R.M. Motility of vinculin-deficient F9 embryonic carcinoma cells analyzed by video, laser confocal, and reflection interference contrast microscopy. Exp. Cell Res. 1995, 221, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, W.H.; Ezzell, R.M. Viscoelasticity in wild-type and vinculin-deficient (5.51) mouse F9 embryonic carcinoma cells examined by atomic force microscopy and rheology. Exp. Cell Res. 1996, 226, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Thoumine, O.; Ott, A. Comparison of the mechanical properties of normal and transformed fibroblasts. Biorheology 1997, 34, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M.; Laidler, P.; Gil, D.; Lekki, J.; Stachura, Z.; Hrynkiewicz, A.Z. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur. Biophys. J. 1999, 28, 312–316. [Google Scholar] [CrossRef]
- Ward, K.A.; Li, W.-I. Viscoelastic Properties of Transformed Cells: Role in Tumor Cell Progression and Metastasis Formation. Biorheology 1991, 28, 301–313. [Google Scholar] [CrossRef]
- Li, Q.S.; Lee, G.Y.H.; Ong, C.N.; Lim, C.T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef]
- Plodinec, M.; Loparic, M.; Monnier, C.A.; Obermann, E.C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J.T.; Aebi, U.; Bentires-Alj, M.; Lim, R.Y.H.; et al. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Jiang, N.; Zheng, L.; Zeng, J.; Qiu, C.; Yang, H.; Xie, S. Quantitative analysis of the cell-surface roughness and viscoelasticity for breast cancer cells discrimination using atomic force microscopy. Scanning 2016, 38, 558–563. [Google Scholar] [CrossRef] [Green Version]
- Dumitru, A.C.; Mohammed, D.; Maja, M.; Yang, J.; Verstraeten, S.; del Campo, A.; Mingeot-Leclercq, M.P.; Tyteca, D.; Alsteens, D. Label-Free Imaging of Cholesterol Assemblies Reveals Hidden Nanomechanics of Breast Cancer Cells. Adv. Sci. 2020, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Palmer, X.L.; Ortega-Rodas, J.; Balraj, V.; Dastider, I.G.; Chandra, S. Biomechanical and Biophysical Properties of Breast Cancer Cells Under Varying Glycemic Regimens. Breast Cancer Basic Clin. Res. 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Weder, G.; Hendriks-Balk, M.C.; Smajda, R.; Rimoldi, D.; Liley, M.; Heinzelmann, H.; Meister, A.; Mariotti, A. Increased plasticity of the stiffness of melanoma cells correlates with their acquisition of metastatic properties. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.R.; Pabijan, J.; Garcia, R.; Lekka, M. The softening of human bladder cancer cells happens at an early stage of the malignancy process. Beilstein J. Nanotechnol. 2014, 5, 447–457. [Google Scholar] [CrossRef]
- Watanabe, T.; Kuramochi, H.; Takahashi, A.; Imai, K.; Katsuta, N.; Nakayama, T.; Fujiki, H.; Suganuma, M. Higher cell stiffness indicating lower metastatic potential in B16 melanoma cell variants and in (2)-epigallocatechin gallate-treated cells. J. Cancer Res. Clin. Oncol. 2012, 138, 859–866. [Google Scholar] [CrossRef]
- Pogoda, K.; Pięta, E.; Roman, M.; Piergies, N.; Liberda, D.; Wróbel, T.P.; Janmey, P.A.; Paluszkiewicz, C.; Kwiatek, W.M. In search of the correlation between nanomechanical and biomolecular properties of prostate cancer cells with different metastatic potential. Arch. Biochem. Biophys. 2021, 697, 108718. [Google Scholar] [CrossRef]
- Chivukula, V.K.; Krog, B.L.; Nauseef, J.T.; Henry, M.D.; Vigmostad, S.C. Alterations in cancer cell mechanical properties after fluid shear stress exposure: A micropipette aspiration study. Cell Health Cytoskelet. 2015, 7, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Faria, E.C.; Ma, N.; Gazi, E.; Gardner, P.; Brown, M.; Clarke, N.W.; Snook, R.D. Measurement of elastic properties of prostate cancer cells using AFM. Analyst 2008, 133, 1498–1500. [Google Scholar] [CrossRef]
- Darling, E.M.; Zauscher, S.; Block, J.A.; Guilak, F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: Do cell properties reflect metastatic potential? Biophys. J. 2007, 92, 1784–1791. [Google Scholar] [CrossRef] [Green Version]
- Swaminathan, V.; Mythreye, K.; O’Brien, E.T.; Berchuck, A.; Blobe, G.C.; Superfine, R. Mechanical Stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011, 71, 5075–5080. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Lu, Q.; Sharma, S.; Ly, C.; Di Carlo, D.; Rowat, A.C.; Leclaire, M.; Kim, D.; Chow, C.; Gimzewski, J.K.; et al. Single Cell Mechanotype and Associated Molecular Changes in Urothelial Cell Transformation and Progression. Front. Cell. Dev. Biol. 2020, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Li, Y.; Liu, W.; Jin, L.; Jiang, X.; Wang, X.; Ding, Z.; Peng, Y.; Zhou, J.; Fan, J.; et al. The nanomechanical signature of liver cancer tissues and its molecular origin. Nanoscale 2015, 7, 12998–13010. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, A.; Staunton, J.R.; Nandakumar, V.; Banyai, N.; Davies, P.C.W.; Ros, R. AFM stiffness nanotomography of normal, metaplastic and dysplastic human esophageal cells. Phys. Biol. 2011, 8, 015007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.H.; Lin, H.K.; Lin, I.H.; Chiou, Y.W.; Chen, H.W.; Liu, C.Y.; Harn, H.I.C.; Chiu, W.T.; Wang, Y.K.; Shen, M.R.; et al. Mechanical phenotype of cancer cells: Cell softening and loss of stiffness sensing. Oncotarget 2015, 6, 20946–20958. [Google Scholar] [CrossRef] [Green Version]
- Prabhune, M.; Belge, G.; Dotzauer, A.; Bullerdiek, J.; Radmacher, M. Comparison of mechanical properties of normal and malignant thyroid cells. Micron 2012, 43, 1267–1272. [Google Scholar] [CrossRef]
- Zhao, S.; Stamm, A.; Lee, J.S.; Gruverman, A.; Lim, J.Y.; Gu, L. Elasticity of Differentiated and Undifferentiated Human Neuroblastoma Cells Characterized by Atomic Force Microscopy. J. Mech. Med. Biol. 2015, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Şahin, M.; Öncü, G.; Yılmaz, M.A.; Özkan, D.; Saybaşılı, H. Transformation of SH-SY5Y cell line into neuron-like cells: Investigation of electrophysiological and biomechanical changes. Neurosci. Lett. 2021, 745, 135628. [Google Scholar] [CrossRef]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Kai, K.; Choi, D.S.; Iwamoto, T.; Nguyen, Y.H.; Wong, H.; Landis, M.D.; Ueno, N.T.; Chang, J.; Qin, L. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2012, 109, 18707–18712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remmerbach, T.W.; Wottawah, F.; Dietrich, J.; Lincoln, B.; Wittekind, C.; Guck, J. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009, 69, 1728–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gullekson, C.; Cojoc, G.; Schürmann, M.; Guck, J.; Pelling, A. Mechanical mismatch between Ras transformed and untransformed epithelial cells. Soft Matter 2017, 13, 8483–8491. [Google Scholar] [CrossRef]
- Ciucci, S.; Ge, Y.; Durán, C.; Palladini, A.; Jiménez-Jiménez, V.; Martínez-Sánchez, L.M.; Wang, Y.; Sales, S.; Shevchenko, A.; Poser, S.W.; et al. Enlightening discriminative network functional modules behind principal component analysis separation in differential-omic science studies. Sci. Rep. 2017, 7, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Wullkopf, L.; West, A.K.V.; Leijnse, N.; Cox, T.R.; Madsen, C.D.; Oddershede, L.B.; Erler, J.T. Cancer cells’ ability to mechanically adjust to extracellular matrix stiffness correlates with their invasive potential. Mol. Biol. Cell 2018, 29, 2378–2385. [Google Scholar] [CrossRef] [PubMed]
- Holenstein, C.N.; Horvath, A.; Schär, B.; Schoenenberger, A.D.; Bollhalder, M.; Goedecke, N.; Bartalena, G.; Otto, O.; Herbig, M.; Guck, J.; et al. The relationship between metastatic potential and in vitro mechanical properties of osteosarcoma cells. Mol. Biol. Cell 2019, 30, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Davis, S.P.; Yang, F.; Paulsen, K.S.; Kumar, M.; DeVaux, R.S.; Wang, X.; Conklin, D.S.; Oberai, A.; Herschkowitz, J.I.; et al. Inertial Microfluidic Cell Stretcher (iMCS): Fully Automated, High-Throughput, and Near Real-Time Cell Mechanotyping. Small 2017, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef]
- Suresh, S. Nanomedicine: Elastic clues in cancer detection. Nat. Nanotechnol. 2007, 2, 748–749. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Tondre, J.; Wong, R.; Rao, J.Y.; Gimzewski, J.K. AFM-based analysis of human metastatic cancer cells. Nanotechnology 2008, 19, 384003. [Google Scholar] [CrossRef] [PubMed]
- Tse, H.T.K.; Gossett, D.R.; Moon, Y.S.; Masaeli, M.; Sohsman, M.; Ying, Y.; Mislick, K.; Adams, R.P.; Rao, J.; Di Carlo, D. Quantitative Diagnosis of Malignant Pleural Effusions by Single-Cell Mechanophenotyping. Sci. Transl. Med. 2013, 5, 212ra163. [Google Scholar] [CrossRef] [Green Version]
- Guck, J.; Chilvers, E.R. Mechanics meets medicine. Sci. Transl. Med. 2013, 5, 5–7. [Google Scholar] [CrossRef]
- Osmulski, P.; Mahalingam, D.; Gaczynska, M.E.; Liu, J.; Huang, S.; Horning, A.M.; Wang, C.M.; Thompson, I.M.; Huang, T.H.M.; Chen, C.L. Nanomechanical biomarkers of single circulating tumor cells for detection of castration resistant prostate cancer. Prostate 2014, 74, 1297–1307. [Google Scholar] [CrossRef] [Green Version]
- Janmey, P.A.; Euteneuer, U.; Traub, P.; Schliwa, M. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J. Cell Biol. 1991, 113, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charrier, E.E.; Janmey, P.A. Mechanical Properties of Intermediate Filament Proteins. Methods Enzymol. 2016, 568, 35–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schliwa, M. Action of Cytochalasin D on Cytoskeletal Networks High-voltage Electron Microscopy Cytochalasin D Applied to Intact Cells. J. Cell Biol. 1982, 92, 79–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotsch, C.; Braet, F.; Wisse, E.; Radmacher, M. AFM imaging and elasticity measurements on living rat liver macrophages. Cell Biol. Int. 1997, 21, 685–696. [Google Scholar] [CrossRef]
- Wu, H.W.; Kuhn, T.; Moy, V.T. Mechanical properties of L929 cells measured by atomic force microscopy: Effects of anticytoskeletal drugs and membrane crosslinking. Scanning 1998, 20, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Bushell, G.R.; Cahill, C.; Clarke, F.M.; Gibson, C.T.; Myhra, S.; Watson, G.S. Imaging and force-distance analysis of human fibroblasts in vitro by atomic force microscopy. Cytometry 1999, 36, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Rotsch, C.; Radmacher, M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: An atomic force microscopy study. Biophys. J. 2000, 78, 520–535. [Google Scholar] [CrossRef] [Green Version]
- Schulze, C.; Müller, K.; Käs, J.A.; Gerdelmann, J.C. Compaction of cell shape occurs before decrease of elasticity in CHO-K1 cells treated with actin cytoskeleton disrupting drug cytochalasin D. Cell Motil. Cytoskelet. 2009, 66, 193–201. [Google Scholar] [CrossRef]
- Liu, Y.; Mollaeian, K.; Shamim, M.H.; Ren, J. Effect of F-actin and microtubules on cellular mechanical behavior studied using atomic force microscope and an image recognition-based cytoskeleton quantification approach. Int. J. Mol. Sci. 2020, 21, 392. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Zhang, L.; Kang, H.; Zhang, B.; Bao, G.; Wang, J. Mechanical, nanomorphological and biological reconstruction of early-stage apoptosis in HeLa cells induced by cytochalasin B. Oncol. Rep. 2019, 41, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M.; Laidler, P.; Ignacak, J.J.; Labd, M.; Lekki, J.; Struszczyk, H.; Stachura, Z.; Hrynkiewicz, A.Z. The effect of chitosan on stiffness and glycolytic activity of human bladder cells. Biochim. Biophys. Acta Mol. Cell Res. 2001, 1540, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.; Shao, Y.; Wineman, A.; Fu, J.; Waas, A. Atomic force microscopy indentation and inverse analysis for non-linear viscoelastic identification of breast cancer cells. Math. Biosci. 2016, 277, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Rudzka, D.A.; Spennati, G.; McGarry, D.J.; Chim, Y.H.; Neilson, M.; Ptak, A.; Munro, J.; Kalna, G.; Hedley, A.; Moralli, D.; et al. Migration through physical constraints is enabled by MAPK-induced cell softening via actin cytoskeleton re-organization. J. Cell Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S.; Cho, C.H.; Park, E.K.; Jung, M.H.; Yoon, K.S.; Park, H.K. AFM-Detected apoptotic changes in morphology and biophysical property caused by paclitaxel in Ishikawa and HeLa cells. PLoS ONE 2012, 7, e30066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.C.K.; Yang, C.H.; Ju, M.S. Cytotoxic and biomechanical effects of clinical dosing schemes of paclitaxel on neurons and cancer cells. Cancer Chemother. Pharmacol. 2020, 86, 245–255. [Google Scholar] [CrossRef]
- Raudenska, M.; Kratochvilova, M.; Vicar, T.; Gumulec, J.; Balvan, J.; Polanska, H.; Pribyl, J.; Masarik, M. Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, W.; Li, L.; Zhu, X.; Liu, J.; Wang, X.; Song, Z.; Xu, H.; Wang, Z. Biomechanical measurement and analysis of colchicine-induced effects on cells by nanoindentation using an atomic force microscope. J. Biomech. 2018, 67, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Ketene, A.N.; Roberts, P.C.; Shea, A.A.; Schmelz, E.M.; Agah, M. Actin filaments play a primary role for structural integrity and viscoelastic response in cells. Integr. Biol. 2012, 4, 540–549. [Google Scholar] [CrossRef]
- Suresh, S.; Spatz, J.; Mills, J.P.; Micoulet, A.; Dao, M.; Lim, C.T.; Beil, M.; Seufferlein, T. Connections between single-cell biomechanics and human disease states: Gastrointestinal cancer and malaria. Acta Biomater. 2005, 23, S3–S15. [Google Scholar] [CrossRef]
- Seltmann, K.; Roth, W.; Kröger, C.; Loschke, F.; Lederer, M.; Hüttelmaier, S.; Magin, T.M. Keratins mediate localization of hemidesmosomes and repress cell motility. J. Investig. Dermatol. 2013, 133, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathje, L.S.Z.; Nordgren, N.; Pettersson, T.; Rönnlund, D.; Widengren, J.; Aspenström, P.; Gad, A.K.B. Oncogenes induce a vimentin filament collapse mediated by HDAC6 that is linked to cell stiffness. Proc. Natl. Acad. Sci. USA 2014, 111, 1515–1520. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Koch, D.; Cardenas, R.; Kas, J.; Shin, C.K. Cell motility and local viscoelasticity of fibroblasts. Biophys. J. 2005, 89, 4330–4342. [Google Scholar] [CrossRef] [Green Version]
- Johnson-Lyles, D.N.; Peifley, K.; Lockett, S.; Neun, B.W.; Hansen, M.; Clogston, J.; Stern, S.T.; McNeil, S.E. Fullerenol cytotoxicity in kidney cells is associated with cytoskeleton disruption, autophagic vacuole accumulation, and mitochondrial dysfunction. Toxicol. Appl. Pharmacol. 2010, 248, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Mrđanović, J.; Šolajić, S.; Bogdanović, V.; Stankov, K.; Bogdanović, G.; Djordjevic, A. Effects of fullerenol C60(OH)24 on the frequency of micronuclei and chromosome aberrations in CHO-K1 cells. Mutat. Res. Toxicol. Environ. Mutagen. 2009, 680, 25–30. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Wang, X. AFM-Based Study of Fullerenol (C 60 (OH) 24)-Induced Changes of Elasticity in Living SMCC-7721 Cells. J. Mech. Behav. Biomed. Mater. 2015, 45, 65–74. [Google Scholar] [CrossRef]
- Luo, L. RHO GTPASES in neuronal morphogenesis. Nat. Rev. Neurosci. 2000, 1, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Iu, C.Y.Y.; Lui, C.N.P.; Zou, Y.; Fung, C.K.M.; Li, H.W.; Xi, N.; Yung, K.K.L.; Lai, K.W.C. Investigating dynamic structural and mechanical changes of neuroblastoma cells associated with glutamate-mediated neurodegeneration. Sci. Rep. 2014, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pierzchała, K.; Lekka, M.; Magrez, A.; Kulik, A.J.; Forró, L.; Sienkiewicz, A. Photocatalytic and phototoxic properties of TiO2-based nanofilaments: ESR and AFM assays. Nanotoxicology 2012, 6, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 2005, 60, 24–34. [Google Scholar] [CrossRef]
- Byfield, F.J.; Reen, R.K.; Shentu, T.P.; Levitan, I.; Gooch, K.J. Endothelial actin and cell stiffness is modulated by substrate stiffness in 2D and 3D. J. Biomech. 2009, 42, 1114–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, J.; Wang, N.; Wang, Y. Mechanotransduction at focal adhesions: From physiology to cancer development. J. Cell. Mol. Med. 2013, 17, 597–604. [Google Scholar] [CrossRef]
- Lorenzen, J.; Sinkus, R.; Lorenzen, M.; Dargatz, M.; Leussler, C.; Röschmann, P.; Adam, G. MR elastography of the breast: Preliminary clinical results. RoFo 2002, 174, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.K.; Yin, M.; Glockner, J.F.; Takahashi, N.; Araoz, P.A.; Talwalkar, J.A.; Ehman, R.L. MR elastography of liver tumors: Preliminary results. Am. J. Roentgenol. 2008, 190, 1534–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenner, J.; Stacer, A.C.; Winterroth, F.; Johnson, T.D.; Luker, K.E.; Luker, G.D. Macroscopic stiffness of breast tumors predicts metastasis. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef]
- Ciasca, G.; Sassun, T.E.; Minelli, E.; Antonelli, M.; Papi, M.; Santoro, A.; Giangaspero, F.; Delfini, R.; De Spirito, M. Nano-mechanical signature of brain tumours. Nanoscale 2016, 8, 19629–19643. [Google Scholar] [CrossRef] [PubMed]
- Cieśluk, M.; Pogoda, K.; Deptuła, P.; Werel, P.; Kułakowska, A.; Kochanowicz, J.; Mariak, Z.; Łysoń, T.; Reszeć, J.; Bucki, R. Nanomechanics and Histopathology as Diagnostic Tools to Characterize Freshly Removed Human Brain Tumors. Int. J. Nanomed. 2020, 15, 7509–7521. [Google Scholar] [CrossRef] [PubMed]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Brossel, R.; Yahi, A.; David, S.; Velasquez, L.M.; Guinebretière, J.M. Mechanical signals inhibit growth of a grafted tumor in vivo: Proof of concept. PLoS ONE 2016, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rianna, C.; Radmacher, M.; Kumar, S. Direct evidence that tumor cells soften when navigating confined spaces. Mol. Biol. Cell 2020, 31, 1726–1734. [Google Scholar] [CrossRef] [PubMed]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.L.; Bonnecaze, R.T.; Zaman, M.H. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys. J. 2009, 97, 1013–1021. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Q.; Lan, H.Y.; Wu, Y.C.; Yang, W.H.; Chiou, A.; Yang, M.H. Epithelial-mesenchymal transition softens head and neck cancer cells to facilitate migration in 3D environments. J. Cell. Mol. Med. 2018, 22, 3837–3846. [Google Scholar] [CrossRef]
- Staunton, J.R.; Doss, B.L.; Lindsay, S.; Ros, R. Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen i matrices. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Tavares, S.; Vieira, A.F.; Taubenberger, A.V.; Araújo, M.; Martins, N.P.; Brás-Pereira, C.; Polónia, A.; Herbig, M.; Barreto, C.; Otto, O.; et al. Actin stress fiber organization promotes cell stiffening and proliferation of pre-invasive breast cancer cells. Nat. Commun. 2017, 8, 15237. [Google Scholar] [CrossRef]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef]
- Haeger, A.; Krause, M.; Wolf, K.; Friedl, P. Cell jamming: Collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2386–2395. [Google Scholar] [CrossRef]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the collagen matrix in aging skin promotes melanoma metastasis and affects immune cell motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.L.; Pegoraro, A.F.; Li, H.; Li, K.; Yuan, Y.; Xu, G.; Gu, Z.; Sun, J.; Hao, Y.; Gupta, S.K.; et al. Cell swelling, softening and invasion in a three-dimensional breast cancer model. Nat. Phys. 2020, 16, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gensbittel, V.; Kräter, M.; Harlepp, S.; Busnelli, I.; Guck, J.; Goetz, J.G. Mechanical Adaptability of Tumor Cells in Metastasis. Dev. Cell 2021, 56, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.K.; Lin, H.H.; Harn, H.I.C.; Hughes, M.W.; Tang, M.J.; Yang, C.C. Mechanical forces in skin disorders. J. Dermatol. Sci. 2018, 90, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Dean, D. Mechanical Properties of Stem Cells from Different Sources During Vascular Smooth Muscle Cell Differentiation. MCB Mol. Cell. Biomech. 2017, 14, 153–169. [Google Scholar] [CrossRef]
- Sharma, S.; Santiskulvong, C.; Rao, J.; Gimzewski, J.K.; Dorigo, O. The role of Rho GTPase in cell stiffness and cisplatin resistance in ovarian cancer cells. Integr. Biol. 2014, 6, 611–617. [Google Scholar] [CrossRef]
- Seo, Y.H.; Jo, Y.N.; Oh, Y.J.; Park, S. Nano-mechanical reinforcement in drug-resistant ovarian cancer cells. Biol. Pharm. Bull. 2015, 38, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Park, S. Mechanical Alteration Associated with Chemotherapeutic Resistance of Breast Cancer Cells. J. Cancer Prev. 2018, 23, 87–92. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Runel, G.; Lopez-Ramirez, N.; Chlasta, J.; Masse, I. Biomechanical Properties of Cancer Cells. Cells 2021, 10, 887. https://doi.org/10.3390/cells10040887
Runel G, Lopez-Ramirez N, Chlasta J, Masse I. Biomechanical Properties of Cancer Cells. Cells. 2021; 10(4):887. https://doi.org/10.3390/cells10040887
Chicago/Turabian StyleRunel, Gaël, Noémie Lopez-Ramirez, Julien Chlasta, and Ingrid Masse. 2021. "Biomechanical Properties of Cancer Cells" Cells 10, no. 4: 887. https://doi.org/10.3390/cells10040887
APA StyleRunel, G., Lopez-Ramirez, N., Chlasta, J., & Masse, I. (2021). Biomechanical Properties of Cancer Cells. Cells, 10(4), 887. https://doi.org/10.3390/cells10040887





