Precision Oncology Beyond Genomics: The Future Is Here—It Is Just Not Evenly Distributed
Abstract
1. Introduction
2. Intra-Tumor Heterogeneity—The Challenge of Treating “Many Cancers in One”
3. Integration of Intra-Tumor Heterogeneity into Multi-Layered Personalized Cancer Therapy
3.1. Current Approaches to Analyze ITH from Tumor Samples
3.1.1. Genomic Approaches to ITH and Precision Oncology
3.1.2. Proteomic Approaches
3.1.3. Metabolomic Approaches
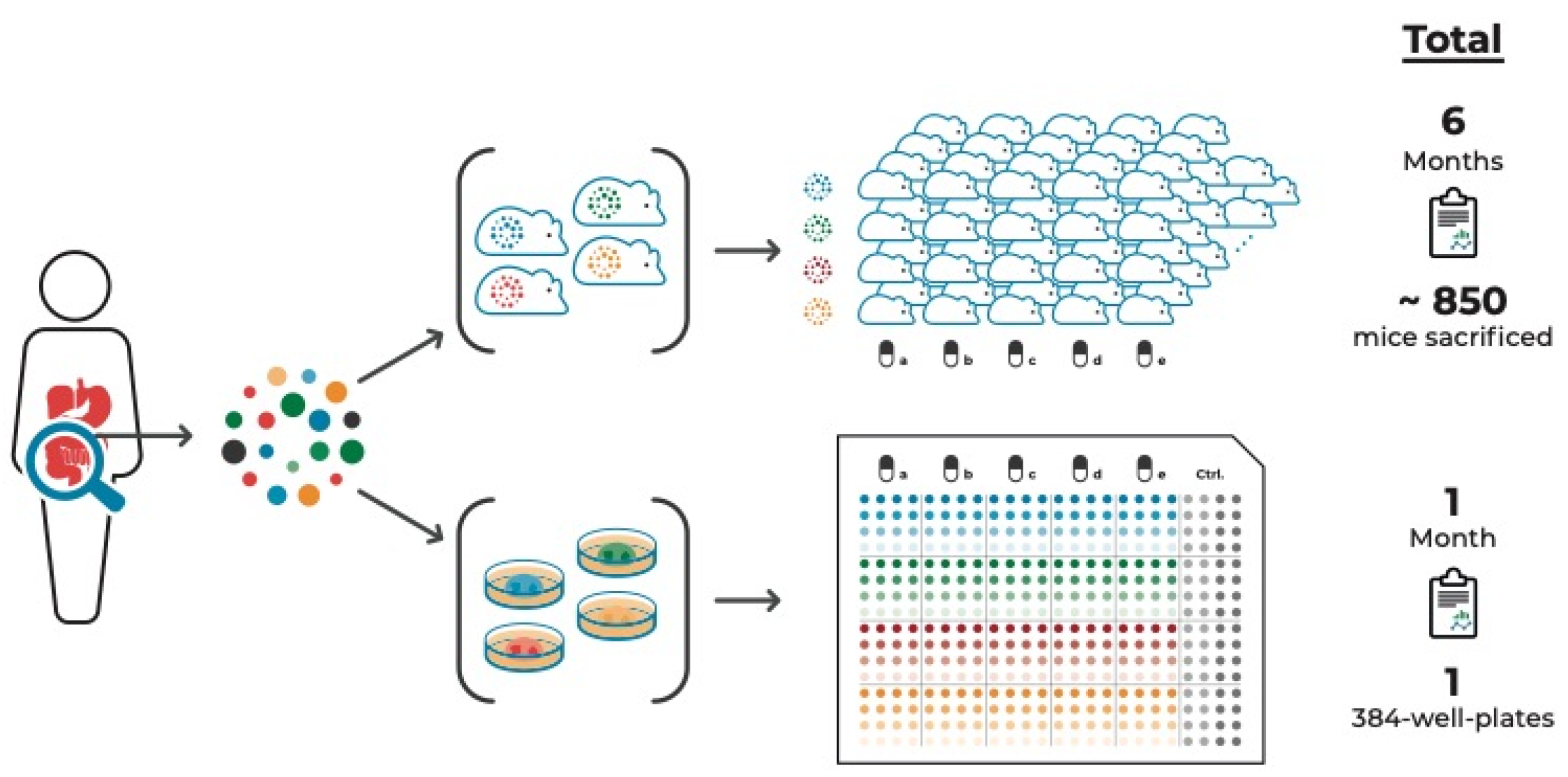
3.2. Suitable In Vitro Strategies for Modeling Intra-Tumor Heterogeneity
4. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Report on Cancer: Setting Priorities, Investing Wisely and Providing Care for All; World Health Organization: Geneve, Switzerland, 2020. [Google Scholar]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Shankaran, V.; Obel, J.; Benson, A.B. Predicting response to EGFR inhibitors in metastatic colorectal cancer: Current practice and future directions. Oncologist 2010, 15, 157–167. [Google Scholar] [CrossRef][Green Version]
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for cancer: A trigger for metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef]
- Roser, M.; Ritchie, H. Cancer. Available online: https://ourworldindata.org/cancer (accessed on 14 October 2020).
- Pich, O.; Muiños, F.; Lolkema, M.P.; Steeghs, N.; Gonzalez-Perez, A.; Lopez-Bigas, N. The mutational footprints of cancer therapies. Nat. Genet. 2019, 51, 1732–1740. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2019, 6, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Joe, A.; Felsher, D. Oncogene addiction. Cancer Res. 2008, 68, 3077–3080. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Nicolantonio, F.D.; Sartore-Bianchi, A.; Siena, S.; Bardelli, A. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014, 4, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Emburgh, B.O.V.; Sartore-Bianchi, A.; Nicolantonio, F.D.; Siena, S.; Bardelli, A. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol. Oncol. 2014, 8, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Abramson, R. Overview of Targeted Therapies for Cancer. My Cancer Genome. Available online: https://www.mycancergenome.org/content/molecular-medicine/overview-of-targeted-therapies-for-cancer/ (accessed on 30 January 2021).
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; IARC Publications: Lyon, France, 2020; ISBN 978-92-832-0448-0. [Google Scholar]
- Caravagna, G.; Giarratano, Y.; Ramazzotti, D.; Tomlinson, I.; Graham, T.A.; Sanguinetti, G.; Sottoriva, A. Detecting repeated cancer evolution from multi-region tumor sequencing data. Nat. Methods 2018, 15, 707–714. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer heterogeneity: Implications for targeted therapeutics. Brit. J. Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta BBA Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.W.; Loeb, L.A.; Salk, J.J. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2016, 13, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Raphael, B.J.; Chen, F.; Wendl, M.C. Advances for studying clonal evolution in cancer. Cancer Lett. 2013, 340, 212–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garattini, S.; Nerini, I.F.; D’Incalci, M. Not only tumor but also therapy heterogeneity. Annals of Oncology 2017, 29, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Kurebayashi, Y.; Ojima, H.; Tsujikawa, H.; Kubota, N.; Maehara, J.; Abe, Y.; Kitago, M.; Shinoda, M.; Kitagawa, Y.; Sakamoto, M. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018, 68, 1025–1041. [Google Scholar] [CrossRef]
- Zhang, S.-C.; Hu, Z.-Q.; Long, J.-H.; Zhu, G.-M.; Wang, Y.; Jia, Y.; Zhou, J.; Ouyang, Y.; Zeng, Z. Clinical implications of tumor-infiltrating immune cells in breast cancer. J. Cancer 2019, 10, 6175–6184. [Google Scholar] [CrossRef]
- Vareki, S.M. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J. Immunother. Cancer 2018, 6, 157. [Google Scholar] [CrossRef]
- Zhang, Q.; Lou, Y.; Yang, J.; Wang, J.; Feng, J.; Zhao, Y.; Wang, L.; Huang, X.; Fu, Q.; Ye, M.; et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 2019, 68, 2019. [Google Scholar] [CrossRef]
- Sowalsky, A.G.; Ye, H.; Bhasin, M.; Allen, E.M.V.; Loda, M.; Lis, R.T.; Montaser-Kouhsari, L.; Calagua, C.; Ma, F.; Russo, J.W.; et al. Neoadjuvant-Intensive Androgen Deprivation Therapy Selects for Prostate Tumor Foci with Diverse Subclonal Oncogenic Alterations. Cancer Res. 2018, 78, 4716–4730. [Google Scholar] [CrossRef]
- Nowell, P. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Kwon, S.M.; Budhu, A.; Woo, H.G.; Chaisaingmongkol, J.; Dang, H.; Forgues, M.; Harris, C.C.; Zhang, G.; Auslander, N.; Ruppin, E.; et al. Functional genomic complexity defines intratumor heterogeneity and tumor aggressiveness in liver cancer. Sci. Rep. 2019, 9, 16930. [Google Scholar] [CrossRef]
- Sun, X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. A practical approach to tumor heterogeneity in clinical research and diagnostics. Pathobiology 2018, 85, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Strickaert, A.; Saiselet, M.; Dom, G.; Deken, X.D.; Dumont, J.E.; Feron, O.; Sonveaux, P.; Maenhaut, C. Cancer heterogeneity is not compatible with one unique cancer cell metabolic map. Oncogene 2017, 36, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Erkan, M.; Erkan, M.; Hausmann, S.; Michalski, C.W.; Fingerle, A.A.; Dobritz, M.; Kleeff, J.; Friess, H. The role of stroma in pancreatic cancer: Diagnostic and therapeutic implications. Nat. Rev. Gastroentero 2012, 9, 454. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; Groot, A.E.D.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Stanta, G.; Bonin, S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef]
- Loeb, L.A. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat. Rev. Cancer 2011, 11, 450–457. [Google Scholar] [CrossRef]
- Losic, B.; Craig, A.J.; Villacorta-Martin, C.; Martins-Filho, S.N.; Akers, N.; Chen, X.; Ahsen, M.E.; Von Felden, J.; Labgaa, I.; DʹAvola, D.; et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020, 11, 291. [Google Scholar] [CrossRef]
- Swanton, C. Intratumor heterogeneity: Evolution through space and time. Cancer Res. 2012, 72, 4875–4882. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Tan, Y.; Li, S.; Zhou, J.; Wang, Q.; Wang, Y.; Xie, Y.; Chen, L.; Li, J.; Fang, C.; et al. Genomic landscapes by multiregion sequencing combined with circulation tumor DNA detection contribute to molecular diagnosis in glioblastomas. Aging 2019, 11, 11224–11243. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, D.; Andrieux, G.; Boehnke, K.; Keil, M.; Silvestri, A.; Silvestrov, M.; Keilholz, U.; Haybaeck, J.; Erdmann, G.; Sachse, C.; et al. Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures. PLoS Genet. 2019, 15, e1008076. [Google Scholar] [CrossRef]
- Lim, Z.-F.; Ma, P.C. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Wogsland, C.E.; Greenplate, A.R.; Kolstad, A.; Myklebust, J.H.; Irish, J.M.; Huse, K. Mass cytometry of follicular lymphoma tumors reveals intrinsic heterogeneity in proteins including HLA-DR and a deficit in nonmalignant plasmablast and germinal center B-cell populations. Cytom. Part B Clin. Cytom. 2017, 92, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Araf, S.; Wang, J.; Korfi, K.; Pangault, C.; Kotsiou, E.; Rio-Machin, A.; Rahim, T.; Heward, J.; Clear, A.; Iqbal, S.; et al. Correction: Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2019, 33, 1540. [Google Scholar] [CrossRef] [PubMed]
- Gawad, C.; Koh, W.; Quake, S.R. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. P. Natl. Acad. Sci. USA 2014, 111, 17947–17952. [Google Scholar] [CrossRef]
- Farmanbar, A.; Firouzi, S.; Makałowski, W.; Kneller, R.; Iwanaga, M.; Utsunomiya, A.; Nakai, K.; Watanabe, T. Mutational intratumor heterogeneity is a complex and early event in the development of adult T-cell leukemia/lymphoma. Neoplasia 2018, 20, 883–893. [Google Scholar] [CrossRef]
- Stanta, G.; Jahn, S.W.; Bonin, S.; Hoefler, G. Tumour heterogeneity: Principles and practical consequences. Virchows Arch. 2016, 469, 371–384. [Google Scholar] [CrossRef]
- Gambara, G.; Gaebler, M.; Keilholz, U.; Regenbrecht, C.R.A.; Silvestri, A. From chemotherapy to combined targeted therapeutics: In vitro and in vivo models to decipher intra-tumor heterogeneity. Front. Pharmacol. 2018, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, A.; Laere, B.D.; Peeters, D.; Peeters, M.; Salgado, R.; Dirix, L.; Laere, S.V. Evaluation and consequences of heterogeneity in the circulating tumor cell compartment. Oncotarget 2015, 7, 48625–48643. [Google Scholar] [CrossRef]
- Inde, Z.; Dixon, S.J. The impact of non-genetic heterogeneity on cancer cell death. Crit. Rev. Biochem. Mol. 2017, 53, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hinohara, K.; Polyak, K. Intratumoral heterogeneity: More than just mutations. Trends Cell Biol. 2019, 29, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.; Huang, S. Precision oncology: Between vaguely right and precisely wrong. Cancer Res. 2017, 77, 6473–6479. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- MacConaill, L.E. Existing and emerging technologies for tumor genomic profiling. J. Clin. Oncol. 2013, 31, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Creighton, C.J.; Davis, C.; Donehower, L.; Drummond, J.; Wheeler, D.; Ally, A.; Balasundaram, M.; Birol, I.; Butterfield, Y.S.N.; et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Janiszewska, M. The microcosmos of intratumor heterogeneity: The space-time of cancer evolution. Oncogene 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fujimoto, J.; Zhang, J.; Wedge, D.C.; Song, X.; Zhang, J.; Seth, S.; Chow, C.-W.; Cao, Y.; Gumbs, C.; et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014, 346, 256–259. [Google Scholar] [CrossRef]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V. Perspective: The precision-oncology illusion. Nature 2016, 537, S63. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Soria, J.-C.; Berger, R.; Miller, W.H.; Rubin, E.; Kugel, A.; Tsimberidou, A.; Saintigny, P.; Ackerstein, A.; Braña, I.; et al. Genomic and transcriptomic profiling expands precision cancer medicine: The WINTHER trial. Nat. Med. 2019, 25, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Karapetis, C.S.; Khambata-Ford, S.; Jonker, D.J.; O’Callaghan, C.J.; Tu, D.; Tebbutt, N.C.; Simes, R.J.; Chalchal, H.; Shapiro, J.D.; Robitaille, S.; et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008, 359, 1757–1765. [Google Scholar] [CrossRef]
- Soeda, H.; Shimodaira, H.; Gamoh, M.; Ando, H.; Isobe, H.; Suto, T.; Takahashi, S.; Kakudo, Y.; Amagai, K.; Mori, T.; et al. Phase II trial of cetuximab plus irinotecan for oxaliplatin- and irinotecan-based chemotherapy-refractory patients with advanced and/or metastatic colorectal cancer: Evaluation of efficacy and safety based on KRAS mutation status (T-CORE0801). Oncology 2014, 87, 7–20. [Google Scholar] [CrossRef]
- Amado, R.G.; Wolf, M.; Peeters, M.; Cutsem, E.V.; Siena, S.; Freeman, D.J.; Juan, T.; Sikorski, R.; Suggs, S.; Radinsky, R.; et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 1626–1634. [Google Scholar] [CrossRef]
- Bokemeyer, C.; Cutsem, E.V.; Rougier, P.; Ciardiello, F.; Heeger, S.; Schlichting, M.; Celik, I.; Köhne, C.-H. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: Pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur. J. Cancer 2012, 48, 1466–1475. [Google Scholar] [CrossRef]
- Price, T.J.; Peeters, M.; Kim, T.W.; Li, J.; Cascinu, S.; Ruff, P.; Suresh, A.S.; Thomas, A.; Tjulandin, S.; Zhang, K.; et al. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): A randomised, multicentre, open-label, non-Inferiority phase 3 study. Lancet Oncol. 2014, 15, 569–579. [Google Scholar] [CrossRef]
- Ye, L.-C.; Liu, T.-S.; Ren, L.; Wei, Y.; Zhu, D.-X.; Zai, S.-Y.; Ye, Q.-H.; Yu, Y.; Xu, B.; Qin, X.-Y.; et al. Randomized controlled trial of cetuximab plus chemotherapy for patients with KRAS wild-type unresectable colorectal liver-limited metastases. J. Clin. Oncol. 2013, 31, 1931–1938. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.-Y.; Siena, S.; Cassidy, J.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Randomized phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: The PRIME Study. J. Clin. Oncol. 2010, 28, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Arnold, D.; Lueza, B.; Douillard, J.-Y.; Peeters, M.; Lenz, H.-J.; Venook, A.; Heinemann, V.; Cutsem, E.V.; Pignon, J.-P.; Tabernero, J.; et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann. Oncol. 2017, 28, 1713–1729. [Google Scholar] [CrossRef]
- García-Foncillas, J.; Sunakawa, Y.; Aderka, D.; Wainberg, Z.; Ronga, P.; Witzler, P.; Stintzing, S. Distinguishing features of cetuximab and panitumumab in colorectal cancer and other solid tumors. Front. Oncol. 2019, 9, 849. [Google Scholar] [CrossRef]
- Goldberg, R.M.; Montagut, C.; Wainberg, Z.A.; Ronga, P.; Audhuy, F.; Taieb, J.; Stintzing, S.; Siena, S.; Santini, D. Optimising the use of cetuximab in the continuum of care for patients with metastatic colorectal cancer. ESMO Open 2018, 3, e000353. [Google Scholar] [CrossRef]
- Lu, M.; Zhan, X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018, 9, 77–102. [Google Scholar] [CrossRef]
- Stunnenberg, H.G.; Hubner, N.C. Genomics meets proteomics: Identifying the culprits in disease. Hum. Genet. 2014, 133, 689–700. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2016, 19, 114. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef]
- Akbani, R.; Ng, P.K.S.; Werner, H.M.J.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.-Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on the cancer genome atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 2020, 183, 1436–1456. [Google Scholar] [CrossRef]
- Leong, A.S.-Y.; Zhuang, Z. The changing role of pathology in breast cancer diagnosis and treatment. Pathobiology 2011, 78, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Li, H.; Peng, X.-X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2014, 8, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Sanchez-Espiridion, B.; Lin, M.; White, L.; Mishra, L.; Raju, G.S.; Kopetz, S.; Eng, C.; Hildebrandt, M.A.T.; Chang, D.W.; et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer 2017, 123, 4066–4074. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Cruz, M.M.A.; Jyotsana, N.; Sharma, A.; Yun, H.; Görlich, K.; Wichmann, M.; Schwarzer, A.; Preller, M.; Thol, F.; et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood 2013, 122, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, C.; Qiu, L.; Wang, Y.; Qin, X.; Liu, Y.; Li, Z. Serum unsaturated free fatty acids: A potential biomarker panel for early-stage detection of colorectal cancer. J. Cancer 2016, 7, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Cekanova, M.; Rathore, K. Animal models and therapeutic molecular targets of cancer: Utility and limitations. Drug Des. Dev. Ther. 2014, 8, 1911–1922. [Google Scholar] [CrossRef]
- Tratar, U.L.; Horvat, S.; Cemazar, M. Transgenic mouse models in cancer research. Front. Oncol. 2018, 8, 268. [Google Scholar] [CrossRef]
- Day, C.-P.; Merlino, G.; Van Dyke, T. Preclinical mouse cancer models: A maze of opportunities and challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Politi, K.; Pao, W. How genetically engineered mouse tumor models provide insights into human cancers. J. Clin. Oncol. 2011, 29, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Van Staveren, W.C.G.; Solís, D.Y.W.; Hébrant, A.; Detours, V.; Dumont, J.E.; Maenhaut, C. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim. Biophys. Acta BBA Rev. Cancer 2009, 1795, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Techn. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Clevers, H.; Tuveson, D.A. Organoid models for cancer research. Annu. Rev. Cancer Biol. 2019, 3, 223–234. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Bock, C.; Boutros, M.; Camp, J.G.; Clarke, L.; Clevers, H.; Knoblich, J.A.; Liberali, P.; Regev, A.; Rios, A.C.; Stegle, O.; et al. The organoid cell atlas. Nat. Biotechnol. 2020, 1–5. [Google Scholar] [CrossRef]
- Schütte, M.; Risch, T.; Abdavi-Azar, N.; Boehnke, K.; Schumacher, D.; Keil, M.; Yildiriman, R.; Jandrasits, C.; Borodina, T.; Amstislavskiy, V.; et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 2017, 8, 14262. [Google Scholar] [CrossRef]
- Byrne, A.T.; Alférez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinská, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; Jong, S.D.; Jonkers, J.; Maelandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef]
- Hynds, R.E.; Vladimirou, E.; Janes, S.M. The secret lives of cancer cell lines. Dis. Model. Mech. 2018, 11, dmm037366. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.; Marshall, D.; Reid, Y.; Parkes, H.; Gelber, C. The costs of using unauthenticated, over-passaged cell lines: How much more data do we need? Biotechniques 2007, 43, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, B.A.; Camp, F.; Miknyoczki, S. Animal models of disease: Pre-clinical animal models of cancer and their applications and utility in drug discovery. Biochem. Pharmacol. 2014, 87, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Bibby, M.C. Orthotopic models of cancer for preclinical drug evaluation advantages and disadvantages. Eur. J. Cancer 2004, 40, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Huynh, A.S.; Abrahams, D.F.; Torres, M.S.; Baldwin, M.K.; Gillies, R.J.; Morse, D.L. Development of an orthotopic human pancreatic cancer xenograft model using ultrasound guided injection of cells. PLoS ONE 2011, 6, e20330. [Google Scholar] [CrossRef]
- Eglen, R.M.; Klein, J.-L. Three-dimensional cell culture: A rapidly emerging approach to cellular science and drug discovery. SLAS Discov. 2017, 22, 453–455. [Google Scholar] [CrossRef][Green Version]
- Sittampalam, S.; Eglen, R.; Ferguson, S.; Maynes, J.T.; Olden, K.; Schrader, L.; Shelper, T.; Ferrer, M. Three-dimensional cell culture assays: Are they more predictive of in vivo efficacy than 2D monolayer cell-based assays? Assay Drug. Dev. Technol. 2015, 13, 254–261. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Mönch, D.; Maaß, A.; Gromoll, C.; Hehr, T.; Leibold, T.; Schlitt, H.J.; Dahlke, M.-H.; Renner, P. Three dimensional cultivation increases chemo- and radioresistance of colorectal cancer cell lines. PLoS ONE 2021, 16, e0244513. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- L’Espérance, S.; Bachvarova, M.; Tetu, B.; Mes-Masson, A.-M.; Bachvarov, D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genom. 2008, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Zietarska, M.; Maugard, C.M.; Filali-Mouhim, A.; Alam-Fahmy, M.; Tonin, P.N.; Provencher, D.M.; Mes-Masson, A. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol. Carcinogen. 2007, 46, 872–885. [Google Scholar] [CrossRef] [PubMed]
- Bogdanowicz, D.R.; Lu, H.H. Studying cell-cell communication in co-culture. Biotechnol. J. 2013, 8, 395–396. [Google Scholar] [CrossRef]
- Betriu, N.; Semino, C.E. Development of a 3D co-culture system as a cancer model using a self-assembling peptide scaffold. Gels 2018, 4, 65. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid modeling of the tumor immune microenvironment. Cell 2018, 175, 1972–1988. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van Haar, J.D.; Fanchi, L.F.; Slagter, M.; Van Velden, D.L.D.; Kaing, S.; Kelderman, S.; et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 2018, 174, 1586–1598. [Google Scholar] [CrossRef] [PubMed]
- Schnalzger, T.E.; Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3D Model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, K.; Mochizuki, W.; Matsumoto, Y.; Matsumoto, T.; Fukuda, M.; Mizutani, T.; Watanabe, M.; Nakamura, T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016, 51, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ephraim, Y.E.; Kretzschmar, K.; Clevers, H. Organoids in immunological research. Nat. Rev. Immunol. 2020, 20, 279–293. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung CANCER. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.-J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Cutsem, E.V.; McDermott, R.; Hill, A.; et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet. Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Klein, E.; Hau, A.-C.; Oudin, A.; Golebiewska, A.; Niclou, S.P. Glioblastoma organoids: Pre-clinical applications and challenges in the context of immunotherapy. Front. Oncol. 2020, 10, 604121. [Google Scholar] [CrossRef] [PubMed]
- Mertz, D.R.; Ahmed, T.; Takayama, S. Engineering cell heterogeneity into organs-on-a-chip. Lab Chip 2018, 18, 2378–2395. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-Y.; Lee, J.-H.; Shin, Y.; Chung, S.; Kuh, H.-J. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS ONE 2016, 11, e0159013. [Google Scholar] [CrossRef] [PubMed]
- Wnorowski, A.; Yang, H.; Wu, J.C. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv. Drug Deliver. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Devarasetty, M.; Forsythe, S.; Atala, A.; Soker, S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. 2016, 113, 2020–2032. [Google Scholar] [CrossRef]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Cajal, S.R.Y.; Sesé, M.; Capdevila, C.; Aasen, T.; Mattos-Arruda, L.D.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Pisco, A.O.; Huang, S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: “What does not kill me strengthens me”. Brit. J. Cancer 2015, 112, 1725–1732. [Google Scholar] [CrossRef] [PubMed]
- Pisco, A.O.; Brock, A.; Zhou, J.; Moor, A.; Mojtahedi, M.; Jackson, D.; Huang, S. Non-darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun. 2013, 4, 2467. [Google Scholar] [CrossRef] [PubMed]
- Lagadec, C.; Vlashi, E.; Donna, L.D.; Dekmezian, C.; Pajonk, F. Radiation-induced reprogramming of breast cancer cells. Stem Cells 2012, 30, 833–844. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic plasticity and the hallmarks of cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef]
- Sandhu, C.; Qureshi, A.; Emili, A. Panomics for precision medicine. Trends Mol. Med. 2018, 24, 85–101. [Google Scholar] [CrossRef]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef]
- Salgia, R.; Mambetsariev, I.; Pharaon, R.; Fricke, J.; Baroz, A.R.; Hozo, I.; Chen, C.; Koczywas, M.; Massarelli, E.; Reckamp, K.; et al. Evaluation of omics-based strategies for the management of advanced lung cancer. JCO Oncol. Pract. 2021, 17, e257–e265. [Google Scholar] [CrossRef]
- Cirillo, D.; Núñez-Carpintero, I.; Valencia, A. Artificial intelligence in cancer research: Learning at different levels of data granularity. Mol. Oncol. 2021, 15, 817–829. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfohl, U.; Pflaume, A.; Regenbrecht, M.; Finkler, S.; Graf Adelmann, Q.; Reinhard, C.; Regenbrecht, C.R.A.; Wedeken, L. Precision Oncology Beyond Genomics: The Future Is Here—It Is Just Not Evenly Distributed. Cells 2021, 10, 928. https://doi.org/10.3390/cells10040928
Pfohl U, Pflaume A, Regenbrecht M, Finkler S, Graf Adelmann Q, Reinhard C, Regenbrecht CRA, Wedeken L. Precision Oncology Beyond Genomics: The Future Is Here—It Is Just Not Evenly Distributed. Cells. 2021; 10(4):928. https://doi.org/10.3390/cells10040928
Chicago/Turabian StylePfohl, Ulrike, Alina Pflaume, Manuela Regenbrecht, Sabine Finkler, Quirin Graf Adelmann, Christoph Reinhard, Christian R. A. Regenbrecht, and Lena Wedeken. 2021. "Precision Oncology Beyond Genomics: The Future Is Here—It Is Just Not Evenly Distributed" Cells 10, no. 4: 928. https://doi.org/10.3390/cells10040928
APA StylePfohl, U., Pflaume, A., Regenbrecht, M., Finkler, S., Graf Adelmann, Q., Reinhard, C., Regenbrecht, C. R. A., & Wedeken, L. (2021). Precision Oncology Beyond Genomics: The Future Is Here—It Is Just Not Evenly Distributed. Cells, 10(4), 928. https://doi.org/10.3390/cells10040928





