Pathophysiological Significance of Neutrophilic Transfer RNA-Derived Small RNAs in Asymptomatic Moyamoya Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. Library Preparation and tsRNA Sequencing
Sequencing Data Analysis
2.3. Bioinformatics Analysis
2.4. Validation by Quantitative Reverse Transcription PCR (qRT-PCR)
3. Results
3.1. Differential Expression Analysis
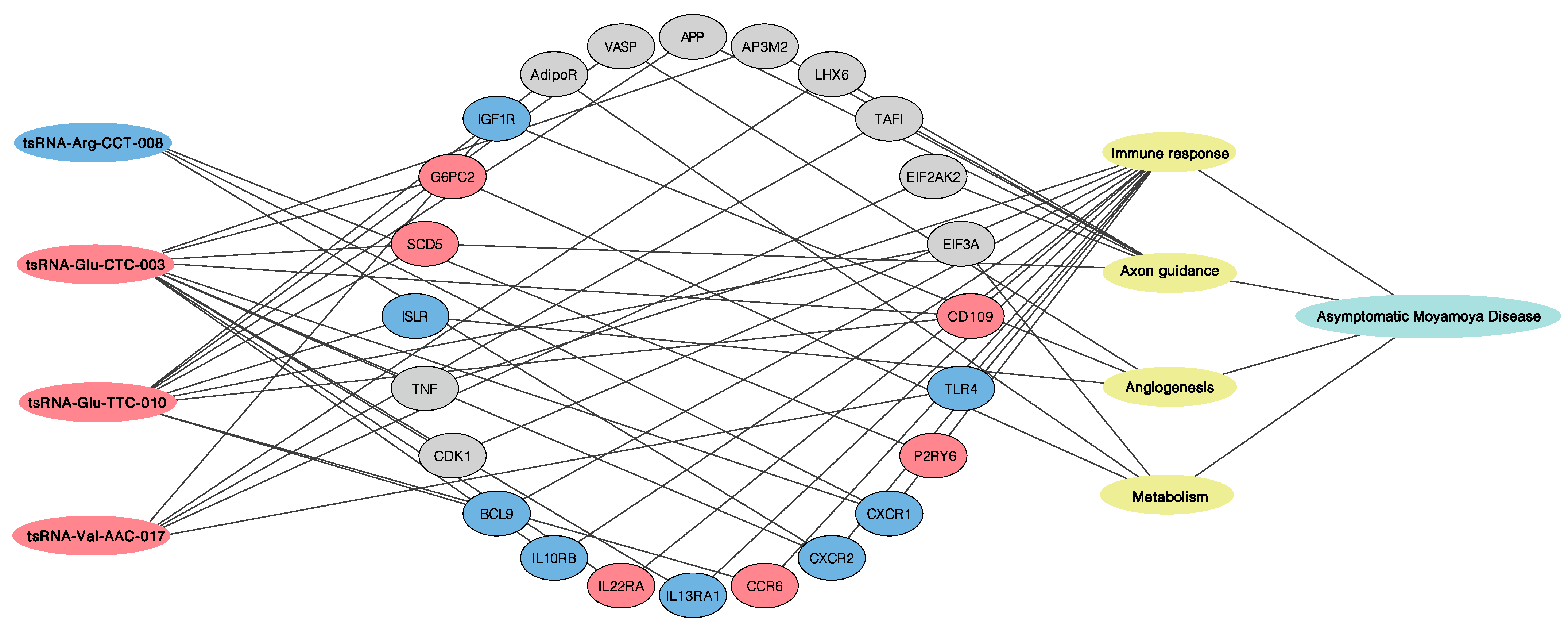
3.2. Functional Pathway Analysis
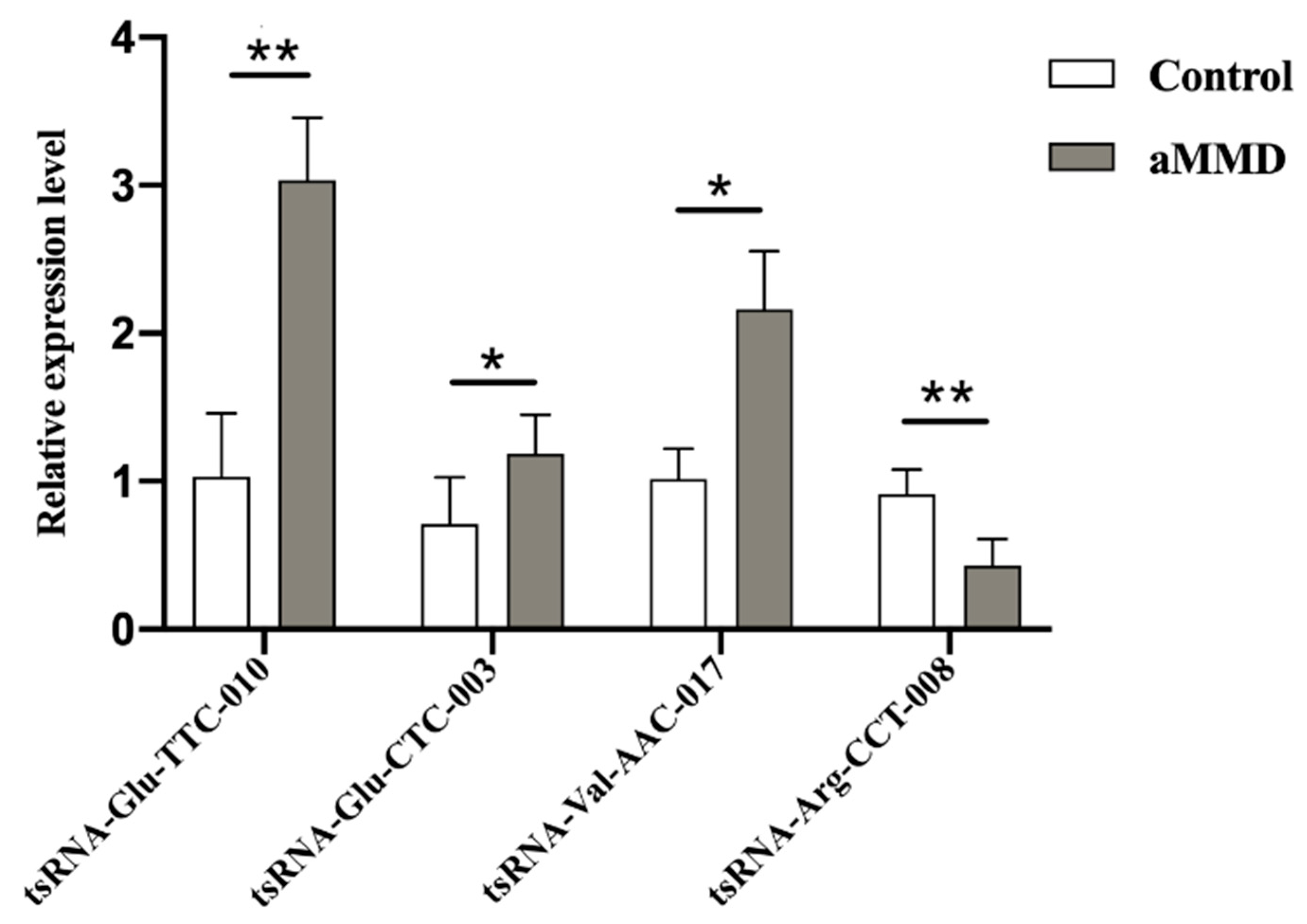
3.3. qRT-PCR Validation
4. Discussion
4.1. Immune Response
4.2. Cell Proliferation and Axon Guidance
4.3. Angiogenesis and Metabolism Adjustment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuroda, S.; Houkin, K. Moyamoya disease: Current concepts and future perspectives. Lancet Neurol. 2008, 7, 1056–1066. [Google Scholar] [CrossRef]
- Kim, J.S. Moyamoya Disease: Epidemiology, Clinical Features, and Diagnosis. J. Stroke 2016, 18, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Ahn, I.M.; Park, D.H.; Hann, H.J.; Kim, K.H.; Kim, H.J.; Ahn, H.S. Incidence, prevalence, and survival of moyamoya disease in Korea: A nationwide, population-based study. Stroke 2014, 45, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Research Committee on the Pathology and Treatment of Spontaneous Occlusion of the Circle of Willis; Health Labour Sciences Research Grant for Research on Measures for Infractable Diseases. Guidelines for diagnosis and treatment of moyamoya disease (spontaneous occlusion of the circle of Willis). Neurol. Med. Chir. 2012, 52, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.-Y.; Wang, Q.-N.; Zhang, Y.; Zhang, Q.; Li, D.-S.; Yang, W.-Z.; Zhang, Z.-S.; Zong, R.; Han, C.; Duan, L. Epidemiology of Moyamoya Disease in China: Single-Center, Population-Based Study. World Neurosurg. 2019, 122, e917–e923. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.M.; Smith, E.R. Moyamoya Disease and Moyamoya Syndrome. N. Engl. J. Med. 2009, 360, 1226–1237. [Google Scholar] [CrossRef] [PubMed]
- Jo, K.-I.; Yeon, J.Y.; Hong, S.-C.; Kim, J.-S. Clinical Course of Asymptomatic Adult Moyamoya Disease. Cerebrovasc. Dis. 2014, 37, 94–101. [Google Scholar] [CrossRef]
- Bang, O.Y.; Fujimura, M.; Kim, S.-K. The Pathophysiology of Moyamoya Disease: An Update. J. Stroke 2016, 18, 12–20. [Google Scholar] [CrossRef]
- Huang, S.; Guo, Z.-N.; Shi, M.; Yang, Y.; Rao, M. Etiology and pathogenesis of Moyamoya Disease: An update on disease prevalence. Int. J. Stroke 2017, 12, 246–253. [Google Scholar] [CrossRef]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.S.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Ni, Y.-Q.; Lin, X.; Zhan, J.-K.; Liu, Y.-S. Roles and Functions of Exosomal Non-coding RNAs in Vascular Aging. Aging Dis. 2020, 11, 164–178. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, Z.; Sheng, J. TRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Yim, D.; Lakshmanan, V.; Tirumalai, V.; Koh, J.; Park, J.; Cheong, J.; Low, J.; Lim, M.; Sze, S.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef]
- Elkordy, A.; Rashad, S.; Shehabeldeen, H.; Mishima, E.; Niizuma, K.; Abe, T.; Tominaga, T. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res. 2019, 1714, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, W.; Zheng, F.; Tang, D.; Dai, W.; Huang, S.; Zhang, C.; Zeng, J.; Wang, G.; Dai, Y. The potential role of tRNAs and small RNAs derived from tRNAs in the occurrence and development of systemic lupus erythematosus. Biochem. Biophys. Res. Commun. 2020, 527, 561–567. [Google Scholar] [CrossRef]
- Peng, Y.; Zou, J.; Wang, J.-H.; Zeng, H.; Tan, W.; Yoshida, S.; Zhang, L.; Li, Y.; Zhou, Y. Small RNA Sequencing Reveals Transfer RNA-derived Small RNA Expression Profiles in Retinal Neovascularization. Int. J. Med. Sci. 2020, 17, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Qin, Y.; Clark, W.C.; Dai, Q.; Yi, C.; He, C.; Lambowitz, A.M.; Pan, T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 2015, 12, 835–837. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef]
- Han, Z.; Li, L.; Liu, P.; Huang, Y.; Zhang, S.; Li, G.; Li, F.; Zhao, H.; Tao, Z.; Wang, R.; et al. Metabolic Adjustments by LncRNAs in Peripheral Neutrophils Partly Account for the Complete Compensation of Asymptomatic MMD Patients. CNS Neurol. Disord. Drug Targets 2020, 19, 306–317. [Google Scholar] [CrossRef]
- Masuda, J.; Ogata, J.; Yutani, C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke 1993, 24, 1960–1967. [Google Scholar] [CrossRef] [PubMed]
- Soriano, S.; Cowan, D.; Proctor, M.; Scott, R. Levels of soluble adhesion molecules are elevated in the cerebrospinal fluid of children with moyamoya syndrome. Neurosurgery 2002, 50, 544–549. [Google Scholar] [PubMed]
- Fujimura, M.; Fujimura, T.; Kakizaki, A.; Sato-Maeda, M.; Niizuma, K.; Tomata, Y.; Aiba, S.; Tominaga, T. Increased serum production of soluble CD163 and CXCL5 in patients with moyamoya disease: Involvement of intrinsic immune reaction in its pathogenesis. Brain Res. 2018, 1679, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Pais, V.; Danaila, L.; Pais, E. Ultrastructural Patterns of the Activated Cell Death Programs in the Human Brain. Ultrastruct. Pathol. 2013, 37, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Z.; Dong, Z.; Ma, M.; Yang, W.; Han, C.; Du, M.; Liu, Y.; Yang, H.; Liu, W.; et al. Increased thyroid function and elevated thyroid autoantibodies in pediatric patients with moyamoya disease: A case-control study. Stroke 2011, 42, 1138–1139. [Google Scholar] [CrossRef]
- Shetty, A.K.; Zanirati, G. The Interstitial System of the Brain in Health and Disease. Aging Dis. 2020, 11, 200–211. [Google Scholar] [CrossRef]
- Hara, S.; Hori, M.; Murata, S.; Ueda, R.; Tanaka, Y.; Inaji, M.; Maehara, T.; Aoki, S.; Nariai, T. Microstructural Damage in Normal-Appearing Brain Parenchyma and Neurocognitive Dysfunction in Adult Moyamoya Disease. Stroke 2018, 49, 2504–2507. [Google Scholar] [CrossRef]
- Adav, S.S.; Sze, S.K. Hypoxia-Induced Degenerative Protein Modifications Associated with Aging and Age-Associated Disorders. Aging Dis. 2020, 11, 341–364. [Google Scholar] [CrossRef]
- Hosoda, C.; Nariai, T.; Ishiwata, K.; Ishii, K.; Matsushima, Y.; Ohno, K. Correlation between Focal Brain Metabolism and Higher Brain Function in Patients with Moyamoya Disease. Int. J. Stroke 2010, 5, 367–373. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence | Tm (°C) | Product Length (bp) |
|---|---|---|---|
| U6 | F: 5′GCTTCGGCAGCACATATACTAAAAT3′ R: 5′CGCTTCACGAATTTGCGTGTCAT3′ | 60 | 89 |
| tsRNA-Glu-TTC-010 | F: 5′AGTCCGACGATCTCCCACAT3′ R: 5′TCCGATCTAGGAATCCTAACCG3′ | 60 | 49 |
| tsRNA-Glu-CTC-003 | F: 5′TCTACAGTCCGACGATCTCCCT3′ R: 5′GCTCTTCCGATCTACTAGACCACC 3′ | 60 | 46 |
| tsRNA-Val-AAC-017 | F: 5′GACGATCGTTTCCGTAGTGTAGTG3′ R: 5′TGCTCTTCCGATCTAAACGTGA3′ | 60 | 50 |
| tsRNA-Arg-CCT-008 | F: 5′CTACAGTCCGACGATCGCCT3′ R: 5′TGTGCTCTTCCGATCTCAAAAGT3′ | 60 | 46 |
| TsRNA_ID | Type | Length | FC (abs) | FDR-Adjusted p-Value | Regulation | TsRNA Sequence |
|---|---|---|---|---|---|---|
| tsRNA-Glu-TTC-003 | tRF-1 | 22 | 122.3993273 | 0.00029169 | up | ATGATGTATGCTTTGTTTCTGTT |
| tsRNA-Leu-TAA-037 | tRF-5b | 22 | 92.58353218 | 0.00043738 | up | GTTAAGATGGCAGAGCCCGGTA |
| tsRNA-Gly-GCC-022 | tRF-1 | 16 | 89.6665225 | 0.01625079 | up | GCACGCCCTCCCATTT |
| tsRNA-Gly-TCC-035 | tRF-1 | 15 | 83.29000089 | 0.00332883 | up | GCGGGCGGACCTTTT |
| tsRNA-Leu-TAA-012 | tRF-1 | 16 | 81.97503369 | 0.01469735 | up | AAGAGGAGTTGTTTTT |
| tsRNA-Gln-TTG-047 | tRF-1 | 18 | 81.75096387 | 0.00300796 | up | TTCAAAGGTGAACGTTTT |
| tsRNA-Val-CAC-007 | tRF-5c | 28 | 76.86797098 | 0.00077968 | up | GCTTCTGTAGTGTAGTGGTTATCACGTT |
| tsRNA-Gly-CCC-050 | tRF-1 | 31 | 74.16577619 | 0.00836043 | up | AAAGGGTCTTTTTCACCCCGCTGTTGCTCTT |
| tsRNA-Ala-TGC-001 | tRF-1 | 14 | 72.54554601 | 0.03955729 | up | AACGGTGACTTTTT |
| tsRNA-Glu-TTC-003 | tRF-5b | 22 | 64.48624603 | 0.00029169 | up | AGTAAGGTCAGCTAAATAAGCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Liu, P.; Wang, R.; Huang, Y.; Luo, J.; Jiao, L.; Tao, Z.; Zheng, Y.; Fan, J.; Zhao, H.; et al. Pathophysiological Significance of Neutrophilic Transfer RNA-Derived Small RNAs in Asymptomatic Moyamoya Disease. Cells 2021, 10, 1086. https://doi.org/10.3390/cells10051086
Li L, Liu P, Wang R, Huang Y, Luo J, Jiao L, Tao Z, Zheng Y, Fan J, Zhao H, et al. Pathophysiological Significance of Neutrophilic Transfer RNA-Derived Small RNAs in Asymptomatic Moyamoya Disease. Cells. 2021; 10(5):1086. https://doi.org/10.3390/cells10051086
Chicago/Turabian StyleLi, Lingzhi, Ping Liu, Rongliang Wang, Yuyou Huang, Jichang Luo, Liqun Jiao, Zhen Tao, Yangmin Zheng, Junfen Fan, Haiping Zhao, and et al. 2021. "Pathophysiological Significance of Neutrophilic Transfer RNA-Derived Small RNAs in Asymptomatic Moyamoya Disease" Cells 10, no. 5: 1086. https://doi.org/10.3390/cells10051086
APA StyleLi, L., Liu, P., Wang, R., Huang, Y., Luo, J., Jiao, L., Tao, Z., Zheng, Y., Fan, J., Zhao, H., Han, Z., & Luo, Y. (2021). Pathophysiological Significance of Neutrophilic Transfer RNA-Derived Small RNAs in Asymptomatic Moyamoya Disease. Cells, 10(5), 1086. https://doi.org/10.3390/cells10051086





