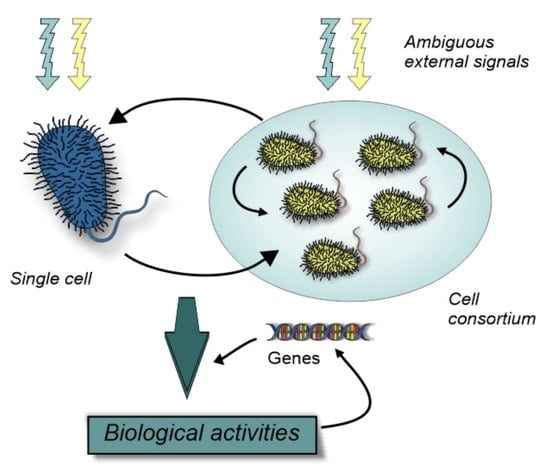
Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution
Abstract
:1. Introduction
2. Traditional Views on the Sources of Biological Variation
3. Unicellular Insights into Biological Variation
4. Variations in Multicellular Organisms
4.1. General Mechanisms
4.2. The Tools of Viral-Cellular Variation
5. The Origins and Extent of Natural Cellular Engineering
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Futuyma, D.J. Evolutionary Biology Today and the Call for an Extended Synthesis. Interface Focus 2017, 7, 20160145. [Google Scholar] [CrossRef]
- Koonin, E.V. The Logic of Chance: The Nature and Origin of Biological Evolution; Pearson Education: Upper Saddle River, NJ, USA, 2012; ISBN 978-0-13-254249-4. [Google Scholar]
- Miller, W.B. Cognition, Information Fields and Hologenomic Entanglement: Evolution in Light and Shadow. Biology 2016, 5, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, W.B. Biological Information Systems: Evolution as Cognition-Based Information Management. Prog. Biophys. Mol. Biol. 2018, 134, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.B.; Torday, J.S. A Systematic Approach to Cancer: Evolution beyond Selection. Clin. Transl. Med. 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, W.B.; Torday, J.S. Four Domains: The Fundamental Unicell and Post-Darwinian Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2018, 140, 49–73. [Google Scholar] [CrossRef]
- Miller, W.B., Jr.; Torday, J.S.; Baluška, F. Biological Evolution as Defense of “Self”. Prog. Biophys. Mol. Biol. 2019, 142, 54–74. [Google Scholar] [CrossRef]
- Miller, W.; Torday, J.; Baluška, F. The N-Space Episenome Unifies Cellular Information Space-Time within Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2020, 150. [Google Scholar] [CrossRef]
- Miller, W.B.; Baluška, F.; Torday, J.S. Cellular Senomic Measurements in Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2020, 156, 20–33. [Google Scholar] [CrossRef]
- Torday, J.; Miller, W., Jr. Cellular-Molecular Mechanisms in Epigenetic Evolutionary Biology; Springer Nature: Cham, Switzerland, 2020. [Google Scholar]
- Leitão, A.L.; Costa, M.C.; Gabriel, A.F.; Enguita, F.J. Interspecies Communication in Holobionts by Non-Coding RNA Exchange. Int. J. Mol. Med. Sci. 2020, 21, 2333. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, J.A. Evolution: A View from the 21st Century; FT Press Science: Saddle River, NJ, USA, 2011; ISBN 978-0-13-278093-3. [Google Scholar]
- Baluška, F.; Levin, M. On Having No Head: Cognition throughout Biological Systems. Front. Psychol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Reber, A.S. The First Minds: Caterpillars, Karyotes, and Consciousness; Oxford University Press: Oxford, UK, 2018; ISBN 978-0-1-085415-7. [Google Scholar]
- Slijepcevic, P. Natural Intelligence and Anthropic Reasoning. Biosemiotics 2020, 13, 285–307. [Google Scholar] [CrossRef]
- Ford, B.J. Are Cells Ingenious? Microscope 2004, 52, 135–144. [Google Scholar]
- Ford, B.J. On Intelligence in Cells: The Case for Whole Cell Biology. Interdiscip. Sci. Rev. 2009, 34, 350–365. [Google Scholar] [CrossRef] [Green Version]
- Ford, B.J. Cellular Intelligence: Microphenomenology and the Realities of Being. Prog. Biophys. Mol. Biol. 2017, 131, 273–287. [Google Scholar] [CrossRef]
- Dodig-Crnkovic, G. Modeling Life as Cognitive Info-Computation. In CiE 2014: Language, Life, Limits; Beckmann, A., Csuhaj-Varjú, E., Meer, K., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 153–162. [Google Scholar]
- Lyon, P. The Cognitive Cell: Bacterial Behavior Reconsidered. Front. Microbiol. 2015, 6, 264. [Google Scholar] [CrossRef]
- Baluška, F.; Reber, A. Sentience and Consciousness in Single Cells: How the First Minds Emerged in Unicellular Species. Bioessays 2019, 41, 1800229. [Google Scholar] [CrossRef]
- De Loof, A. From Darwin’s On the Origin of Species by Means of Natural Selection...to the Evolution of Life with Communication Activity as its Very Essence and Driving Force (=Mega-Evolution). Life Excit. Biol. 2015, 3, 153–187. [Google Scholar] [CrossRef]
- Zakirov, B.; Charalambous, G.; Aspalter, I.M.; van-Vuuren, K.; Mead, T.; Harrington, K.; Thuret, R.; Regan, E.R.; Herbert, S.P.; Bentley, K. Active Perception during Angiogenesis: Filopodia Speed Up Notch Selection of Tip Cells in Silico and in Vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing Cognition: Getting Down to Biological Basics. Phil. Trans. R. Soc. B. 2021, 376. [Google Scholar] [CrossRef] [PubMed]
- Shettleworth, S.J. Cognition, Evolution, and Behavior; Oxford University Press: Oxford, UK, 1998; ISBN 978-0-19-988638. [Google Scholar]
- Slijepcevic, P. Principles of Information Processing and Natural Learning in Biological Systems. J. Gen. Philos. Sci. 2019. [Google Scholar] [CrossRef] [Green Version]
- Pigliucci, M. Do We Need an Extended Evolutionary Synthesis? Evolution 2007, 61, 2743–2749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laland, K.; Uller, T.; Feldman, M.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling Smee, J.; Wray, G.A.; Hoekstra, H.E.; et al. Does Evolutionary Theory Need a Rethink? Nature 2014, 514, 161–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlesworth, D.; Barton, N.H.; Charlesworth, B. The Sources of Adaptive Variation. Proc. R. Soc. B. 2017, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, W.F. Too Much Eukaryote LGT. Bioessays 2017, 39. [Google Scholar] [CrossRef]
- Gould, S.J. The Return of Hopeful Monsters. Nat. Hist. 1977, 86, 23–30. Available online: http://www.somosbacteriasyvirus.com/monsters.pdf (accessed on 21 February 2021).
- Gould, S.J.; Eldredge, N. Punctuated Equilibria: The Tempo and Mode of Evolution Reconsidered. Paleobiology 1977, 3, 115–151. Available online: https://www.jstor.org/stable/2400177 (accessed on 14 January 2021). [CrossRef] [Green Version]
- Laland, K.N.; Uller, T.; Feldman, M.W.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling-Smee, J. The Extended Evolutionary Synthesis: Its Structure, Assumptions and Predictions. Proc. R. Soc. B. 2015, 282. [Google Scholar] [CrossRef]
- Moczek, A.P. An Evolutionary Biology for the 21st Century. In Perspectives on Evolutionary and Developmental Biology: Essays for Alessandro Minelli; Giuseppe, F., Ed.; Padova University Press: Padova, Italy, 2019; pp. 23–27. ISBN 978-88-6938-140-9. [Google Scholar]
- Trerotola, M.; Relli, V.; Simeone, P.; Alberti, S. Epigenetic Inheritance and the Missing Heritability. Hum. Genom. 2015, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Asfahl, K.L.; Schuster, M. Social Interactions in Bacterial Cell-Cell Signaling. FEMS Microbiol. Rev. 2017, 41, 92–107. [Google Scholar] [CrossRef] [Green Version]
- Brown-Jaque, M.; Calero-Cáceres, W.; Muniesa, M. Transfer of Antibiotic-Resistance Genes via Phage-Related Mobile Elements. Plasmid 2015, 79, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Balcazar, J.L. Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [Green Version]
- Popat, R.; Cornforth, D.M.; McNally, L.; Brown, S.P. Collective Sensing and Collective Responses in Quorum-Sensing Bacteria. J. R. Soc. Interface 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial quorum sensing. Cell 2018, 174, 1328–1328e1. [Google Scholar] [CrossRef]
- Goldenfeld, N.; Woese, C. Life is Physics: Evolution as a Collective Phenomenon Far from Equilibrium. Annu. Rev. Condens. Matter Phys. 2011, 2, 375–399. [Google Scholar] [CrossRef] [Green Version]
- Zatyka, M.; Thomas, C.M. Control of Genes for Conjugative Transfer of Plasmids and Other Mobile Elements. FEMS Microbiol. Rev. 1998, 21, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Pathways of DNA Transfer to Plants from Agrobacterium Tumefaciens and Related Bacterial Species. Annu. Rev. Phytopathol. 2019, 57, 231–251. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Otten, L. Widespread Occurrence of Natural Genetic Transformation of Plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.B.; Mulley, G.; Dills, A.H.; Alsohim, A.S.; McGuffin, L.J.; Studholme, D.J.; Silby, M.W.; Brockhurst, M.A.; Johnson, L.J.; Jackson, R.W. Evolutionary Resurrection of Flagellar Motility via Rewiring of the Nitrogen Regulation System. Science 2015, 347, 1014–1017. [Google Scholar] [CrossRef] [Green Version]
- Ponder, R.G.; Fonville, N.C.; Rosenberg, S.M. A Switch from High-Fidelity to Error-Prone DNA Double-Strand Break Repair Underlies Stress-Induced Mutation. Mol. Cell 2005, 19, 791–804. [Google Scholar] [CrossRef]
- Shee, C.; Gibson, J.L.; Darrow, M.C.; Gonzalez, C.; Rosenberg, S.M. Impact of a Stress-Inducible Switch to Mutagenic Repair of DNA Breaks on Mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 13659–13664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlodek, A.; Kendrew, S.G.; Coates, N.J.; Hold, A.; Pogwizd, J.; Rudder, S.; Sheehan, L.S.; Higginbotham, S.J.; Stanley-Smith, A.E.; Warneck, T.; et al. Diversity Oriented Biosynthesis via Accelerated Evolution of Modular Gene Clusters. Nat. Commun. 2017, 8, 1206. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Ishida, K.; Sugimoto, Y.; Jenke-Kodama, H.; Hertweck, C. Emulating Evolutionary Processes to Morph Aureothin-Type Modular Polyketide Synthases and Associated Oxygenases. Nat. Commun. 2019, 10, 3918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gestel, J.; Vlamakis, H.; Kolter, R. From Cell Differentiation to Cell Collectives: Bacillus Subtilis Uses Division of Labor to Migrate. PLoS Biol. 2015, 13, e1002141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Jacob, E.; Levine, H. Self-Engineering Capabilities of Bacteria. J. R. Soc. Interface 2006, 3, 197–214. [Google Scholar] [CrossRef] [Green Version]
- Ben-Jacob, E. Social Behavior of Bacteria: From Physics to Complex Organization. Eur. Phys. J. B 2008, 65, 315–322. [Google Scholar] [CrossRef]
- Ben-Jacob, E. Learning from Bacteria about Natural Information Processing. Ann. N. Y. Acad. Sci. 2009, 1178, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Baluška, F.; Miller, W.B., Jr. Senomic View of the Cell: Senome versus Genome. Commun. Integr. Biol. 2018, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Villarreal, L.P.; Ryan, F. Viruses in the Origin of Life and its Subsequent Diversification. In Handbook of Astrobiology; Kolb, V.M., Ed.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar] [CrossRef]
- Broecker, F.; Moelling, K. Evolution of Immune Systems from Viruses and Transposable Elements. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration Site Selection by Retroviruses and Transposable Elements in Eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.N. Functional Dualism of Transposon Transcripts in Evolution of Eukaryotic Genomes. Russ. J. Dev. Biol. 2018, 49, 339–355. [Google Scholar] [CrossRef]
- Caporale, L.H.; Doyle, J. In Darwinian Evolution, Feedback from Natural Selection Leads to Biased Mutations. Ann. N. Y. Acad. Sci. 2013, 1305, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Gerhart, J.; Kirschner, M. The Theory of Facilitated Variation. Pnas 2007, 104, 8582–8589. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, J.A. How Life Changes Itself: The Read-Write (RW) Genome. Phys. Life Rev. 2013, 10, 287–323. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Living Organisms Author Their Read-Write Genomes in Evolution. Biology 2017, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Witzany, G. Natural Genome Editing from a Biocommunicative Perspective. Biosemiotics 2011, 4, 349–368. [Google Scholar] [CrossRef]
- De Loof, A. The Evolution of “Life”: A Metadarwinian Integrative Approach. Commun. Integr. Biol. 2017, 10, e1301335. [Google Scholar] [CrossRef] [PubMed]
- Lupien, L.E.; Bloch, K.; Dehairs, J.; Feng, W.W.; Davis, W.L.; Dennis, T.; Swinnen, J.V.; Wells, W.A.; Smits, N.C.; Kuemmerle, N.B.; et al. Endocytosis of Very Low-Density Lipoprotein Particles: An Unexpected Mechanism for Lipid Acquisition by Breast Cancer Cells. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Sobhy, H. A Comparative Review of Viral Entry and Attachment during Large and Giant DsDNA Virus Infections. Arch. Virol. 2017, 162, 3567–3585. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic Theories for Eukaryote Origin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140330. [Google Scholar] [CrossRef] [Green Version]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, 911–921. [Google Scholar] [CrossRef] [Green Version]
- Nowack, E.C.M.; Price, D.C.; Bhattacharya, D.; Singer, A.; Melkonian, M.; Grossman, A.R. Gene Transfers from Diverse Bacteria Compensate for Reductive Genome Evolution in the Chromatophore of Paulinella chromatophora. Proc. Natl. Acad. Sci. USA 2016, 113, 12214–12219. [Google Scholar] [CrossRef] [Green Version]
- Overholtzer, M.; Mailleux, A.A.; Mouneimne, G.; Normand, G.; Schnitt, S.J.; King, R.W.; Cibas, E.S.; Brugge, J.S. A Nonapoptotic Cell Death Process, Entosis, That Occurs by Cell-in-Cell Invasion. Cell 2007, 131, 966–979. [Google Scholar] [CrossRef] [Green Version]
- Janssen, A.; Medema, R.H. Entosis: Aneuploidy by Invasion. Nat. Cell. Biol. 2011, 13, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Vogel, D.; Dussutour, A. Direct Transfer of Learned Behaviour via Cell Fusion in Non-Neural Organisms. Proc. R. Soc. B 2016, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkhipova, I.R.; Yushenova, I.A. Giant Transposons in Eukaryotes: Is Bigger Better? Genome Biol. Evol. 2019, 11, 906–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, C.; Cordaux, R. Viruses as Vectors of Horizontal Transfer of Genetic Material in Eukaryotes. Curr. Opin. Virol. 2017, 25, 16–22. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Gilbert, C. Endogenous Viruses: Insights into Viral Evolution and Impact on Host Biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Villarreal, L.P. Viruses and the Placenta: The Essential Virus First View. APMIS 2016, 124, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Roberts, R.M.; Ezashi, T.; Schulz, L.C.; Sugimoto, J.; Schust, D.J.; Khan, T.; Zhou, J. Syncytins Expressed in Human Placental Trophoblast. Placenta 2021. [Google Scholar] [CrossRef]
- Koonin, E.V. Darwinian Evolution in the Light of Genomics. Nucleic Acids Res. 2009, 37, 1011–1034. [Google Scholar] [CrossRef]
- Ryan, F. Virolution; Harper-Collins: New York, NY, USA, 2009; ISBN 978-0-00-731512-3. [Google Scholar]
- Black, S.G.; Arnaud, F.; Palmarini, M.; Spencer, T.E. Endogenous Retroviruses in Trophoblast Differentiation and Placental Development. Am. J. Reprod. Immunol. 2010, 64, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enard, D.; Cai, L.; Gwennap, C.; Petrov, D.A. Viruses Are a Dominant Driver of Protein Adaptation in Mammals. Elife 2016, 5, e12469. [Google Scholar] [CrossRef] [PubMed]
- Popov, M.; Kolotova, T.; Davidenko, M. Endogenous Retroviruses as Genetic Modules that Shape the Genome Regulatory Networks during Evolution. J. VN Karazin Kharkiv Natl. Univ. Ser. Med. 2019, 36, 80–95. [Google Scholar]
- Martinez, G.; Castellano, M.; Tortosa, M.; Pallas, V.; Gomez, G. A Pathogenic Non-Coding RNA Induces Changes in Dynamic DNA Methylation of Ribosomal RNA Genes in Host Plants. Nucleic Acids Res. 2013, 42, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional Activation of Retrotransposons Alters the Expression of Adjacent Genes in Wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B. Retroviruses Facilitate the Rapid Evolution of the Mammalian Placenta. Bioessays 2013, 35, 853–861. [Google Scholar] [CrossRef]
- Moon, C.; Baldridge, M.T.; Wallace, M.A.; Burnham, C.-A.; Virgin, H.W.; Stappenbeck, T.S. Vertically Transmitted Faecal IgA Levels Determine Extra-Chromosomal Phenotypic Variation. Nature 2015, 521, 90–93. [Google Scholar] [CrossRef]
- Wilke, C.O.; Sawyer, S.L. At the Mercy of Viruses. eLife 2016, 5, e16758. [Google Scholar] [CrossRef]
- Wallau, G.L.; Vieira, C.; Loreto, É.L.S. Genetic Exchange in Eukaryotes through Horizontal Transfer: Connected by the Mobilome. Mob. DNA 2018, 9, 6. [Google Scholar] [CrossRef]
- Wei, B.; Liu, H.; Liu, X.; Xiao, Q.; Wang, Y.; Zhang, J.; Hu, Y.; Liu, Y.; Yu, G.; Huang, Y. Genome-Wide Characterization of Non-Reference Transposons in Crops Suggests Non-Random Insertion. BMC Genom. 2016, 17, 536. [Google Scholar] [CrossRef] [Green Version]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten Things You Should Know about Transposable Elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. Noncoding RNAs: Persistent Viral Agents as Modular Tools for Cellular Needs. Ann. N. Y. Acad. Sci. 2009, 1178, 244–267. [Google Scholar] [CrossRef]
- Brandt, J.; Schrauth, S.; Veith, A.-M.; Froschauer, A.; Haneke, T.; Schultheis, C.; Gessler, M.; Leimeister, C.; Volff, J.-N. Transposable Elements as a Source of Genetic Innovation: Expression and Evolution of a Family of Retrotransposon-Derived Neogenes in Mammals. Gene 2005, 345, 101–111. [Google Scholar] [CrossRef]
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal Transfer of Transposable Elements and Why It Matters for Eukaryotic Evolution. Trends Ecol. Evol. 2010, 25, 537–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saier, M.H.; Kukita, C.; Zhang, Z. Transposon-Mediated Directed Mutation in Bacteria and Eukaryotes. Front. Biosci. 2017, 22, 1458–1468. [Google Scholar] [CrossRef] [Green Version]
- Schrader, L.; Schmitz, J. The Impact of Transposable Elements in Adaptive Evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of Transposable Elements on Polyploid Plant Genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Rodriguez, F.; Arkhipova, I.R. Transposable Elements and Polyploid Evolution in Animals. Curr. Opin. Genet. Dev. 2018, 49, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable Elements Contribute to Fungal Genes and Impact Fungal Lifestyle. Sci. Rep. 2019, 9, 4307. [Google Scholar] [CrossRef]
- Jangam, D.; Feschotte, C.; Betrán, E. Transposable Element Domestication as an Adaptation to Evolutionary Conflicts. Trends Genet. 2017, 33, 817–831. [Google Scholar] [CrossRef]
- Belyayev, A. Bursts of Transposable Elements as an Evolutionary Driving Force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R. Neutral Theory, Transposable Elements, and Eukaryotic Genome Evolution. Mol. Biol. Evol. 2018, 35, 1332–1337. [Google Scholar] [CrossRef] [Green Version]
- Oliver, K.R.; Greene, W.K. Mobile DNA and the TE-Thrust Hypothesis: Supporting Evidence from the Primates. Mob. DNA 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapley, J.; Santure, A.W.; Dennis, S.R. Transposable Elements as Agents of Rapid Adaptation May Explain the Genetic Paradox of Invasive Species. Mol. Ecol. 2015, 24, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.-E. Chromatin Signatures and Retrotransposon Profiling in Mouse Embryos Reveal Regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. Nat. 2013, 20, 332–338. [Google Scholar] [CrossRef]
- Suarez, N.A.; Macia, A.; Muotri, A.R. LINE-1 Retrotransposons in Healthy and Diseased Human Brain. Dev. Neurobiol. 2018, 78, 434–455. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.N. Hypothesis on the Origin of Viruses from Transposons. Mol. Genet. Microbiol. Virol. 2018, 33, 223–232. [Google Scholar] [CrossRef]
- Sundaram, V.; Wysocka, J. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos. Trans. R. Soc. Lond. B 2020, 30, 375. [Google Scholar] [CrossRef] [Green Version]
- Moschetti, R.; Palazzo, A.; Lorusso, P.; Viggiano, L.; Massimiliano Marsano, R. “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology 2020, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Masonbrink, R.E.; Shan, W.; Song, F.; Zhang, J.; Yu, W.; Wang, K.; Wu, Y.; Tang, H.; Wendel, J.F.; et al. Rapid Proliferation and Nucleolar Organizer Targeting Centromeric Retrotransposons in Cotton. Plant J. 2016, 88, 992–1005. [Google Scholar] [CrossRef]
- Villarreal, L.; Witzany, G. That is Life: Communicating RNA Networks from Viruses and Cells in Continuous Interaction. Ann. N. Y. Acad. Sci. 2019, 1447, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Smalheiser, N.R. The RNA-Centred View of the Synapse: Non-Coding RNAs and Synaptic Plasticity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [Green Version]
- Palmer, W.H.; Hadfield, J.D.; Obbard, D.J. RNA-Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates. Genetics 2018, 208, 1585–1599. [Google Scholar] [CrossRef] [Green Version]
- Villarreal, L.P.; Witzany, G. Editorial: Genome Invading RNA Networks. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Shapiro, J.A. Nothing in Evolution Makes Sense Except in the Light of Genomics: Read–Write Genome Evolution as an Active Biological Process. Biology 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamai, L. Unveiling Human Non-Random Genome Editing Mechanisms Activated in Response to Chronic Environmental Changes: I. Where Might These Mechanisms Come from and What Might They Have Led To? Cells 2020, 9, 2362. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, T.; Shibata, Y.; Kumar, P.; Dillon, L.; Dutta, A. Extrachromosomal Circular DNA, MicroDNA, without Canonical Promoters Produce Short Regulatory RNAs that Suppress Gene Expression. bioRxiv 2019. [Google Scholar] [CrossRef]
- Lanciano, S.; Carpentier, M.C.; Llauro, C.; Jobet, E.; Robakowska-Hyzorek, D.; Lasserre, E.; Ghesquière, A.; Panaud, O.; Mirouze, M. Sequencing the Extrachromosomal Circular Mobilome Reveals Retrotransposon Activity in Plants. PLoS Genet. 2017, 13, e1006630. [Google Scholar] [CrossRef] [Green Version]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of Form and Function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [Green Version]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-Coding RNA Regulatory Networks. Biochim. Biophys. Acta Gene. Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs Function as CeRNAs to Regulate and Control Human Cancer Progression. Mole. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Ecco, G.; Imbeault, M.; Trono, D. KRAB Zinc Finger Proteins. Development 2017, 144, 2719–2729. [Google Scholar] [CrossRef] [Green Version]
- Lupo, A.; Cesaro, E.; Montano, G.; Zurlo, D.; Izzo, P.; Costanzo, P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genom. 2013, 14, 268–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, K.L.; Palmer, N.D.; Johnson, D.T.; Medina, S.J.; Yan, S.J.; Li, V.; Burmeister, A.R.; Meyer, J.R. Destabilizing Mutations Encode Nongenetic Variation that Drives Evolutionary Innovation. Science 2018, 359, 1542–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, J.R.; Jung, I.; Selvaraj, S.; Shen, Y.; Antosiewicz-Bourget, J.E.; Lee, A.Y.; Ye, Z.; Kim, A.; Rajagopal, N.; Xie, W.; et al. Chromatin Architecture Reorganization during Stem Cell Differentiation. Nature 2015, 518, 331–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waszak, S.M.; Delaneau, O.; Gschwind, A.R.; Kilpinen, H.; Raghav, S.K.; Witwicki, R.M.; Orioli, A.; Wiederkehr, M.; Panousis, N.I.; Yurovsky, A.; et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell 2015, 162, 1039–1050. [Google Scholar] [CrossRef] [Green Version]
- Wellenreuther, M.; Mérot, C.; Berdan, E.; Bernatchez, L. Going beyond SNPs: The Role of Structural Genomic Variants in Adaptive Evolution and Species Diversification. Mol. Ecol. 2019, 28, 1203–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shorter, J.; Lindquist, S. Prions as Adaptive Conduits of Memory and Inheritance. Nat. Rev. Genet. 2005, 6, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Jarosz, D.F.; Jones, S.K.; Chang, A.; Lancaster, A.K.; Lindquist, S. Prions Are a Common Mechanism for Phenotypic Inheritance in Wild Yeasts. Nature 2012, 482, 363–368. [Google Scholar] [CrossRef] [Green Version]
- Miller, W.B., Jr. The Eukaryotic Microbiome: Origins and Implications for Fetal and Neonatal Life. Front. Pediatr. 2016, 4, 96. [Google Scholar] [CrossRef] [Green Version]
- Cho, I.; Blaser, M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.R.; Proctor, L.M.; Surette, M.G.; Suchodolski, J.S. The Microbiome: The Trillions of Microorganisms that Maintain Health and Cause Disease in Humans and Companion Animals. Vet. Pathol. 2016, 53, 10–21. [Google Scholar] [CrossRef]
- Gilbert, S.F.; Bosch, T.C.G.; Ledón-Rettig, C. Eco-Evo-Devo: Developmental Symbiosis and Developmental Plasticity as Evolutionary Agents. Nat. Rev. Genet. 2015, 16, 611–622. [Google Scholar] [CrossRef]
- Malik, S.S.; Azem-e-Zahra, S.; Kim, K.M.; Caetano-Anollés, G.; Nasir, A. Do Viruses Exchange Genes across Superkingdoms of Life? Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Jobson, M.A.; Jordan, J.M.; Sandrof, M.A.; Hibshman, J.D.; Lennox, A.L.; Baugh, L.R. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis Elegans. Genetics 2015, 201, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.J.; Rupert, J.L. Hypoxia and Environmental Epigenetics. High Alt. Med. Biol. 2014, 15, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, R.; Buchner, D.A. Transgenerational Inheritance of Metabolic Disease. Semin. Cell Dev. Biol. 2015, 43, 131–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, M.K. Environmental Stress and Epigenetic Transgenerational Inheritance. BMC Med. 2014, 12, 153. [Google Scholar] [CrossRef]
- Milstein, J.N.; Meiners, J.-C. On the Role of DNA Biomechanics in the Regulation of Gene Expression. J. R. Soc. Interface 2011, 8, 1673–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, J.A. Exploring the Read-Write Genome: Mobile DNA and Mammalian Adaptation. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, K.; Greene, W. Transposable Elements: Powerful Facilitators of Evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Schrader, L.; Kim, J.W.; Ence, D.; Zimin, A.; Klein, A.; Wyschetzki, K.; Weichselgartner, T.; Kemena, C.; Stökl, J.; Schultner, E.; et al. Transposable Element Islands Facilitate Adaptation to Novel Environments in an Invasive Species. Nat. Commun. 2014, 5, 5495. [Google Scholar] [CrossRef] [Green Version]
- Mita, P.; Boeke, J.D. How Retrotransposons Shape Genome Regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Torday, J.S.; Miller, W.B., Jr. The Resolution of Ambiguity as the Basis for Life: A Cellular Bridge Between Western Reductionism and Eastern Holism. Prog. Biophys. Mol. Biol. 2017, 131, 288–297. [Google Scholar] [CrossRef]
- Baverstock, K. The Role of Information in Cell Regulation. Prog. Biophys. Mol. Biol. 2013, 111, 141–143. [Google Scholar] [CrossRef]
- Torday, J.S.; Rehan, V.K. Lung Evolution as a Cipher for Physiology. Physiol. Genom. 2009, 38, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Cárdenas-García, J.F. The Process of Info-Autopoiesis—The Source of All Information. Biosemiotics 2020, 13, 199–221. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B. Phenotype as Agent for Epigenetic Inheritance. Biology 2016, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.I.; Kim, H.; Davies, P.C.W. The Informational Architecture of the Cell. Phil. Trans. R. Soc. A 2016, 374, 20150057. [Google Scholar] [CrossRef]
- Martinez, J.; Klasson, L.; Welch, J.J.; Jiggins, F.M. Life and Death of Selfish Genes: Comparative Genomics Reveals the Dynamic Evolution of Cytoplasmic Incompatibility. Mol. Biol. Evol. 2021, 38, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Chain, E.B. Social Responsibility and the Scientist in Modern Western Society. Perspect. Biol. Med. 1971, 14, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Central Dogma or Central Debate? Physiology 2018, 33, 246–249. [Google Scholar] [CrossRef]
- Turner, P.; Nottale, L.; Zhao, J.; Pesquet, E. New Insights into the Physical Processes that Underpin Cell Division and the Emergence of Different Cellular and Multicellular Structures. Prog. Biophys. Mol. Biol. 2020, 150, 13–42. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Evolution Viewed from Physics, Physiology and Medicine. Interface Focus 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Rudkin, D.M.; Young, G.A. Horseshoe Crabs—An Ancient Ancestry Revealed. In Biology and Conservation of Horseshoe Crabs; Tanacredi, J.T., Botton, M.L., Smith, D., Eds.; Springer US: Boston, MA, USA, 2009; pp. 25–44. ISBN 978-0-387-89959-6. [Google Scholar]
- Stockdale, M.T.; Benton, M.J. Environmental Drivers of Body Size Evolution in Crocodile-Line Archosaurs. Commun. Bio. 2021, 4, 38. [Google Scholar] [CrossRef]
- Witzany, G. Evolution of Genetic Information without Error Replication. In Theoretical Information Studies; World Scientific Series in Information Studies: London, UK, 2018; Volume 11, pp. 295–320. ISBN 978-981-327-748-9. [Google Scholar]
- Kiliç, A.M.; Plasencia, P.; Ishida, K.; Guex, J.; Hirsch, F. Proteromorphosis of Neospathodus (Conodonta) during the Permian–Triassic Crisis and Recovery. Rev. Micropaléontol. 2016, 59, 33–39. [Google Scholar] [CrossRef]
- Guex, J. The Controversial Cope’s, Haeckel’s and Dollo’s Evolutionary Rules: The Role of Evolutionary Retrogradation; Springer International Publishing: Cham, Switzerland, 2020; pp. 13–22. ISBN 978-3-030-47279-5. [Google Scholar] [CrossRef]
- Guex, J. Retrograde Evolution during Major Extinction Crises; Springer Briefs in Evolutionary Biology; Springer International Publishing: Lausanne, Switzerland, 2016; ISBN 978-3-319-27916-9. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B., Jr. Terminal Addition in a Cellular World. Prog. Biophys. Mol. Biol. 2018, 135. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. No Genome Is an Island: Toward a 21st Century Agenda for Evolution. Ann. N. Y. Acad. Sci. 2019, 1447, 21–52. [Google Scholar] [CrossRef]
- González Plaza, J.J. Small RNAs as Fundamental Players in the Transference of Information during Bacterial Infectious Diseases. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Force for Ancient and Recent Life: Viral and Stem-Loop RNA Consortia Promote Life. Ann. N. Y. Acad. Sci. 2015, 1341, 25–34. [Google Scholar] [CrossRef]
- Villarreal, L.P. Viral Ancestors of Antiviral Systems. Viruses 2011, 3, 1933–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witzany, G. RNA Sociology: Group Behavioral Motifs of RNA Consortia. Life 2014, 4, 800–818. [Google Scholar] [CrossRef] [Green Version]
- Ambrosi, A.; Cattoglio, C.; Serio, C.D. Retroviral Integration Process in the Human Genome: Is It Really Non-Random? A New Statistical Approach. PLoS Comput. Biol. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Rabadan, R.; Levine, A.J.; Krasnitz, M. Non-Random Reassortment in Human Influenza A Viruses. Influenza Other Respir. Viruses 2008, 2, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, M.; Driesch, C.; Jansen, L.; Runnebaum, I.B.; Dürst, M. Non-Random Integration of the HPV Genome in Cervical Cancer. PLoS ONE 2012, 7, e39632. [Google Scholar] [CrossRef] [Green Version]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Activities of Transposable Elements: From Conflicts to Benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Klimenko, O.V. Small non-coding RNAs as regulators of structural evolution and carcinogenesis. Non-Coding RNA Res. 2017, 2, 88–92. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Bazán, E.R. Symbiosis: Viruses as Intimate Partners. Annu. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F. Virusphere: Explains the Science Behind the Coronavirus Outbreak; HarperCollins Publishers: New York, NY, USA, 2019; ISBN 978-0-00-829669-8. [Google Scholar]
- Burke, G.R.; Simmonds, T.J.; Sharanowski, B.J.; Geib, S.M. Rapid Viral Symbiogenesis via Changes in Parasitoid Wasp Genome Architecture. Mol. Biol. Evol. 2018, 35, 2463–2474. [Google Scholar] [CrossRef]
- Venner, S.; Feschotte, C.; Biémont, C. Dynamics of Transposable Elements: Towards a Community Ecology of the Genome. Trends Genet. 2009, 25, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venner, S.; Miele, V.; Terzian, C.; Biémont, C.; Daubin, V.; Feschotte, C.; Pontier, D. Ecological Networks to Unravel the Routes to Horizontal Transposon Transfers. PLoS Biol. 2017, 15, e2001536. [Google Scholar] [CrossRef]
- Auboeuf, D. Physicochemical Foundations of Life That Direct Evolution: Chance and Natural Selection Are Not Evolutionary Driving Forces. Life 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, M.T.; Loreto, E.L.S. The Catenulida Flatworm Can Express Genes from its Microbiome or from the DNA It Ingests. Sci. Rep. 2019, 9, 19045. [Google Scholar] [CrossRef] [Green Version]
- Annila, A.; Baverstock, K. Genes without Prominence: A Reappraisal of the Foundations of Biology. J. R. Soc. Interface 2014, 11. [Google Scholar] [CrossRef]
- Torday, J.S. The Cell as the First Niche Construction. Biology 2016, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- ElMaghraby, M.F.; Andersen, P.R.; Pühringer, F.; Hohmann, U.; Meixner, K.; Lendl, T.; Tirian, L.; Brennecke, J. A Heterochromatin-Specific RNA Export Pathway Facilitates PiRNA Production. Cell 2019, 178, 964–979.e20. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by MicroRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Chen, W.L.; Kung, W.-H.; Huang, H.-D. Plant MiRNAs Found in Human Circulating System Provide Evidences of Cross Kingdom RNAi. BMC Genom. 2017, 18, 112. [Google Scholar] [CrossRef] [Green Version]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L.; et al. Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice from DSS-Induced Colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torday, J.S.; Miller, W.B. Life is determined by its environment. Int. J. Astrobiol. 2016, 1, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noble, R.; Noble, D. Harnessing Stochasticity: How Do Organisms Make Choices? Chaos 2018, 28, 106309. [Google Scholar] [CrossRef] [Green Version]
- Noble, R.; Noble, D. Was the Watchmaker Blind? Or Was She One-Eyed? Biology 2017, 6, 47. [Google Scholar] [CrossRef] [Green Version]
- Kitano, H. Biological Robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- De Visser, J.A.G.M.; Hermisson, J.; Wagner, G.P.; Ancel Meyers, L.; Bagheri-Chaichian, H.; Blanchard, J.L.; Chao, L.; Cheverud, J.M.; Elena, S.F.; Fontana, W.; et al. Perspective: Evolution and Detection of Genetic Robustness. Evolution 2003, 57, 1959–1972. [Google Scholar] [CrossRef]
- Siegal, M.L.; Bergman, A. Waddington’s Canalization Revisited: Developmental Stability and Evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 10528–10532. [Google Scholar] [CrossRef] [Green Version]
- Loison, L. Canalization and Genetic Assimilation: Reassessing the Radicality of the Waddingtonian Concept of Inheritance of Acquired Characters. Semin. Cell Dev. Biol. 2019, 88, 4–13. [Google Scholar] [CrossRef]
- Flatt, T. The Evolutionary Genetics of Canalization. Q. Rev. Biol. 2005, 80, 287–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornstein, E.; Shomron, N. Canalization of Development by MicroRNAs. Nat. Genet. 2006, 38, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Stelling, J.; Sauer, U.; Szallasi, Z.; Doyle, F.J.; Doyle, J. Robustness of Cellular Functions. Cell 2004, 118, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, P. Evolution 2.0: Breaking the Deadlock between Darwin and Design; BenBella Books: Dallas, TX, USA, 2015; ISBN 978-1-940363-90-5. [Google Scholar]
- Shakhnovich, B.E.; Koonin, E.V. Origins and Impact of Constraints in Evolution of Gene Families. Genome Res. 2006, 16, 1529–1536. [Google Scholar] [CrossRef] [Green Version]
- Futuyma, D.J. Evolutionary Constraint and Ecological Consequences. Evolution 2010, 64, 1865–1884. [Google Scholar] [CrossRef] [PubMed]
- Murren, C.J.; Auld, J.R.; Callahan, H.; Ghalambor, C.K.; Handelsman, C.A.; Heskel, M.A.; Kingsolver, J.G.; Maclean, H.J.; Masel, J.; Maughan, H.; et al. Constraints on the Evolution of Phenotypic Plasticity: Limits and Costs of Phenotype and Plasticity. Heredity 2015, 115, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Babloyantz, I.A.; Kaczmarek, L.K. Self-Organization in Biological Systems with Multiple Cellular Contacts. Bull. Math. Biol. 1979, 41, 193–201. [Google Scholar] [CrossRef]
- Vijver, G.; Salthe, S.N.; Delpos, M. (Eds.) Evolutionary Systems: Biological and Epistemological Perspectives on Selection and Self-Organization; Springer-Science+Business Media: Dordrecht, The Netherlands, 2013; ISBN 978-94-017-1510-2. [Google Scholar] [CrossRef]
- Heylighen, F. Stigmergy as a Universal Coordination Mechanism I: Definition and Components. Cognit. Syst. Res. 2016, 38, 4–13. [Google Scholar] [CrossRef]
- Kauffman, S.A. Investigations; Oxford University Press: New York, NY, USA, 2000; ISBN 978-0-19-972894-7. [Google Scholar]
- Coelho, M.T.P.; Diniz-Filho, J.A.; Rangel, T.F. A Parsimonious View of the Parsimony Principle in Ecology and Evolution. Ecography 2019, 42, 968–976. [Google Scholar] [CrossRef] [Green Version]
- Noble, D. A theory of biological relativity: No privileged level of causation. Interface Focus 2012, 2, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Kassen, R. Experimental Evolution of Innovation and Novelty. Trends Ecol. Evol. 2019, 34, 712–722. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, W.B., Jr.; Enguita, F.J.; Leitão, A.L. Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells 2021, 10, 1125. https://doi.org/10.3390/cells10051125
Miller WB Jr., Enguita FJ, Leitão AL. Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells. 2021; 10(5):1125. https://doi.org/10.3390/cells10051125
Chicago/Turabian StyleMiller, William B., Jr., Francisco J. Enguita, and Ana Lúcia Leitão. 2021. "Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution" Cells 10, no. 5: 1125. https://doi.org/10.3390/cells10051125
APA StyleMiller, W. B., Jr., Enguita, F. J., & Leitão, A. L. (2021). Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells, 10(5), 1125. https://doi.org/10.3390/cells10051125