Automated Analysis of Cerebrospinal Fluid Cells Using Commercially Available Blood Cell Analysis Devices—A Critical Appraisal
Abstract
1. Introduction
2. The Challenge
3. CSF White and Red Blood Cell Count
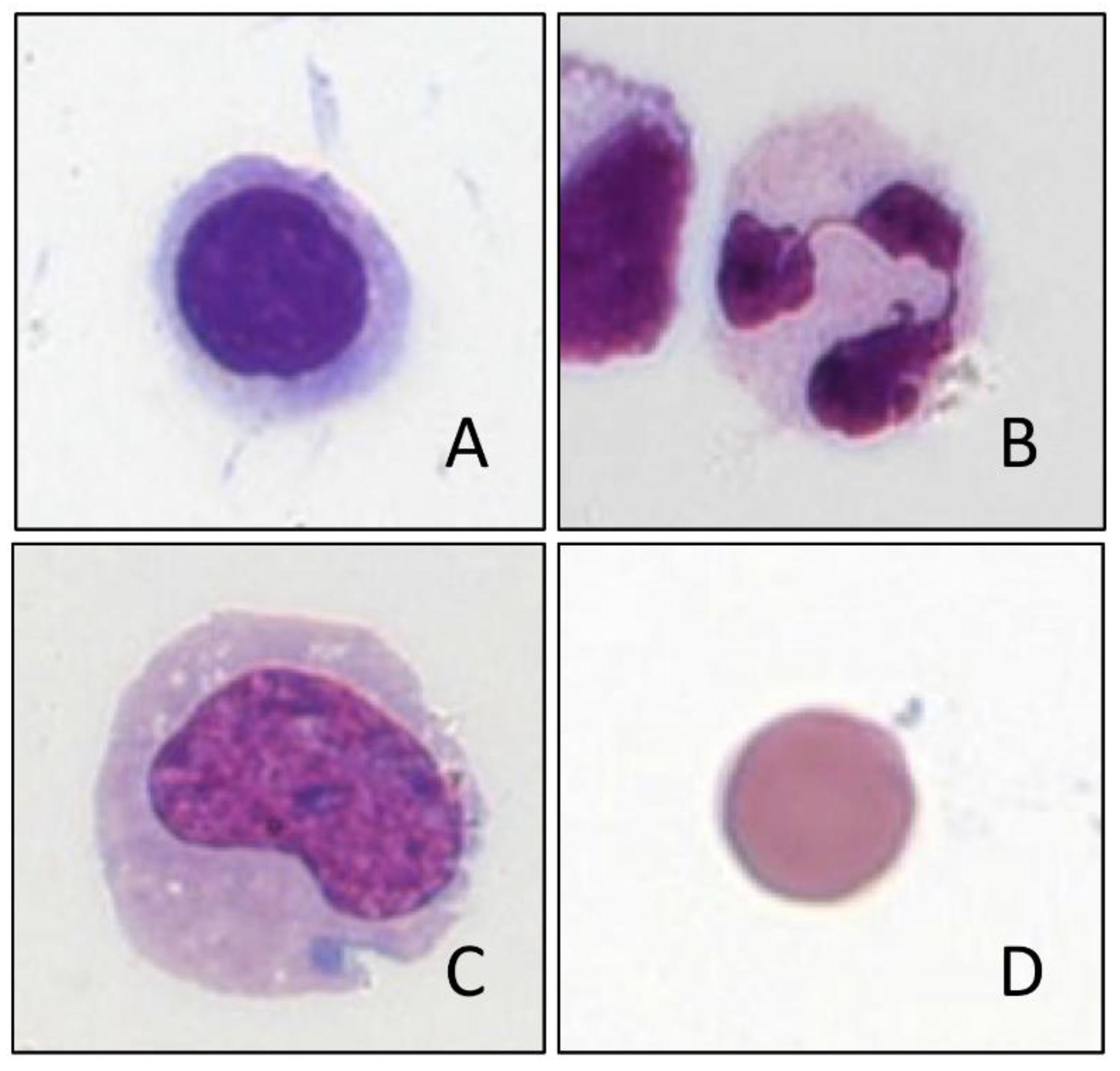
4. Cell Differentiation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Jonge, R.; Brouwer, R.; de Graaf, M.T.; Luitwieler, R.L.; Fleming, C.; de Frankrijker-Merkestijn, M.; Sillevis Smitt, P.A.; Boonstra, J.G.; Lindemans, J. Evaluation of the new body fluid mode on the Sysmex XE-5000 for counting leukocytes and erythrocytes in cerebrospinal fluid and other body fluids. Clin. Chem. Lab. Med. 2010, 48, 665–675. [Google Scholar] [CrossRef]
- Kleine, T.O.; Nebe, C.T.; Lower, C.; Geilenkeuser, W.J.; Dorn-Beineke, A. Cell analysis in cerebrospinal fluid (CSF) using Sysmex(R) hematology analyzers XT-4000i and XE-5000: Evaluation with CSF controls of the Joint German Society for Clinical Chemistry and Laboratory Medicine (DGKL). Cytometry A 2012, 81, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.U.; Chi, H.S.; Park, S.H.; Jang, S.; Kim, Y.J.; Park, C.J. Body fluid cellular analysis using the Sysmex XN-2000 automatic hematology analyzer: Focusing on malignant samples. Int. J. Lab. Hematol. 2015, 37, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Fleming, C.; Brouwer, R.; Lindemans, J.; de Jonge, R. Validation of the body fluid module on the new Sysmex XN-1000 for counting blood cells in cerebrospinal fluid and other body fluids. Clin. Chem Lab. Med. 2012, 50, 1791–1798. [Google Scholar] [CrossRef]
- Aguadero, V.; Cano-Corres, R.; Berlanga, E.; Torra, M. Evaluation of biological fluid analysis using the sysmex XN automatic Hematology analyzer. Cytometry B Clin. Cytom. 2018, 94, 680–688. [Google Scholar] [CrossRef]
- Cho, J.; Oh, J.; Lee, S.G.; Lee, Y.H.; Song, J.; Kim, J.H. Performance Evaluation of Body Fluid Cellular Analysis Using the Beckman Coulter UniCel DxH 800, Sysmex XN-350, and UF-5000 Automated Cellular Analyzers. Ann. Lab. Med. 2020, 40, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Buoro, S.; Seghezzi, M.; Dominoni, P.; Moioli, V.; Manenti, B.; Previtali, G.; Ottomano, C.; Lippi, G. Lack of harmonization in high fluorescent cell automated counts with body fluids mode in ascitic, pleural, synovial, and cerebrospinal fluids. Int. J. Lab. Hematol. 2019, 41, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Takemura, H.; Ai, T.; Kimura, K.; Nagasaka, K.; Takahashi, T.; Tsuchiya, K.; Yang, H.; Konishi, A.; Uchihashi, K.; Horii, T.; et al. Evaluation of cell count and classification capabilities in body fluids using a fully automated Sysmex XN equipped with high-sensitive Analysis (hsA) mode and DI-60 Hematology analyzer system. PLoS ONE 2018, 13, e0195923. [Google Scholar] [CrossRef]
- Zelazowska-Rutkowska, B.; Zak, J.; Wojtkowska, M.; Zaworonek, J.; Cylwik, B. Use of the Sysmex XT-4000i Hematology analyzer in the differentiation of cerebrospinal fluid cells in children. J. Clin. Lab. Anal. 2019, 33, e22822. [Google Scholar] [CrossRef]
- Ziebig, R.; Lun, A.; Sinha, P. Leukocyte counts in cerebrospinal fluid with the automated Hematology analyzer CellDyn 3500 and the urine flow cytometer UF-100. Clin. Chem. 2000, 46, 242–247. [Google Scholar] [CrossRef] [PubMed]
- De Smet, D.; Van Moer, G.; Martens, G.A.; Nanos, N.; Smet, L.; Jochmans, K.; De Waele, M. Use of the Cell-Dyn Sapphire Hematology analyzer for automated counting of blood cells in body fluids. Am. J. Clin. Pathol. 2010, 133, 291–299. [Google Scholar] [CrossRef]
- Hod, E.A.; Brugnara, C.; Pilichowska, M.; Sandhaus, L.M.; Luu, H.S.; Forest, S.K.; Netterwald, J.C.; Reynafarje, G.M.; Kratz, A. Automated cell counts on CSF samples: A multicenter performance evaluation of the GloCyte system. Int. J. Lab. Hematol. 2018, 40, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, J.T.; Delanghe, J.R.; Langlois, M.R.; Taes, Y.E.; De Buyzere, M.L.; Verstraete, A.G. Automated flow cytometric analysis of cerebrospinal fluid. Clin. Chem. 2001, 47, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Buoro, S.; Apassiti Esposito, S.; Alessio, M.; Crippa, A.; Ottomano, C.; Lippi, G. Automated Cerebrospinal Fluid Cell Counts Using the New Body Fluid Mode of Sysmex UF-1000i. J. Clin. Lab. Anal. 2016, 30, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Tanada, H.; Ikemoto, T.; Masutani, R.; Tanaka, H.; Takubo, T. Evaluation of the automated Hematology analyzer ADVIA(R) 120 for cerebrospinal fluid analysis and usage of unique hemolysis reagent. Int. J. Lab. Hematol. 2014, 36, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Mahieu, S.; Vertessen, F.; Van der Planken, M. Evaluation of ADVIA 120 CSF assay (Bayer) vs. chamber counting of cerebrospinal fluid specimens. Clin. Lab. Haematol. 2004, 26, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Bremell, D.; Mattsson, N.; Wallin, F.; Henriksson, J.; Wall, M.; Blennow, K.; Zetterberg, H.; Hagberg, L. Automated cerebrospinal fluid cell count--new reference ranges and evaluation of its clinical use in central nervous system infections. Clin. Biochem. 2014, 47, 25–30. [Google Scholar] [CrossRef]
- Brown, W.; Keeney, M.; Chin-Yee, I.; Johnson, K.; Lantis, K.; Finn, W.; Wolfe, N.; Kaplan, S. Validation of body fluid analysis on the Coulter LH 750. Lab. Hematol. 2003, 9, 155–159. [Google Scholar]
- Glasser, L.; Murphy, C.A.; Machan, J.T. The clinical reliability of automated cerebrospinal fluid cell counts on the Beckman-Coulter LH750 and Iris iQ200. Am. J. Clin. Pathol. 2009, 131, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.O.; Nebe, C.T.; Lower, C.; Lehmitz, R.; Geilenkeuser, W.J.; Kruse, R.; Dorn-Beineke, A. Evaluation of cell counting and leukocyte differentiation in cerebrospinal fluid controls using Hematology analyzers by the German Society for Clinical Chemistry and Laboratory Medicine. Clin. Chem. Lab. Med. 2010, 48, 839–848. [Google Scholar] [CrossRef]
- Strik, H.; Luthe, H.; Nagel, I.; Ehrlich, B.; Bahr, M. Automated cerebrospinal fluid cytology: Limitations and reasonable applications. Anal. Quant. Cytol. Histol. 2005, 27, 167–173. [Google Scholar] [PubMed]
- Kresie, L.; Benavides, D.; Bollinger, P.; Walters, J.; Pierson, D.; Richmond, T.; Issa-Dyer, K.; Fahs, M. Performance evaluation of the application of body fluids on the Sysmex XE-2100 series automated Hematology analyzer. Lab. Hematol. 2005, 11, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.O. Mechanisierte Zählung und Differenzierung von Liquorzellen. Lab. Med. 1991, 15, 51–59. [Google Scholar] [CrossRef]
- Fleming, C.; Russcher, H.; Lindemans, J.; de Jonge, R. Clinical relevance and contemporary methods for counting blood cells in body fluids suspected of inflammatory disease. Clin. Chem. Lab. Med. 2015, 53, 1689–1706. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S49–S52. [Google Scholar]
- Boer, K.; Deufel, T.; Reinhoefer, M. Evaluation of the XE-5000 for the automated analysis of blood cells in cerebrospinal fluid. Clin. Biochem. 2009, 42, 684–691. [Google Scholar] [CrossRef]
- Li, A.; Gronlund, E.; Brattsand, G. Automated white blood cell counts in cerebrospinal fluid using the body fluid mode on the platform Sysmex XE-5000. Scand. J. Clin. Lab. Investig. 2014, 74, 673–680. [Google Scholar] [CrossRef]
- Liang, X.; Chen, J.; Xiao, X.; Yu, Y.; Li, W.; Zhang, Z. Automated cell analysis of cerebrospinal fluid with XE-5000. Clin. Lab. 2014, 60, 1785–1793. [Google Scholar] [CrossRef]
- Zimmermann, M.; Ruprecht, K.; Kainzinger, F.; Heppner, F.L.; Weimann, A. Automated vs. manual cerebrospinal fluid cell counts: A work and cost analysis comparing the Sysmex XE-5000 and the Fuchs-Rosenthal manual counting chamber. Int. J. Lab. Hematol. 2011, 33, 629–637. [Google Scholar] [CrossRef]
- Sandhaus, L.M.; Ciarlini, P.; Kidric, D.; Dillman, C.; O’Riordan, M. Automated cerebrospinal fluid cell counts using the Sysmex XE-5000: Is it time for new reference ranges? Am. J. Clin. Pathol. 2010, 134, 734–738. [Google Scholar] [CrossRef]
- Heller, T.; Nagel, I.; Ehrlich, B.; Bahr, M.; Strik, H. Automated cerebrospinal fluid cytology. Anal. Quant. Cytol. Histol. 2008, 30, 139–144. [Google Scholar] [PubMed]
- Zur, B.; Eichhorn, L.; Albers, E.; Stoffel-Wagner, B. Evaluation of 2 Hematology analyzers in body fluid mode versus flow cytometry immunophenotyping of mainly neurosurgical cerebrospinal fluid samples. J. Neurol. Surg. A Cent. Eur. Neurosurg. 2012, 73, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Fleming, C.; Russcher, H.; Brouwer, R.; Lindemans, J.; de Jonge, R. Evaluation of Sysmex XN-1000 High-Sensitive Analysis (hsA) Research Mode for Counting and Differentiating Cells in Cerebrospinal Fluid. Am. J. Clin. Pathol. 2016, 145, 299–307. [Google Scholar] [CrossRef]
- Buoro, S.; Peruzzi, B.; Fanelli, A.; Seghezzi, M.; Manenti, B.; Lorubbio, M.; Biagioli, T.; Nannini, S.; Ottomano, C.; Lippi, G. Two-site evaluation of the diagnostic performance of the Sysmex XN Body Fluid (BF) module for cell count and differential in Cerebrospinal Fluid. Int. J. Lab. Hematol. 2018, 40, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Wienefoet, C. Automated Body Fluid Analysis—Sysmex Scientific Customer Information. Available online: https://docplayer.nl/28823938-Automated-body-fluid-analysis-6-october-2016-gebruikersdag-vlaanderen-claudia-wienefoet.html (accessed on 6 October 2016).
- Isenmann, S.; Strik, H.; Wick, M.; Gross, C.C. Liquorzytologie: Methoden und Möglichkeiten. Fortschr. Neurol. Psychiatr. 2017, 85, 616–630. [Google Scholar] [CrossRef]
- Harris, N.; Kunicka, J.; Kratz, A. The ADVIA 2120 Hematology system: Flow cytometry-based analysis of blood and body fluids in the routine Hematology laboratory. Lab. Hematol. 2005, 11, 47–61. [Google Scholar] [CrossRef]
- Zimmermann, M.; Otto, C.; Gonzalez, J.B.; Prokop, S.; Ruprecht, K. Cellular origin and diagnostic significance of high-fluorescent cells in cerebrospinal fluid detected by the XE-5000 Hematology analyzer. Int. J. Lab. Hematol. 2013, 35, 580–588. [Google Scholar] [CrossRef]
- Bonig, L.; Mohn, N.; Ahlbrecht, J.; Wurster, U.; Raab, P.; Puppe, W.; Suhs, K.W.; Stangel, M.; Skripuletz, T.; Schwenkenbecher, P. Leptomeningeal Metastasis: The Role of Cerebrospinal Fluid Diagnostics. Front. Neurol 2019, 10, 839. [Google Scholar] [CrossRef]
- Wick, M. Ausgewählte Methoden der Liquordiagnostik und klinischen Neurochemie; Deutsche Gesellschaft für Liquordiagnostik und Klinische Neurochemie e.V.: Ulm, Germany, 2020; Instand Schriftenreihe Volume II, Düsseldorf. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wick, M.; Gross, C.C.; Tumani, H.; Wildemann, B.; Stangel, M.; on behalf of the German Society of CSF Diagnostics and Clinical Neurochemistry, DGLN e.V. Automated Analysis of Cerebrospinal Fluid Cells Using Commercially Available Blood Cell Analysis Devices—A Critical Appraisal. Cells 2021, 10, 1232. https://doi.org/10.3390/cells10051232
Wick M, Gross CC, Tumani H, Wildemann B, Stangel M, on behalf of the German Society of CSF Diagnostics and Clinical Neurochemistry, DGLN e.V. Automated Analysis of Cerebrospinal Fluid Cells Using Commercially Available Blood Cell Analysis Devices—A Critical Appraisal. Cells. 2021; 10(5):1232. https://doi.org/10.3390/cells10051232
Chicago/Turabian StyleWick, Manfred, Catharina C. Gross, Hayrettin Tumani, Brigitte Wildemann, Martin Stangel, and on behalf of the German Society of CSF Diagnostics and Clinical Neurochemistry, DGLN e.V. 2021. "Automated Analysis of Cerebrospinal Fluid Cells Using Commercially Available Blood Cell Analysis Devices—A Critical Appraisal" Cells 10, no. 5: 1232. https://doi.org/10.3390/cells10051232
APA StyleWick, M., Gross, C. C., Tumani, H., Wildemann, B., Stangel, M., & on behalf of the German Society of CSF Diagnostics and Clinical Neurochemistry, DGLN e.V. (2021). Automated Analysis of Cerebrospinal Fluid Cells Using Commercially Available Blood Cell Analysis Devices—A Critical Appraisal. Cells, 10(5), 1232. https://doi.org/10.3390/cells10051232





