Extracellular Matrix Components Regulate Bone Sialoprotein Expression in MDA-MB-231 Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Sample Preparation
2.3. Antibodies and Dyes
2.4. Data Acquisition and Analysis
3. Results
3.1. BME and Collagen 1 Enhance BSP Expression in MDA-MB-231 Spheroids
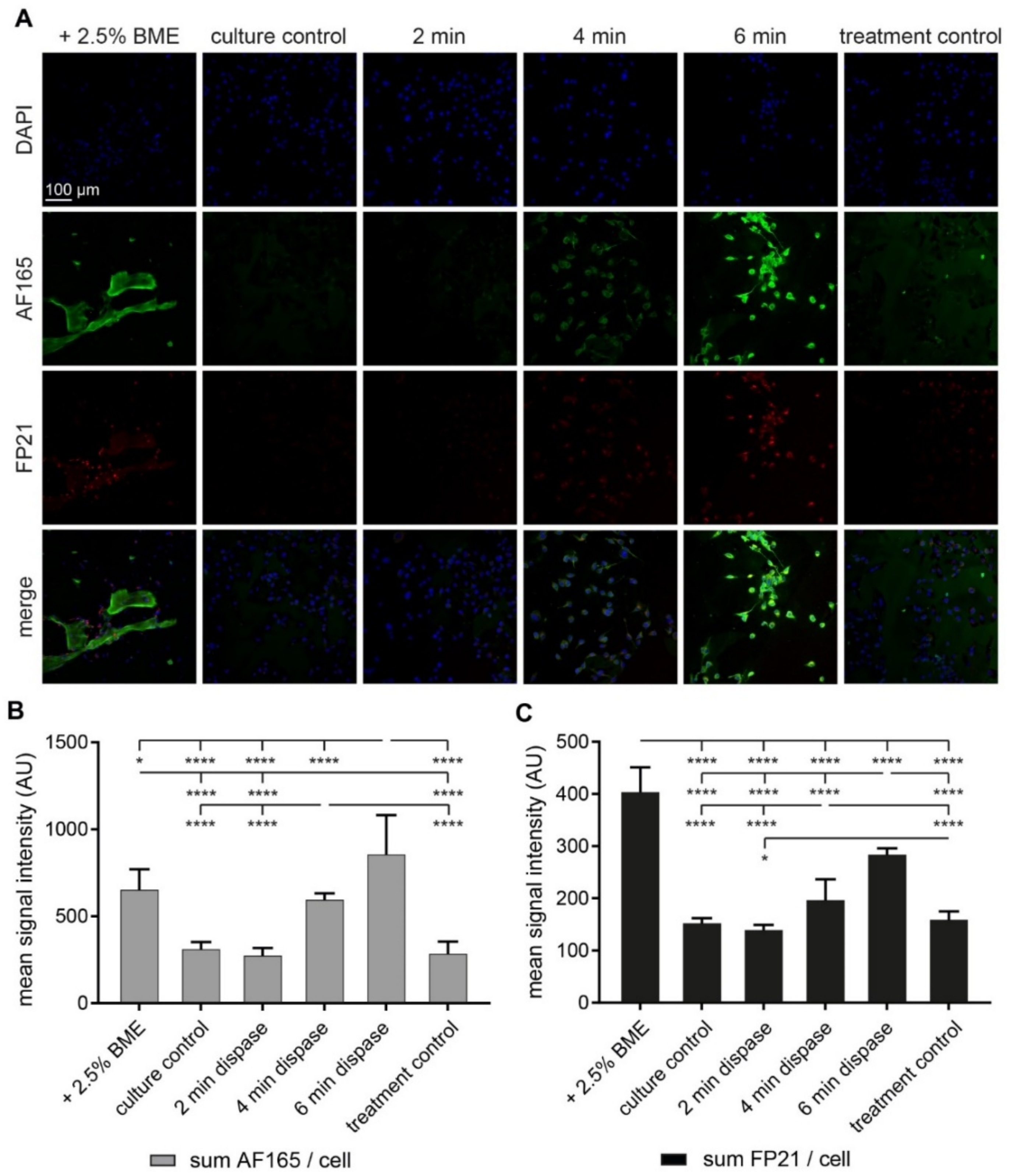
3.2. Both BME and Short-Term Protease Treatment Enhance BSP Immunofluorescence
3.3. Dispase Appears to Induce BSP Biosynthesis in MDA-MB-231 Cells
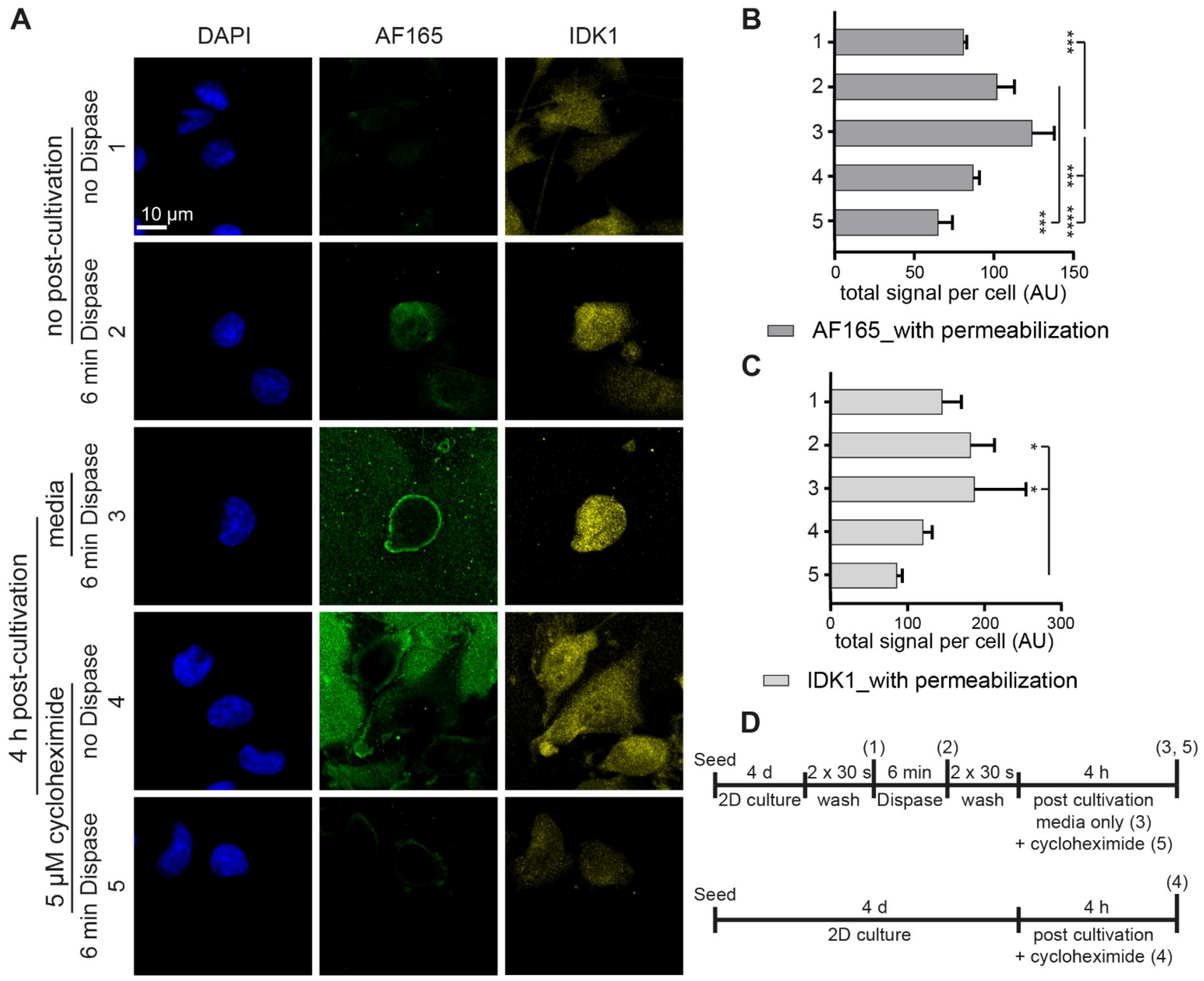
3.4. Regulatory Markers Are Consistent with Dispase-Induced BSP Expression
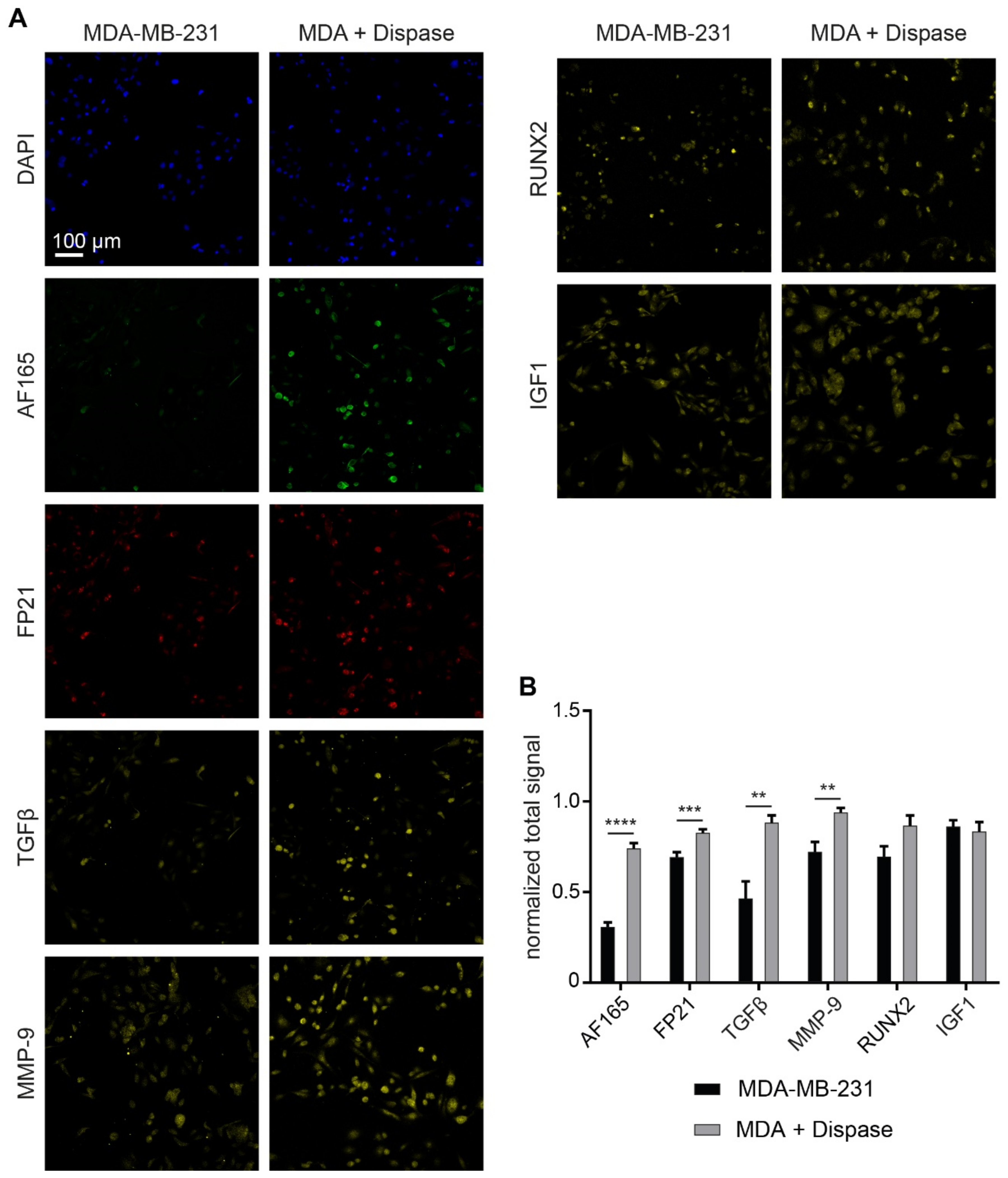
3.5. Dispase Leads to an Increase of BSP Levels in MDA-MB-231 Spheroids
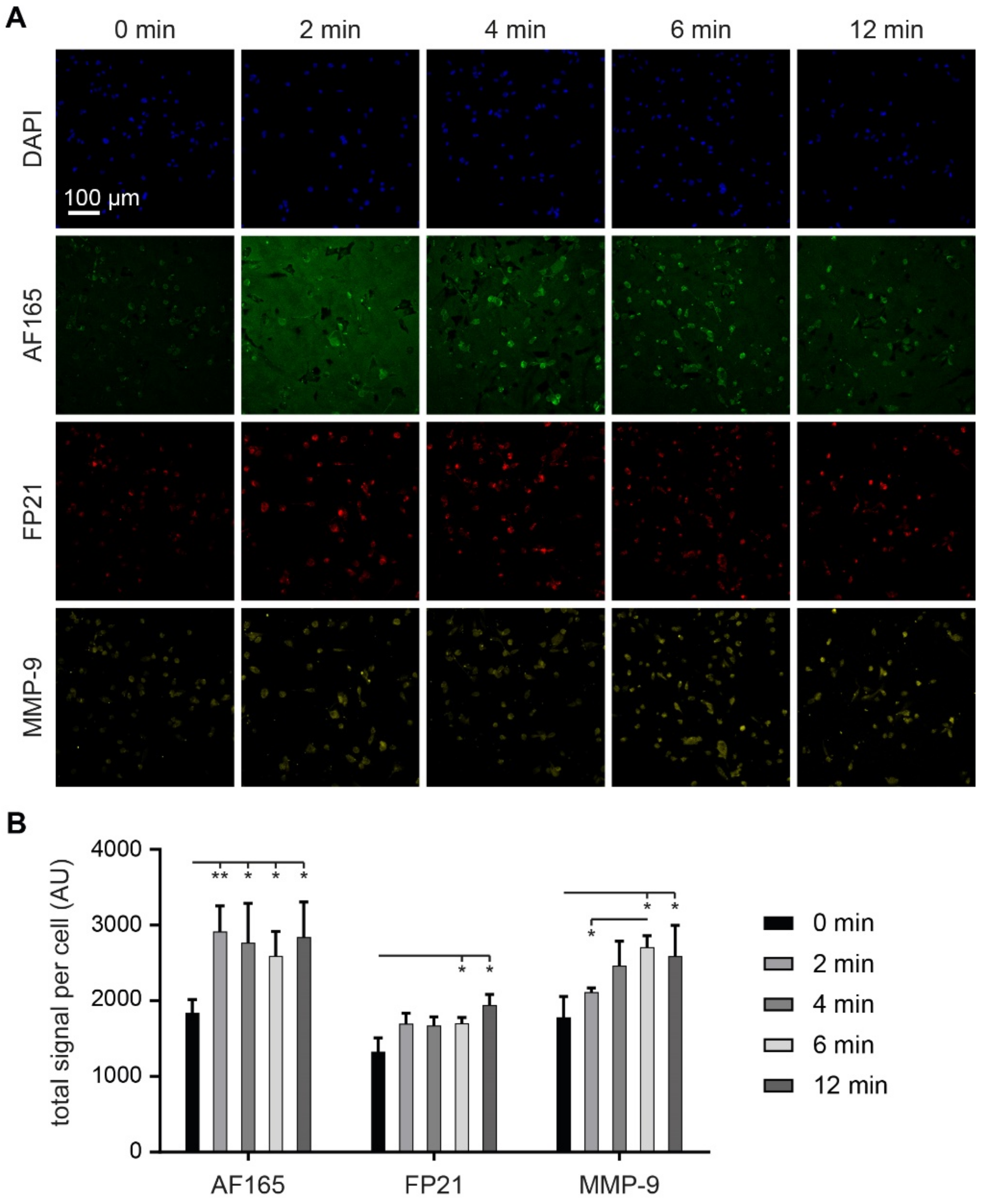
3.6. MMP-9 Exposure Increases BSP Signals in Adherent MDA-MB-231 Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review 1975–2017. Natl. Cancer Inst. 2020, 4, 1975–2006. [Google Scholar]
- Macedo, F.; Ladeira, K.; Pinho, F.; Saraiva, N.; Bonito, N.; Pinto, L.; Gonçalves, F. Bone metastases: An overview. Oncol. Rev. 2017, 11, 321. [Google Scholar] [CrossRef]
- Selvaggi, G.; Scagliotti, G.V. Management of bone metastases in cancer: A review. Crit. Rev. Oncol. Hematol. 2005, 56, 365–378. [Google Scholar] [CrossRef]
- Taube, T.; Elomaa, I.; Blomqvist, C.; Beneton, M.N.C.; Kanis, J.A. Histomorphometric evidence for osteoclast-mediated bone resorption in metastatic breast cancer. Bone 1994, 15, 161–166. [Google Scholar] [CrossRef]
- Bussard, K.M.; Gay, C.V.; Mastro, A.M. The bone microenvironment in metastasis; What is special about bone? Cancer Metastasis Rev. 2008, 27, 41–55. [Google Scholar] [CrossRef]
- Capietto, A.H.; Chan, S.R.; Ricci, B.; Allen, J.A.; Su, X.; Novack, D.V.; Schreiber, R.D.; Faccio, R. Novel ERα positive breast cancer model with estrogen independent growth in the bone microenvironment. Oncotarget 2016, 7, 49751–49764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allocca, G.; Hughes, R.; Wang, N.; Brown, H.K.; Ottewell, P.D.; Brown, N.J.; Holen, I. The bone metastasis niche in breast cancer-potential overlap with the haematopoietic stem cell niche in vivo. J. Bone Oncol. 2019, 17, 100244. [Google Scholar] [CrossRef] [PubMed]
- Reznikov, N.; Chase, H.; Brumfeld, V.; Shahar, R.; Weiner, S. The 3D structure of the collagen fibril network in human trabecular bone: Relation to trabecular organization. Bone 2015, 71, 189–195. [Google Scholar] [CrossRef]
- Lin, X.; Patil, S.; Gao, Y.G.; Qian, A. The Bone Extracellular Matrix in Bone Formation and Regeneration. Front. Pharmacol. 2020, 11, 757. [Google Scholar] [CrossRef]
- Li, J.; Bao, Q.; Chen, S.; Liu, H.; Feng, J.; Qin, H.; Li, A.; Liu, D.; Shen, Y.; Zhao, Y.; et al. Different bone remodeling levels of trabecular and cortical bone in response to changes in Wnt/β-catenin signaling in mice. J. Orthop. Res. 2017, 35, 812–819. [Google Scholar] [CrossRef]
- McCrea, P.D.; Gu, D. The catenin family at a glance. J. Cell Sci. 2010, 123, 637–642. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Yang, S.; Shao, J.; Li, Y.P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. 2007, 12, 3068–3092. [Google Scholar] [CrossRef] [Green Version]
- Kirkham, G.R.; Cartmell, S.H. Genes and Proteins Involved in the Regulation of Osteogenesis. Top. Tissue Eng. 2007, 3, 67–84. [Google Scholar]
- Shupp, A.B.; Kolb, A.D.; Mukhopadhyay, D.; Bussard, K.M. Cancer metastases to bone: Concepts, mechanisms, and interactions with bone osteoblasts. Cancers 2018, 10, 182. [Google Scholar] [CrossRef] [Green Version]
- Owen, R.; Reilly, G.C. In vitro models of bone remodelling and associated disorders. Front. Bioeng. Biotechnol. 2018, 6, 134. [Google Scholar] [CrossRef]
- Hardy, E.; Fernandez-Patron, C. Destroy to Rebuild: The Connection Between Bone Tissue Remodeling and Matrix Metalloproteinases. Front. Physiol. 2020, 11, 47. [Google Scholar] [CrossRef]
- Bellahcène, A.; Castronovo, V.; Ogbureke, K.U.E.; Fisher, L.W.; Fedarko, N.S. Small Integrin-Binding LIgand N-linked Glycoproteins (SIBLINGs): Multifunctional proteins in cancer Akeila. Nat. Rev. Cancer 2008, 8, 212–226. [Google Scholar] [CrossRef] [Green Version]
- Hattar, S.; Asselin, A.; Greenspan, D.; Oboeuf, M.; Berdal, A.; Sautier, J.M. Potential of biomimetic surfaces to promote in vitro osteoblast-like cell differentiation. Biomaterials 2005, 26, 839–848. [Google Scholar] [CrossRef]
- Polyak, K.; Haviv, I.; Campbell, I.G. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009, 25, 30–38. [Google Scholar] [CrossRef]
- Prestwich, G.D. Simplifying the extracellular matrix for 3-D cell culture and tissue engineering: A pragmatic approach. J. Cell. Biochem. 2007, 101, 1370–1383. [Google Scholar] [CrossRef]
- Nürnberg, E.; Vitacolonna, M.; Klicks, J.; von Molitor, E.; Cesetti, T.; Keller, F.; Bruch, R.; Ertongur-Fauth, T.; Riedel, K.; Scholz, P.; et al. Routine Optical Clearing of 3D-Cell Cultures: Simplicity Forward. Front. Mol. Biosci. 2020, 7, 441. [Google Scholar] [CrossRef] [PubMed]
- Keller, F.; Bruch, R.; Schneider, R.; Meier-hubberten, J.; Hafner, M.; Rudolf, R. A Scaffold-Free 3-D Co-Culture Mimics the Major Features of the Reverse Warburg Effect In Vitro. Cells 2020, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Keller, F.; Rudolf, R.; Hafner, M. Towards optimized breast cancer 3D spheroid mono- and co-culture models for pharmacological research and screening. J. Cell. Biotechnol. 2019, 5, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Loibl, S.; Königs, A.; Kaufmann, M.; Costa, S.D.; Bischoff, J. PTHrP and Bone Sialoprotein as Prognostic Markers for Developing Bone Metastases in Breast Cancer Patients. Zentralbl. Gynakol. 2006, 128, 330–335. [Google Scholar] [CrossRef]
- Benton, G.; Kleinman, H.K.; George, J.; Arnaoutova, I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int. J. Cancer 2011, 128, 1751–1757. [Google Scholar] [CrossRef]
- Kleinman, H.K.; McGarvey, M.L.; Liotta, L.A.; Robey, P.G.; Tryggvason, K.; Martin, G.R. Isolation and Characterization of Type IV Procollagen, Laminin, and Heparan Sulfate Proteoglycan from the EHS Sarcoma. Biochemistry 1982, 21, 6188–6193. [Google Scholar] [CrossRef]
- Vukicevic, S.; Kleinman, H.K.; Luyten, F.P.; Roberts, A.B.; Roche, N.S.; Reddi, A.H. Identification of multiple active growth factors in basement membrane matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp. Cell Res. 1992, 202, 1–8. [Google Scholar] [CrossRef]
- Midha, S.; Murab, S.; Ghosh, S. Osteogenic signaling on silk-based matrices. Biomaterials 2016, 97, 133–153. [Google Scholar] [CrossRef]
- Gillette, K.M.; Forbes, K.; Sehgal, I. Detection of matrix metalloproteinases (MMP), tissue inhibitor of metalloproteinase-2, urokinase and plasminogen activator inhibitor-1 within matrigel and growth factor-reduced Matrigel basement membrane. Tumori J. 2003, 89, 421–425. [Google Scholar] [CrossRef]
- Mishra, A.; Shiozawa, Y.; Pienta, K.J.; Taichman, R.S. Homing of cancer cells to the bone. Cancer Microenviron. 2011, 4, 221–235. [Google Scholar] [CrossRef] [Green Version]
- Huang, H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef] [Green Version]
- Zepp, M.; Kovacheva, M.; Altankhuyag, M.; Westphal, G.; Berger, I.; Gather, K.S.; Hilbig, H.; Neuhaus, J.; Hänsch, G.M.; Armbruster, F.P.; et al. IDK1 is a rat monoclonal antibody against hypoglycosylated bone sialoprotein with application as biomarker and therapeutic agent in breast cancer skeletal metastasis. J. Pathol. Clin. Res. 2018, 4, 55–68. [Google Scholar] [CrossRef] [Green Version]
- Gordon, G.M.; Ledee, D.R.; Feuer, W.J.; Fini, M.E. Cytokines and signaling pathways regulating matrix metalloproteinase-9 (MMP-9) expression in corneal epithelial cellsy. J. Cell. Physiol. 2009, 221, 402–411. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Huang, H.Z. The role of RANKL and MMP-9 in the bone resorption caused by ameloblastoma. J. Oral Pathol. Med. 2010, 39, 592–598. [Google Scholar] [CrossRef]
- Sundaram, K.; Nishimura, R.; Senn, J.; Youssef, R.F.; London, S.D.; Reddy, S.V. RANK ligand signaling modulates the matrix metalloproteinase-9 gene expression during osteoclast differentiation. Exp. Cell Res. 2007, 313, 168–178. [Google Scholar] [CrossRef]
- Manicone, A.M.; McGuire, J.K. Matrix Metalloproteinases as Modulators of Inflammation. Semin. Cell Dev. Biol. 2008, 19, 34–41. [Google Scholar] [CrossRef] [Green Version]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Yu, Q.; Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 2000, 14, 163–176. [Google Scholar] [CrossRef]
- Yao, J.; Xiong, S.; Klos, K.; Nguyen, N.; Grijalva, R.; Li, P.; Yu, D. Multiple signaling pathways involved in activation of matrix metalloproteinase-9 (MMP-9) by heregulin-β1 in human breast cancer cells. Oncogene 2001, 20, 8066–8074. [Google Scholar] [CrossRef] [Green Version]
- Schneider-Poetsch, T.; Ju, J.; Eyler, D.E.; Dang, Y.; Bhat, S.; Merrick, W.C.; Green, R.; Shen, B.; Liu, J. Inhibition of Eukaryotic Translation Elongation by Cycloheximide and Lactimidomycin. Nat. Chem. Biol. 2010, 6, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Stein, K.C.; Frydman, J. The stop-and-go traffic regulating protein biogenesis: How translation kinetics controls proteostasis. J. Biol. Chem. 2019, 294, 2076–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuberger, D.M.; Schuepbach, R.A. Correction to: Protease-activated receptors (PARs): Mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thromb. J. 2019, 17, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, L.; Shenoy, S.K.; Lefkowitz, R.J.; DeFea, K. Constitutive protease-activated receptor-2-mediated migration of MDA MB-231 breast cancer cells requires both β-arrestin-1 and -2. J. Biol. Chem. 2004, 279, 55419–55424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oguma, T.; Asano, K.; Tomomatsu, K.; Kodama, M.; Fukunaga, K.; Shiomi, T.; Ohmori, N.; Ueda, S.; Takihara, T.; Shiraishi, Y.; et al. Induction of Mucin and MUC5AC Expression by the Protease Activity of Aspergillus fumigatus in Airway Epithelial Cells. J. Immunol. 2011, 187, 999–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardi, V.C.; Van den Steen, P.E.; Opdenakker, G.; Schweighofer, B.; Deryugina, E.I.; Quigley, J.P. Neutrophil MMP-9 proenzyme, unencumbered by TIMP-1, undergoes efficient activation in vivo and catalytically induces angiogenesis via a basic fibroblast growth factor (FGF-2)/FGFR-2 pathway. J. Biol. Chem. 2009, 284, 25854–25866. [Google Scholar] [CrossRef] [Green Version]
- Deryugina, E.I.; Quigley, J.P. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: Contrasting, overlapping and compensatory functions. Biochim. Biophys. Acta-Mol. Cell Res. 2010, 1803, 103–120. [Google Scholar] [CrossRef] [Green Version]
- Ochiai, H.; Okada, S.; Saito, A.; Hoshi, K.; Yamashita, H.; Takato, T.; Azuma, T. Inhibition of insulin-like growth factor-1 (IGF-1) expression by prolonged transforming growth factor-β1 (TGF-β1) administration suppresses osteoblast differentiation. J. Biol. Chem. 2012, 287, 22654–22661. [Google Scholar] [CrossRef] [Green Version]
- Ogata, Y. Bone sialoprotein and its transcriptional regulatory mechanism. J. Periodontal Res. 2008, 43, 127–135. [Google Scholar] [CrossRef]
- Shi, M.; Zhu, J.; Wang, R.; Chen, X.; Mi, L.; Walz, T.; Springer, T.A. Latent TGF-β structure and activation. Nature 2016, 474, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Toth, M.; Chvyrkova, I.; Bernardo, M.M.; Hernandez-Barrantes, S.; Fridman, R. Pro-MMP-9 activation by the MT1-MMP/MMP-2 axis and MMP-3: Role of TIMP-2 and plasma membranes. Biochem. Biophys. Res. Commun. 2003, 308, 386–395. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keller, F.; Bruch, R.; Clauder, F.; Hafner, M.; Rudolf, R. Extracellular Matrix Components Regulate Bone Sialoprotein Expression in MDA-MB-231 Breast Cancer Cells. Cells 2021, 10, 1304. https://doi.org/10.3390/cells10061304
Keller F, Bruch R, Clauder F, Hafner M, Rudolf R. Extracellular Matrix Components Regulate Bone Sialoprotein Expression in MDA-MB-231 Breast Cancer Cells. Cells. 2021; 10(6):1304. https://doi.org/10.3390/cells10061304
Chicago/Turabian StyleKeller, Florian, Roman Bruch, Franziska Clauder, Mathias Hafner, and Rüdiger Rudolf. 2021. "Extracellular Matrix Components Regulate Bone Sialoprotein Expression in MDA-MB-231 Breast Cancer Cells" Cells 10, no. 6: 1304. https://doi.org/10.3390/cells10061304





