Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways
Abstract
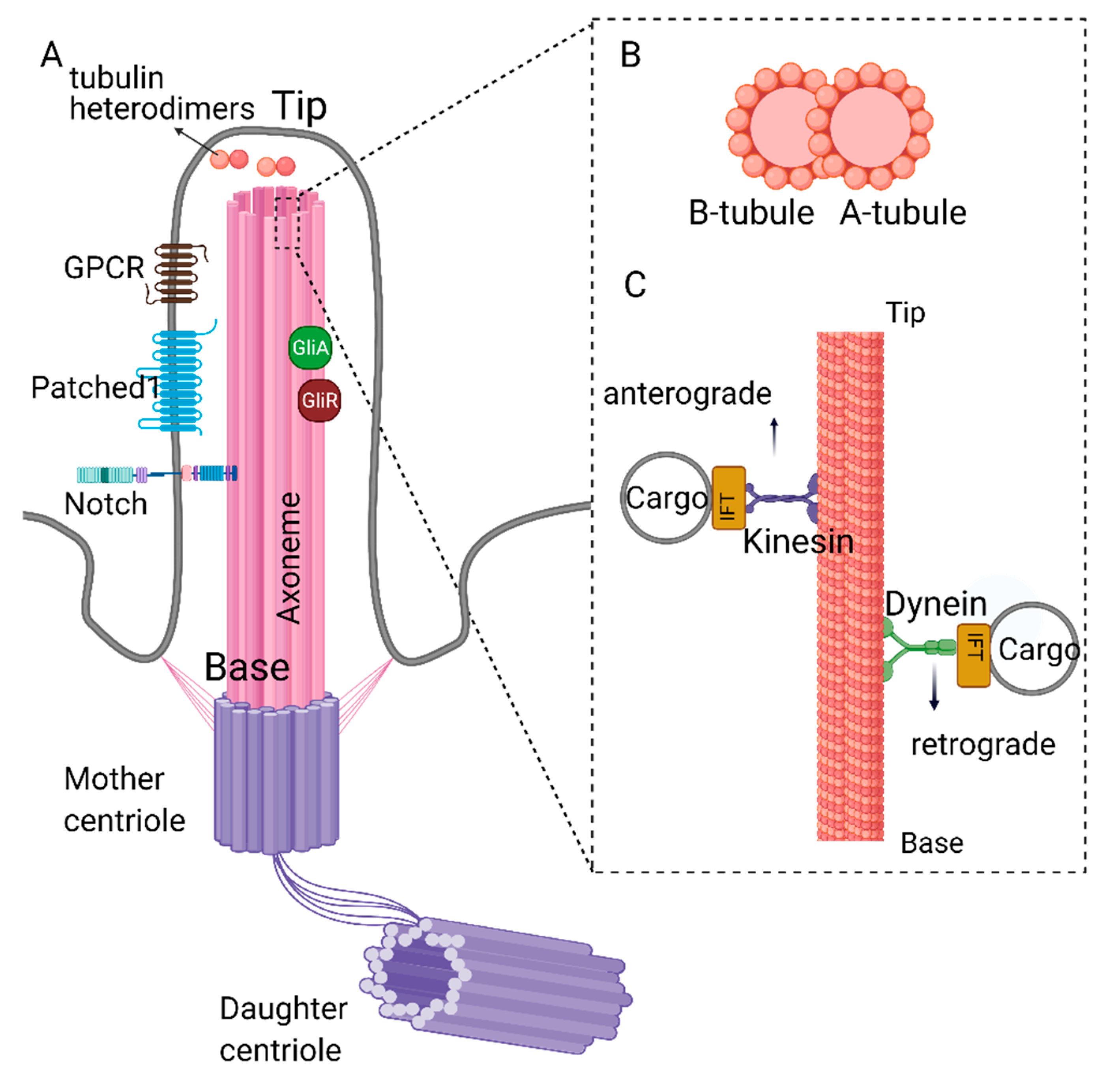
1. Structure and Function of Primary Cilia
2. Primary Cilia and Signal Transduction
3. Stem Cell Biology and Classification
4. Primary Cilia in Embryonic Development: ECSs
5. Primary Cilia in Organ Development/Maintenance: Adult Stem Cells and iPSCs
6. Role of Primary Cilia in Wnt/β-Catenin Regulation and Stem Cell Biology
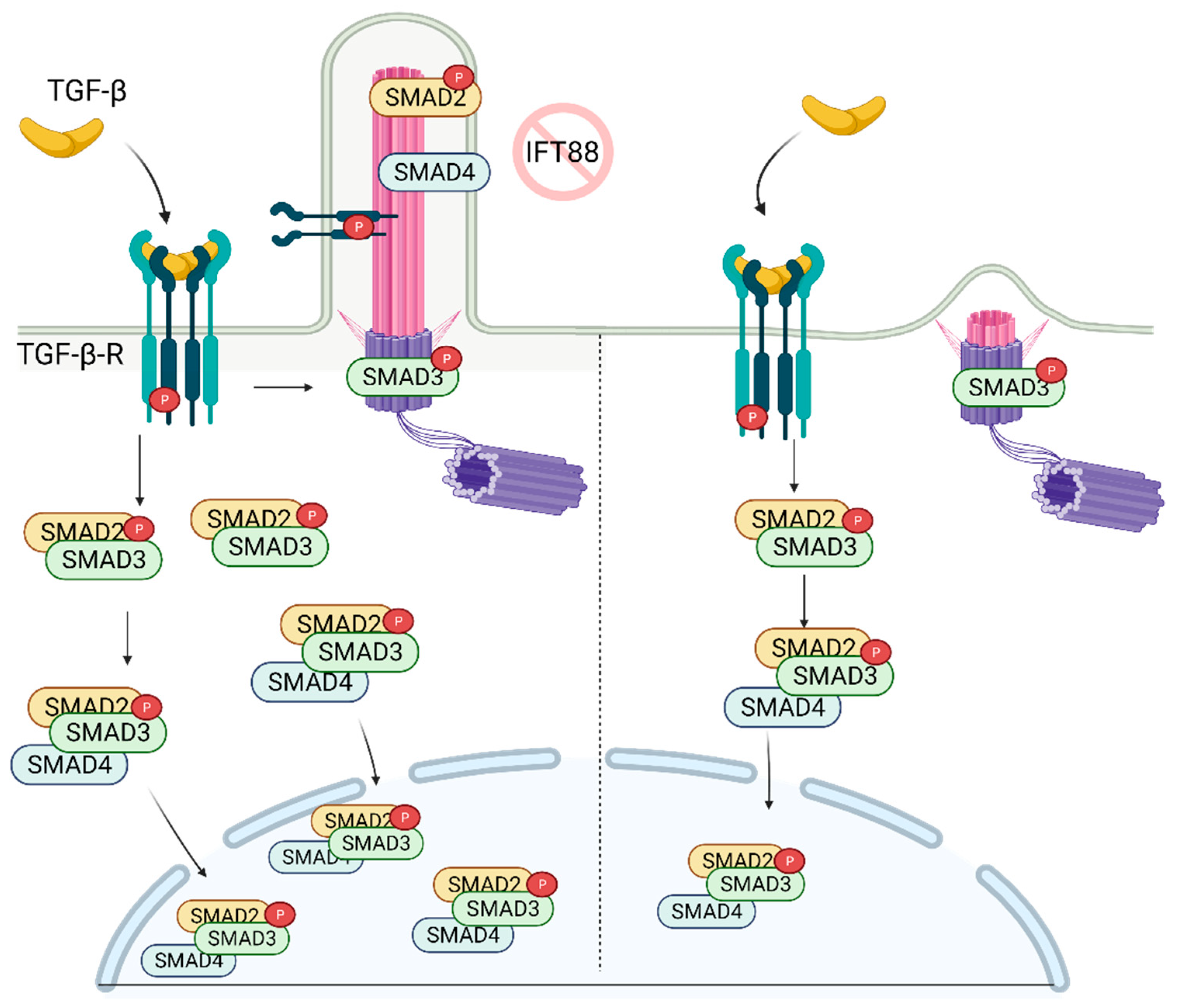
7. Role of Primary Cilia in TGF-β Regulation and Stem Cell Biology
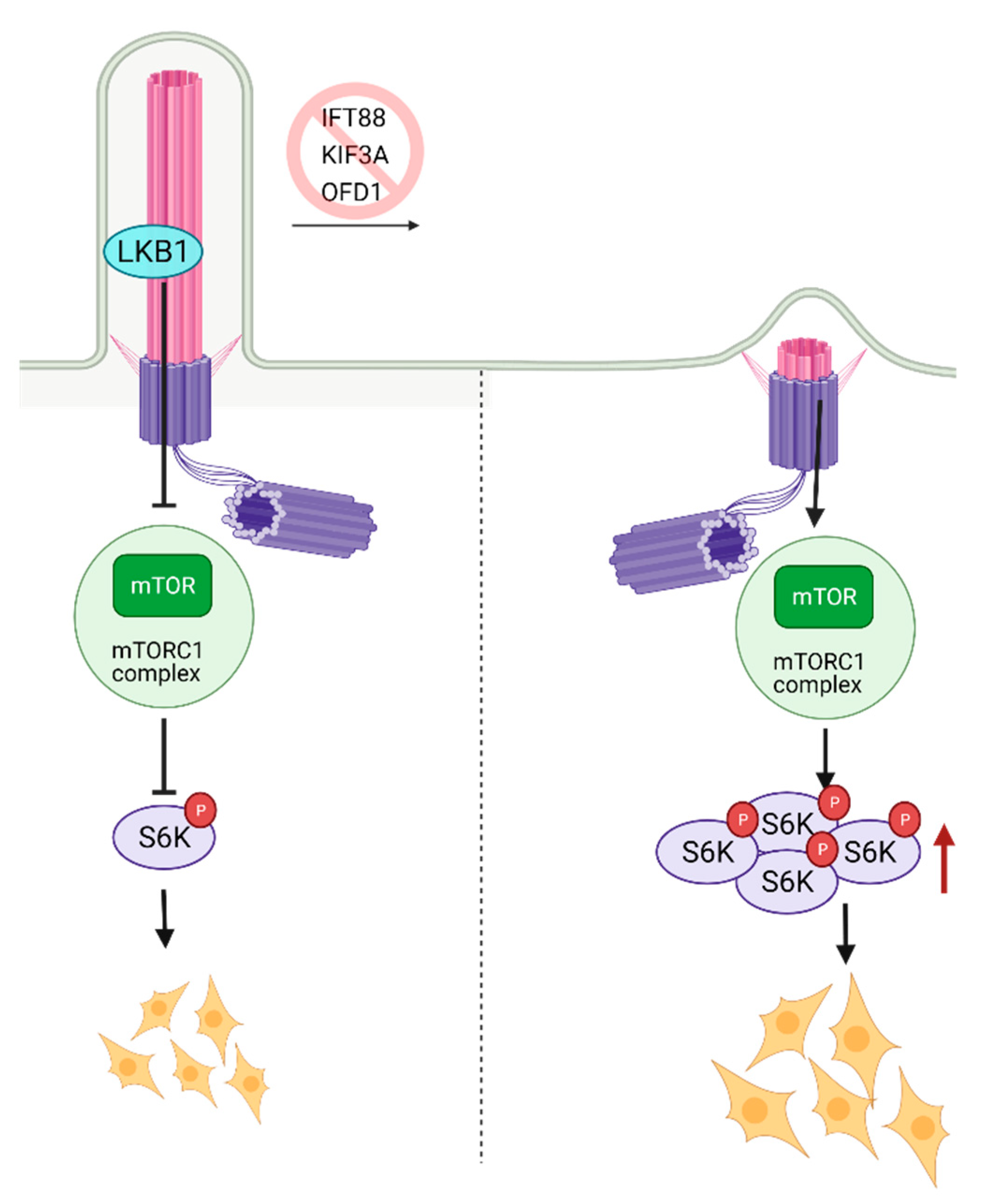
8. Role of Primary Cilia in mTOR Regulation and Stem Cell Biology
9. The Role of Autophagy and Primary Cilia in Embryonic Stem Cells
10. Role of Primary Cilia in Notch Regulation and Stem Cell Biology
11. Primary Cilia in Cancer and Cancer Stem Cells
12. Conclusions and Future Perspectives
13. Clinical Relevance
- Understanding how the primary cilium is involved in stem cell biology is crucial to develop new ciliotherapies [25] to eliminate developmental ciliopathy diseases such as Meckel Syndrome.
- Targeting primary cilia and improving its stability/function in patients suffering from bone diseases might be a new approach to improve the patients’ quality of life.
- Deciphering the role of the primary cilium in cancer stem cell biology will likely improve our knowledge and diversify the cancer treatment options as the majority of the signaling pathways are dysregulated in cancers.
14. Outstanding Questions
- Does the knockout or depletion of essential ciliary genes such as IFT88, IFT20, KIF3A (“builders”) have the same impact on differentiation of the embryonic vs. adult stem cells or iPSCs? The answer to this question might allow for defining differences between signaling in dividing vs. quiescent stem cells.
- Since genes such as IFT88 and others have additional non-ciliary functions, the interpretation of KO results might be more complicated than just lack of cilia. Better tools need to be developed and expression controlled to decipher the cilium dependent vs. independent impact of the knockout of IFT and kinesin proteins.
- Would chemical inhibition of these proteins deliver the same results? The answer will allow us to discriminate between activity vs. protein-protein driven functions of the ciliary “builders” or “disassemblers”.
- Would the utilization of proteasome inhibitors in cilia deficient embryos recover the Notch (NICD) levels and HE to HPSC differentiation? The answer will be needed to understand the role of protein biosynthesis/degradation, including autophagy in ciliary homeostasis.
- Can we reprogram cancer stem cells via manipulation of primary cilia dynamics? The answer to this question will require substantial evidence to be gathered on the driving forces of deciliation in cancer, which includes but is not limited to centrosome defects/amplification, lack of cilia, ”builders”, or overexpression of cilia, “disassemblers”.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loskutov, Y.V.; Griffin, C.L.; Marinak, K.M.; Bobko, A.; Margaryan, N.V.; Geldenhuys, W.J.; Sarkaria, J.N.; Pugacheva, E.N. LPA signaling is regulated through the primary cilium: A novel target in glioblastoma. Oncogene 2018, 37, 1457–1471. [Google Scholar] [CrossRef]
- Sun, S.; Fisher, R.L.; Bowser, S.S.; Pentecost, B.T.; Sui, H. Three-dimensional architecture of epithelial primary cilia. Proc. Natl. Acad. Sci. USA 2019, 116, 9370–9379. [Google Scholar] [CrossRef]
- Plotnikova, O.V.; Golemis, E.A.; Pugacheva, E.N. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008, 68, 2058–2061. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.D. A reinvestigation of cross-sections of cilia. J. Cell Biol. 1968, 37, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Stepanek, L.; Pigino, G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science 2016, 352, 721–724. [Google Scholar] [CrossRef]
- Bhogaraju, S.; Cajanek, L.; Fort, C.; Blisnick, T.; Weber, K.; Taschner, M.; Mizuno, N.; Lamla, S.; Bastin, P.; Nigg, E.A.; et al. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 2013, 341, 1009–1012. [Google Scholar] [CrossRef]
- Kubo, T.; Brown, J.M.; Bellve, K.; Craige, B.; Craft, J.M.; Fogarty, K.; Lechtreck, K.F.; Witman, G.B. Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J. Cell Sci. 2016, 129, 2106–2119. [Google Scholar] [CrossRef]
- Craft Van De Weghe, J.; Harris, J.A.; Kubo, T.; Witman, G.B.; Lechtreck, K.F. Diffusion rather than intraflagellar transport likely provides most of the tubulin required for axonemal assembly in. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Absalon, S.; Blisnick, T.; Kohl, L.; Toutirais, G.; Doré, G.; Julkowska, D.; Tavenet, A.; Bastin, P. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell 2008, 19, 929–944. [Google Scholar] [CrossRef] [PubMed]
- Avidor-Reiss, T.; Maer, A.M.; Koundakjian, E.; Polyanovsky, A.; Keil, T.; Subramaniam, S.; Zuker, C.S. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell 2004, 117, 527–539. [Google Scholar] [CrossRef]
- Rosenbaum, J.L.; Witman, G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002, 3, 813–825. [Google Scholar] [CrossRef]
- Singla, V.; Romaguera-Ros, M.; Garcia-Verdugo, J.M.; Reiter, J.F. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 2010, 18, 410–424. [Google Scholar] [CrossRef]
- Lechtreck, K.F. IFT-Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 2015, 40, 765–778. [Google Scholar] [CrossRef]
- Chávez, M.; Ena, S.; Van Sande, J.; de Kerchove d’Exaerde, A.; Schurmans, S.; Schiffmann, S.N. Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Dev. Cell 2015, 34, 338–350. [Google Scholar] [CrossRef]
- Garcia-Gonzalo, F.R.; Phua, S.C.; Roberson, E.C.; Garcia, G.; Abedin, M.; Schurmans, S.; Inoue, T.; Reiter, J.F. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev. Cell 2015, 34, 400–409. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Wang, G.; Dasgupta, S.; Dinkins, M.; Zhu, G.; Bieberich, E. Characterization of an apical ceramide-enriched compartment regulating ciliogenesis. Mol. Biol. Cell 2012, 23, 3156–3166. [Google Scholar] [CrossRef]
- Janich, P.; Corbeil, D. GM1 and GM3 gangliosides highlight distinct lipid microdomains within the apical domain of epithelial cells. FEBS Lett. 2007, 581, 1783–1787. [Google Scholar] [CrossRef] [PubMed]
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007, 129, 1351–1363. [Google Scholar] [CrossRef]
- Tucker, R.W.; Pardee, A.B.; Fujiwara, K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 1979, 17, 527–535. [Google Scholar] [CrossRef]
- Wheatley, D.N.; Wang, A.M.; Strugnell, G.E. Expression of primary cilia in mammalian cells. Cell Biol. Int. 1996, 20, 73–81. [Google Scholar] [CrossRef]
- Rieder, C.L.; Jensen, C.G.; Jensen, L.C. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J. Ultrastruct. Res. 1979, 68, 173–185. [Google Scholar] [CrossRef]
- Ehler, L.L.; Holmes, J.A.; Dutcher, S.K. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 1995, 141, 945–960. [Google Scholar] [CrossRef]
- Clement, C.A.; Ajbro, K.D.; Koefoed, K.; Vestergaard, M.L.; Veland, I.R.; Henriques de Jesus, M.P.; Pedersen, L.B.; Benmerah, A.; Andersen, C.Y.; Larsen, L.A.; et al. TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 2013, 3, 1806–1814. [Google Scholar] [CrossRef]
- Monnich, M.; Borgeskov, L.; Breslin, L.; Jakobsen, L.; Rogowski, M.; Doganli, C.; Schroder, J.M.; Mogensen, J.B.; Blinkenkjaer, L.; Harder, L.M.; et al. CEP128 Localizes to the Subdistal Appendages of the Mother Centriole and Regulates TGF-beta/BMP Signaling at the Primary Cilium. Cell Rep. 2018, 22, 2584–2592. [Google Scholar] [CrossRef]
- Alvarez-Satta, M.; Matheu, A. Primary cilium and glioblastoma. Ther. Adv. Med. Oncol. 2018, 10, 1169. [Google Scholar] [CrossRef]
- Pan, J. Cilia and ciliopathies: From Chlamydomonas and beyond. Sci. China C Life Sci. 2008, 51, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef]
- Adams, M.; Smith, U.M.; Logan, C.V.; Johnson, C.A. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J. Med. Genet. 2008, 45, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Badano, J.L.; Mitsuma, N.; Beales, P.L.; Katsanis, N. The ciliopathies: An emerging class of human genetic disorders. Annu. Rev. Genom. Hum. Genet. 2006, 7, 125–148. [Google Scholar] [CrossRef]
- Mitchison, H.M.; Valente, E.M. Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 2017, 241, 294–309. [Google Scholar] [CrossRef]
- McConnachie, D.J.; Stow, J.L.; Mallett, A.J. Ciliopathies and the Kidney: A Review. Am. J. Kidney Dis. 2021, 77, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Duong Phu, M.; Bross, S.; Burkhalter, M.D.; Philipp, M. Limitations and opportunities in the pharmacotherapy of ciliopathies. Pharmacol. Ther. 2021, 225, 107841. [Google Scholar] [CrossRef] [PubMed]
- Eggan, K. Short-circuiting epiblast development. Cell Stem Cell 2007, 1, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Kiprilov, E.N.; Awan, A.; Desprat, R.; Velho, M.; Clement, C.A.; Byskov, A.G.; Andersen, C.Y.; Satir, P.; Bouhassira, E.E.; Christensen, S.T.; et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J. Cell Biol. 2008, 180, 897–904. [Google Scholar] [CrossRef]
- Bangs, F.K.; Schrode, N.; Hadjantonakis, A.K.; Anderson, K.V. Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 2015, 17, 113–122. [Google Scholar] [CrossRef]
- Marszalek, J.R.; Ruiz-Lozano, P.; Roberts, E.; Chien, K.R.; Goldstein, L.S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. USA 1999, 96, 5043–5048. [Google Scholar] [CrossRef]
- Nonaka, S.; Tanaka, Y.; Okada, Y.; Takeda, S.; Harada, A.; Kanai, Y.; Kido, M.; Hirokawa, N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 1998, 95, 829–837. [Google Scholar] [CrossRef]
- Murcia, N.S.; Richards, W.G.; Yoder, B.K.; Mucenski, M.L.; Dunlap, J.R.; Woychik, R.P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 2000, 127, 2347–2355. [Google Scholar] [CrossRef]
- Takeda, S.; Yonekawa, Y.; Tanaka, Y.; Okada, Y.; Nonaka, S.; Hirokawa, N. Left-right asymmetry and kinesin superfamily protein KIF3A: New insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 1999, 145, 825–836. [Google Scholar] [CrossRef]
- Schock, E.N.; Brugmann, S.A. Neural crest cells utilize primary cilia to regulate ventral forebrain morphogenesis via Hedgehog-dependent regulation of oriented cell division. Dev. Biol. 2017, 431, 168–178. [Google Scholar] [CrossRef]
- Huang, P.; Schier, A.F. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development 2009, 136, 3089–3098. [Google Scholar] [CrossRef]
- Liu, Z.; Tu, H.; Kang, Y.; Xue, Y.; Ma, D.; Zhao, C.; Li, H.; Wang, L.; Liu, F. Primary cilia regulate hematopoietic stem and progenitor cell specification through Notch signaling in zebrafish. Nat. Commun. 2019, 10, 1839. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Wang, G.; Krishnamurthy, K.; Chiang, Y.W.; Dasgupta, S.; Bieberich, E. Regulation of neural progenitor cell motility by ceramide and potential implications for mouse brain development. J. Neurochem. 2008, 106, 718–733. [Google Scholar] [CrossRef]
- He, Q.; Wang, G.; Wakade, S.; Dasgupta, S.; Dinkins, M.; Kong, J.N.; Spassieva, S.D.; Bieberich, E. Primary cilia in stem cells and neural progenitors are regulated by neutral sphingomyelinase 2 and ceramide. Mol. Biol. Cell 2014, 25, 1715–1729. [Google Scholar] [CrossRef]
- Niwa, H. How is pluripotency determined and maintained? Development 2007, 134, 635–646. [Google Scholar] [CrossRef]
- Jaenisch, R.; Young, R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008, 132, 567–582. [Google Scholar] [CrossRef]
- Guo, G.; von Meyenn, F.; Santos, F.; Chen, Y.; Reik, W.; Bertone, P.; Smith, A.; Nichols, J. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Rep. 2016, 6, 437–446. [Google Scholar] [CrossRef]
- Siu, M.K.; Wong, E.S.; Kong, D.S.; Chan, H.Y.; Jiang, L.; Wong, O.G.; Lam, E.W.; Chan, K.K.; Ngan, H.Y.; Le, X.F.; et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene 2013, 32, 3500–3509. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Ni, A.; Wu, M.J.; Chavala, S.H. Sphere formation permits Oct4 reprogramming of ciliary body epithelial cells into induced pluripotent stem cells. Stem Cells Dev. 2014, 23, 3065–3071. [Google Scholar] [CrossRef]
- Nathwani, B.B.; Miller, C.H.; Yang, T.L.; Solimano, J.L.; Liao, J.C. Morphological differences of primary cilia between human induced pluripotent stem cells and their parental somatic cells. Stem Cells Dev. 2014, 23, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, C.; Akiyama, K.; Chai, Y.; Le, A.D.; Wang, Z.; Shi, S. Gingivae contain neural-crest- and mesoderm-derived mesenchymal stem cells. J. Dent. Res. 2013, 92, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.N.F.B.; Yazid, M.D.; Yunus, M.H.M.; Chowdhury, S.R.; Lokanathan, Y.; Idrus, R.B.H.; Ng, A.M.H.; Law, J.X. Large-Scale Expansion of Human Mesenchymal Stem Cells. Stem Cells Int. 2020, 2020, 9529465. [Google Scholar] [CrossRef]
- Ma, Z.; Qin, M.; Liang, H.; Chen, R.; Cai, S.; Huang, Z.; Tai, G. Primary cilia-dependent signaling is involved in regulating mesenchymal stem cell proliferation and pluripotency maintenance. J. Mol. Histol. 2020, 51, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.; Friemel, A.; Kreis, N.N.; Hoock, S.C.; Roth, S.; Kielland-Kaisen, U.; Brüggmann, D.; Solbach, C.; Louwen, F.; Yuan, J. Primary Cilia Are Dysfunctional in Obese Adipose-Derived Mesenchymal Stem Cells. Stem Cell Rep. 2018, 10, 583–599. [Google Scholar] [CrossRef]
- Ritter, A.; Kreis, N.N.; Roth, S.; Friemel, A.; Jennewein, L.; Eichbaum, C.; Solbach, C.; Louwen, F.; Yuan, J. Restoration of primary cilia in obese adipose-derived mesenchymal stem cells by inhibiting Aurora A or extracellular signal-regulated kinase. Stem Cell Res. Ther. 2019, 10, 255. [Google Scholar] [CrossRef]
- Kabiri, Z.; Greicius, G.; Zaribafzadeh, H.; Hemmerich, A.; Counter, C.M.; Virshup, D.M. Wnt signaling suppresses MAPK-driven proliferation of intestinal stem cells. J. Clin. Investig. 2018, 128, 3806–3812. [Google Scholar] [CrossRef]
- Packard, M.; Koo, E.S.; Gorczyca, M.; Sharpe, J.; Cumberledge, S.; Budnik, V. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell 2002, 111, 319–330. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Liu, H.; Liu, L.; Wang, Y.D.; Chen, W.D. Emerging Roles of Wnt Ligands in Human Colorectal Cancer. Front. Oncol. 2020, 10, 1341. [Google Scholar] [CrossRef] [PubMed]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef]
- Takahashi, M.; Tsunoda, T.; Seiki, M.; Nakamura, Y.; Furukawa, Y. Identification of membrane-type matrix metalloproteinase-1 as a target of the beta-catenin/Tcf4 complex in human colorectal cancers. Oncogene 2002, 21, 5861–5867. [Google Scholar] [CrossRef]
- Lustig, B.; Jerchow, B.; Sachs, M.; Weiler, S.; Pietsch, T.; Karsten, U.; van de Wetering, M.; Clevers, H.; Schlag, P.M.; Birchmeier, W.; et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell Biol. 2002, 22, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.J.; Plescia, J.; Clevers, H.; Fearon, E.R.; Altieri, D.C. Survivin and molecular pathogenesis of colorectal cancer. Lancet 2003, 362, 205–209. [Google Scholar] [CrossRef]
- Bae, Y.K.; Kim, G.H.; Kwon, J.H.; Kim, M.; Choi, S.J.; Oh, W.; Um, S.; Jin, H.J. Primary Cilia Mediate Wnt5a/beta-catenin Signaling to Regulate Adipogenic Differentiation of Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells Following Calcium Induction. Tissue Eng. Regen. Med. 2020, 17, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Corbit, K.C.; Shyer, A.E.; Dowdle, W.E.; Gaulden, J.; Singla, V.; Chen, M.H.; Chuang, P.T.; Reiter, J.F. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 2008, 10, 70–76. [Google Scholar] [CrossRef]
- Ocbina, P.J.; Tuson, M.; Anderson, K.V. Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS ONE 2009, 4, e6839. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, G.; Feng, L.; Jiang, Z.; Yu, M.; Bao, J.; Tian, W. Disruption of kif3a results in defective osteoblastic differentiation in dental mesenchymal stem/precursor cells via the Wnt signaling pathway. Mol. Med. Rep. 2016, 14, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Qiu, N.; Xiao, Z.; Cao, L.; Buechel, M.M.; David, V.; Roan, E.; Quarles, L.D. Disruption of Kif3a in osteoblasts results in defective bone formation and osteopenia. J. Cell Sci. 2012, 125, 1945–1957. [Google Scholar] [CrossRef]
- Gazea, M.; Tasouri, E.; Tolve, M.; Bosch, V.; Kabanova, A.; Gojak, C.; Kurtulmus, B.; Novikov, O.; Spatz, J.; Pereira, G.; et al. Primary cilia are critical for Sonic hedgehog-mediated dopaminergic neurogenesis in the embryonic midbrain. Dev. Biol. 2016, 409, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Dyson, J.M.; Conduit, S.E.; Feeney, S.J.; Hakim, S.; DiTommaso, T.; Fulcher, A.J.; Sriratana, A.; Ramm, G.; Horan, K.A.; Gurung, R.; et al. INPP5E regulates phosphoinositide-dependent cilia transition zone function. J. Cell Biol. 2017, 216, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Gencer, S.; Oleinik, N.; Kim, J.; Panneer Selvam, S.; De Palma, R.; Dany, M.; Nganga, R.; Thomas, R.J.; Senkal, C.E.; Howe, P.H.; et al. TGF-β receptor I/II trafficking and signaling at primary cilia are inhibited by ceramide to attenuate cell migration and tumor metastasis. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Elsafadi, M.; Manikandan, M.; Atteya, M.; Abu Dawud, R.; Almalki, S.; Ali Kaimkhani, Z.; Aldahmash, A.; Alajez, N.M.; Alfayez, M.; Kassem, M.; et al. SERPINB2 is a novel TGFbeta-responsive lineage fate determinant of human bone marrow stromal cells. Sci. Rep. 2017, 7, 10797. [Google Scholar] [CrossRef]
- Labour, M.N.; Riffault, M.; Christensen, S.T.; Hoey, D.A. TGFβ1—Induced recruitment of human bone mesenchymal stem cells is mediated by the primary cilium in a SMAD3-dependent manner. Sci. Rep. 2016, 6, 35542. [Google Scholar] [CrossRef]
- Diguet, N.; Le Garrec, J.F.; Lucchesi, T.; Meilhac, S.M. Imaging and analyzing primary cilia in cardiac cells. Methods Cell Biol. 2015, 127, 55–73. [Google Scholar] [CrossRef]
- Ehnert, S.; Sreekumar, V.; Aspera-Werz, R.H.; Sajadian, S.O.; Wintermeyer, E.; Sandmann, G.H.; Bahrs, C.; Hengstler, J.G.; Godoy, P.; Nussler, A.K. TGF-beta1 impairs mechanosensation of human osteoblasts via HDAC6-mediated shortening and distortion of primary cilia. J. Mol. Med. 2017, 95, 653–663. [Google Scholar] [CrossRef]
- Murakami, M.; Ichisaka, T.; Maeda, M.; Oshiro, N.; Hara, K.; Edenhofer, F.; Kiyama, H.; Yonezawa, K.; Yamanaka, S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell Biol. 2004, 24, 6710–6718. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Garbern, J.C.; Helman, A.; Sereda, R.; Sarikhani, M.; Ahmed, A.; Escalante, G.O.; Ogurlu, R.; Kim, S.L.; Zimmerman, J.F.; Cho, A.; et al. Inhibition of mTOR Signaling Enhances Maturation of Cardiomyocytes Derived From Human-Induced Pluripotent Stem Cells via p53-Induced Quiescence. Circulation 2020, 141, 285–300. [Google Scholar] [CrossRef]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Boyer, L.; Jin, M.; Mertens, J.; Kim, Y.; Ma, L.; Hamm, M.; Gage, F.H.; Hunter, T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 2016, 5. [Google Scholar] [CrossRef]
- Chen, T.; Shen, L.; Yu, J.; Wan, H.; Guo, A.; Chen, J.; Long, Y.; Zhao, J.; Pei, G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell 2011, 10, 908–911. [Google Scholar] [CrossRef] [PubMed]
- Zullo, A.; Iaconis, D.; Barra, A.; Cantone, A.; Messaddeq, N.; Capasso, G.; Dolle, P.; Igarashi, P.; Franco, B. Kidney-specific inactivation of Ofd1 leads to renal cystic disease associated with upregulation of the mTOR pathway. Hum. Mol. Genet. 2010, 19, 2792–2803. [Google Scholar] [CrossRef] [PubMed]
- Boehlke, C.; Kotsis, F.; Patel, V.; Braeg, S.; Voelker, H.; Bredt, S.; Beyer, T.; Janusch, H.; Hamann, C.; Godel, M.; et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 2010, 12, 1115–1122. [Google Scholar] [CrossRef]
- Viau, A.; Bienaimé, F.; Lukas, K.; Todkar, A.P.; Knoll, M.; Yakulov, T.A.; Hofherr, A.; Kretz, O.; Helmstädter, M.; Reichardt, W.; et al. Cilia-localized LKB1 regulates chemokine signaling, macrophage recruitment, and tissue homeostasis in the kidney. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Foerster, P.; Daclin, M.; Asm, S.; Faucourt, M.; Boletta, A.; Genovesio, A.; Spassky, N. mTORC1 signaling and primary cilia are required for brain ventricle morphogenesis. Development 2017, 144, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Pampliega, O.; Orhon, I.; Patel, B.; Sridhar, S.; Diaz-Carretero, A.; Beau, I.; Codogno, P.; Satir, B.H.; Satir, P.; Cuervo, A.M. Functional interaction between autophagy and ciliogenesis. Nature 2013, 502, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Shin, J.H.; Park, S.J.; Jo, Y.K.; Kim, J.S.; Kang, I.H.; Nam, J.B.; Chung, D.Y.; Cho, Y.; Lee, E.H.; et al. Inhibition of autophagy suppresses sertraline-mediated primary ciliogenesis in retinal pigment epithelium cells. PLoS ONE 2015, 10, e0118190. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Wang, Y.; Lalli, M.A.; Guzman, E.; Godshalk, S.E.; Zhou, H.; Kosik, K.S. Primary Cilium-Autophagy-Nrf2 (PAN) Axis Activation Commits Human Embryonic Stem Cells to a Neuroectoderm Fate. Cell 2016, 165, 410–420. [Google Scholar] [CrossRef]
- Trindade, A.; Duarte, A. Notch Signaling Function in the Angiocrine Regulation of Tumor Development. Cells 2020, 9, 2467. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Lowell, S.; Jones, P.; Le Roux, I.; Dunne, J.; Watt, F.M. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 2000, 10, 491–500. [Google Scholar] [CrossRef]
- Takahashi, T.; Shiraishi, A. Stem Cell Signaling Pathways in the Small Intestine. Int. J. Mol. Sci. 2020, 21, 2032. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.V.; Kim, E.J.; Wu, J.; Hynes, M.; Bednar, F.; Proctor, E.; Wang, L.; Dziubinski, M.L.; Simeone, D.M. The Notch pathway is important in maintaining the cancer stem cell population in pancreatic cancer. PLoS ONE 2014, 9, e91983. [Google Scholar] [CrossRef]
- Lake, R.J.; Tsai, P.F.; Choi, I.; Won, K.J.; Fan, H.Y. RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLoS Genet. 2014, 10, e1004204. [Google Scholar] [CrossRef]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef]
- Ezratty, E.J.; Stokes, N.; Chai, S.; Shah, A.S.; Williams, S.E.; Fuchs, E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 2011, 145, 1129–1141. [Google Scholar] [CrossRef]
- Okuyama, R.; Nguyen, B.C.; Talora, C.; Ogawa, E.; Tommasi di Vignano, A.; Lioumi, M.; Chiorino, G.; Tagami, H.; Woo, M.; Dotto, G.P. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism. Dev. Cell 2004, 6, 551–562. [Google Scholar] [CrossRef]
- Kessler, M.; Hoffmann, K.; Brinkmann, V.; Thieck, O.; Jackisch, S.; Toelle, B.; Berger, H.; Mollenkopf, H.J.; Mangler, M.; Sehouli, J.; et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015, 6, 8989. [Google Scholar] [CrossRef] [PubMed]
- Stasiulewicz, M.; Gray, S.D.; Mastromina, I.; Silva, J.C.; Björklund, M.; Seymour, P.A.; Booth, D.; Thompson, C.; Green, R.J.; Hall, E.A.; et al. A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development 2015, 142, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Ultrastructural characterization of primary cilia in pathologically characterized human glioblastoma multiforme (GBM) tumors. BMC Clin. Pathol. 2014, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Kiseleva, A.A.; Nikonova, A.S.; Golemis, E.A. Patterns of Ciliation and Ciliary Signaling in Cancer. Rev. Physiol. Biochem. Pharmacol. 2020. [Google Scholar] [CrossRef]
- Eguether, T.; Hahne, M. Mixed signals from the cell’s antennae: Primary cilia in cancer. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Doornbos, C.; Roepman, R. Moonlighting of mitotic regulators in cilium disassembly. Cell Mol. Life Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Coelho, P.A.; Bury, L.; Shahbazi, M.N.; Liakath-Ali, K.; Tate, P.H.; Wormald, S.; Hindley, C.J.; Huch, M.; Archer, J.; Skarnes, W.C.; et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015, 5, 150209. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Han, Y.G.; Kim, H.J.; Dlugosz, A.A.; Ellison, D.W.; Gilbertson, R.J.; Alvarez-Buylla, A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009, 15, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Bahmad, H.F.; Poppiti, R.J. Medulloblastoma cancer stem cells: Molecular signatures and therapeutic targets. J. Clin. Pathol. 2020, 73, 243–249. [Google Scholar] [CrossRef]
- Kim, J.Y.; Nelson, A.L.; Algon, S.A.; Graves, O.; Sturla, L.M.; Goumnerova, L.C.; Rowitch, D.H.; Segal, R.A.; Pomeroy, S.L. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev. Biol. 2003, 263, 50–66. [Google Scholar] [CrossRef]
- Santos, N.; Reiter, J.F. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J. Cell Sci. 2014, 127, 1500–1510. [Google Scholar] [CrossRef]
- Gate, D.; Danielpour, M.; Bannykh, S.; Town, T. Characterization of cancer stem cells and primary cilia in medulloblastoma. CNS Neurol. Disord. Drug Targets 2015, 14, 600–611. [Google Scholar] [CrossRef]
- Read, T.A.; Fogarty, M.P.; Markant, S.L.; McLendon, R.E.; Wei, Z.; Ellison, D.W.; Febbo, P.G.; Wechsler-Reya, R.J. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell 2009, 15, 135–147. [Google Scholar] [CrossRef]
- Shimada, I.S.; Hwang, S.H.; Somatilaka, B.N.; Wang, X.; Skowron, P.; Kim, J.; Kim, M.; Shelton, J.M.; Rajaram, V.; Xuan, Z.; et al. Basal Suppression of the Sonic Hedgehog Pathway by the G-Protein-Coupled Receptor Gpr161 Restricts Medulloblastoma Pathogenesis. Cell Rep. 2018, 22, 1169–1184. [Google Scholar] [CrossRef]
- Fu, W.; Asp, P.; Canter, B.; Dynlacht, B.D. Primary cilia control hedgehog signaling during muscle differentiation and are deregulated in rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 2014, 111, 9151–9156. [Google Scholar] [CrossRef] [PubMed]
- Hoang-Minh, L.B.; Deleyrolle, L.P.; Siebzehnrubl, D.; Ugartemendia, G.; Futch, H.; Griffith, B.; Breunig, J.J.; De Leon, G.; Mitchell, D.A.; Semple-Rowland, S.; et al. Disruption of KIF3A in patient-derived glioblastoma cells: Effects on ciliogenesis, hedgehog sensitivity, and tumorigenesis. Oncotarget 2016, 7, 7029–7043. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Satta, M.; Moreno-Cugnon, L.; Matheu, A. Primary cilium and brain aging: Role in neural stem cells, neurodegenerative diseases and glioblastoma. Ageing Res. Rev. 2019, 52, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Goranci-Buzhala, G.; Mariappan, A.; Ricci-Vitiani, L.; Josipovic, N.; Pacion, S.; Gottardo, M.; Ptok, J.; Callaini, G.; Rajalingam, K.; Dynlacht, B.; et al. Cilium induction triggers differentiation of glioma stem cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yuan, K.; Frolova, N.; Xie, Y.; Wang, D.; Cook, L.; Kwon, Y.J.; Steg, A.D.; Serra, R.; Frost, A.R. Primary cilia are decreased in breast cancer: Analysis of a collection of human breast cancer cell lines and tissues. J. Histochem. Cytochem. 2010, 58, 857–870. [Google Scholar] [CrossRef]
- Menzl, I.; Lebeau, L.; Pandey, R.; Hassounah, N.B.; Li, F.W.; Nagle, R.; Weihs, K.; McDermott, K.M. Loss of primary cilia occurs early in breast cancer development. Cilia 2014, 3, 7. [Google Scholar] [CrossRef]
- Hassounah, N.B.; Nunez, M.; Fordyce, C.; Roe, D.; Nagle, R.; Bunch, T.; McDermott, K.M. Inhibition of Ciliogenesis Promotes Hedgehog Signaling, Tumorigenesis, and Metastasis in Breast Cancer. Mol. Cancer Res. 2017, 15, 1421–1430. [Google Scholar] [CrossRef]
- García-Zaragoza, E.; Pérez-Tavarez, R.; Ballester, A.; Lafarga, V.; Jiménez-Reinoso, A.; Ramírez, A.; Murillas, R.; Gallego, M.I. Intraepithelial paracrine Hedgehog signaling induces the expansion of ciliated cells that express diverse progenitor cell markers in the basal epithelium of the mouse mammary gland. Dev. Biol. 2012, 372, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Gires, O.; Zhu, L.; Liu, J.; Li, J.; Yang, H.; Ju, G.; Huang, J.; Ge, W.; Chen, Y.; et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat. Commun. 2019, 10, 2863. [Google Scholar] [CrossRef]
- Jeng, K.S.; Chang, C.F.; Lin, S.S. Sonic Hedgehog Signaling in Organogenesis, Tumors, and Tumor Microenvironments. Int. J. Mol. Sci. 2020, 21, 758. [Google Scholar] [CrossRef]
- Guen, V.J.; Chavarria, T.E.; Kröger, C.; Ye, X.; Weinberg, R.A.; Lees, J.A. EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E10532–E10539. [Google Scholar] [CrossRef]
- Kuonen, F.; Huskey, N.E.; Shankar, G.; Jaju, P.; Whitson, R.J.; Rieger, K.E.; Atwood, S.X.; Sarin, K.Y.; Oro, A.E. Loss of Primary Cilia Drives Switching from Hedgehog to Ras/MAPK Pathway in Resistant Basal Cell Carcinoma. J. Investig. Dermatol. 2019, 139, 1439–1448. [Google Scholar] [CrossRef]
- Chippalkatti, R.; Abankwa, D. Promotion of cancer cell stemness by Ras. Biochem. Soc. Trans. 2021, 49, 467–476. [Google Scholar] [CrossRef]
- Vézina, A.; Vaillancourt-Jean, E.; Albarao, S.; Annabi, B. Mesenchymal stromal cell ciliogenesis is abrogated in response to tumor necrosis factor-α and requires NF-κB signaling. Cancer Lett. 2014, 345, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Proulx-Bonneau, S.; Annabi, B. The primary cilium as a biomarker in the hypoxic adaptation of bone marrow-derived mesenchymal stromal cells: A role for the secreted frizzled-related proteins. Biomark. Insights 2011, 6, 107–118. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.; Wang, B.; Xu, Y.; Yang, J.; Zhang, X.; Harten, S.K.; Shukla, D.; Maxwell, P.H.; Pei, D.; et al. VHL inactivation induces HEF1 and Aurora kinase A. J. Am. Soc. Nephrol. 2010, 21, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Hasanov, E.; Chen, G.; Chowdhury, P.; Weldon, J.; Ding, Z.; Jonasch, E.; Sen, S.; Walker, C.L.; Dere, R. Ubiquitination and regulation of AURKA identifies a hypoxia-independent E3 ligase activity of VHL. Oncogene 2017, 36, 3450–3463. [Google Scholar] [CrossRef]
- Carpino, G.; Cardinale, V.; Folseraas, T.; Overi, D.; Grzyb, K.; Costantini, D.; Berloco, P.B.; Di Matteo, S.; Karlsen, T.H.; Alvaro, D.; et al. Neoplastic Transformation of the Peribiliary Stem Cell Niche in Cholangiocarcinoma Arisen in Primary Sclerosing Cholangitis. Hepatology 2019, 69, 622–638. [Google Scholar] [CrossRef]
- Zingg, D.; Debbache, J.; Peña-Hernández, R.; Antunes, A.T.; Schaefer, S.M.; Cheng, P.F.; Zimmerli, D.; Haeusel, J.; Calçada, R.R.; Tuncer, E.; et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 2018, 34, 69–84.e14. [Google Scholar] [CrossRef]
- Kim, E.; Kim, M.; Woo, D.H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef]
- Singer, D.; Thamm, K.; Zhuang, H.; Karbanová, J.; Gao, Y.; Walker, J.V.; Jin, H.; Wu, X.; Coveney, C.R.; Marangoni, P.; et al. Prominin-1 controls stem cell activation by orchestrating ciliary dynamics. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Jászai, J.; Thamm, K.; Karbanová, J.; Janich, P.; Fargeas, C.A.; Huttner, W.B.; Corbeil, D. Prominins control ciliary length throughout the animal kingdom: New lessons from human prominin-1 and zebrafish prominin-3. J. Biol. Chem. 2020, 295, 6007–6022. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanardag, S.; Pugacheva, E.N. Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways. Cells 2021, 10, 1428. https://doi.org/10.3390/cells10061428
Yanardag S, Pugacheva EN. Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways. Cells. 2021; 10(6):1428. https://doi.org/10.3390/cells10061428
Chicago/Turabian StyleYanardag, Sila, and Elena N. Pugacheva. 2021. "Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways" Cells 10, no. 6: 1428. https://doi.org/10.3390/cells10061428
APA StyleYanardag, S., & Pugacheva, E. N. (2021). Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways. Cells, 10(6), 1428. https://doi.org/10.3390/cells10061428





