Abstract
Coronavirus disease 2019 (COVID-19), a global pandemic, is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Angiotensin-converting enzyme 2 (ACE2) is the receptor for SARS-CoV-2 and transmembrane serine protease 2 (TMPRSS2) facilitates ACE2-mediated virus entry. Moreover, the expression of ACE2 in the testes of infertile men is higher than normal, which indicates that infertile men may be susceptible to be infected and SARS-CoV-2 may cause reproductive disorder through the pathway induced by ACE2 and TMPRSS2. Little is known about the pathway regulation of ACE2 and TMPRSS2 expression in male reproductive disorder. Since the regulation of gene expression is mediated by microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) at the post-transcriptional level, the aim of this study was to analyze the dysregulated miRNA–lncRNA interactions of ACE2 and TMPRSS2 in male reproductive disorder. Using bioinformatics analysis, we speculate that the predicted miRNAs including miR-125a-5p, miR-125b-5p, miR-574-5p, and miR-936 as regulators of ACE2 and miR-204-5p as a modulator of TMPRSS2 are associated with male infertility. The lncRNAs with a tissue-specific expression for testis including GRM7-AS3, ARHGAP26-AS1, BSN-AS1, KRBOX1-AS1, CACNA1C-IT3, AC012361.1, FGF14-IT1, AC012494.1, and GS1-24F4.2 were predicted. The identified miRNAs and lncRNAs are proposed as potential biomarkers to study the possible association between COVID-19 and male infertility. This study encourages further studies of miRNA–lncRNA interactions to explain the molecular mechanisms of male infertility in COVID-19 patients.
1. Introduction
In December 2019, a novel coronavirus responsible for coronavirus disease 2019 (COVID-19) was described in Wuhan, China [1,2]. To date, nearly 146,000,000 global confirmed cases of COVID-19, with nearly 3 million deaths, have been observed [3].
SARS-CoV-2, a single-stranded RNA virus belonging to the coronavirus subfamily, is one of the seven different coronaviruses that can infect humans and is the pathogen responsible for COVID-19 [4]. Four coronaviruses, 229E, NL63, OC43, and HKU1, can lead to mild viral symptoms, while the other three, SARS-CoV-1, MERS-CoV, and SARS-CoV-2, can cause more severe respiratory symptoms [4].
The angiotensin-converting enzyme 2 (ACE2) receptor and transmembrane serine protease 2 (TMPRSS2), localized on the host cell membrane, are indispensable for viral proliferation in the infected host [5].
ACE2 and TMPRSS2 are highly expressed in normal human tissues, such as the lung, heart, colon, and testis [6,7]. Overall, the epidemiological findings reported that males are more vulnerable to the infection than females [8]. However, it seems there is not a clear association between the localization of receptors and virus infectivity.
ACE2 and TMPRSS2 are androgen-regulated and expressed in both germ cells and somatic cells [9,10,11].
Previous studies measured hormonal levels in COVID-19-infected male patients and they found a lower expression of testosterone levels and higher luteinizing hormone levels, key mediators in male reproductive health [12,13].
Li et al. indicated that SARS-CoV-2 can be present in the semen of patients with COVID-19 [14]. Ma and colleagues confirmed the nucleic acid sequence of SARS-CoV-2 using RT-qPCR and the presence of the virus by immunohistochemistry in the testes of COVID-19 patients. Furthermore, they demonstrated a higher expression of ACE2 and TMPRSS2 in the testes of infertile men than normal [15]. The SARS-CoV-2 spike protein binds to the ACE2 receptor of the target cells and TMPRSS2 primes cellular protease to cleave the S protein into S1 and S2 subunits. The two subunits have two different roles: S1 is the domain where there is the binding with the ACE2 receptor and S2 is responsible for the fusion with the target cell membrane [5]. Collectively, the previous findings indicate that men with reproductive disorders may be easily infected by SARS-CoV-2 and this virus may cause male reproductive disorders through the pathway activated by ACE2 and TMPRSS2 [16,17].
Despite these recent studies, there are currently no biomarkers able to establish the effects of COVID-19 and elucidate the molecular mechanisms associated with male infertility. Therefore, non-invasive approaches that can diagnose the effects of COVID-19 on male infertility are appealing aspects [18].
The regulation of gene expression is mediated by micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs) at the post-transcriptional level in multiple molecular mechanisms. miRNAs, small non-coding RNAs, are important regulators of gene expression through binding to the 3′ untranslated region of their complementary mRNA sequences and lead to their degradation or inhibition of translation [19,20,21]. Previous studies reported the significant role of miRNAs in spermatogenesis and testicular development [22,23]. As miRNAs are abundant in plasma, serum, and seminal plasma, it makes them appealing potential non-invasive biomarkers [24]. Indeed, miRNAs can modify the host’s transcriptome and modulate viral infection through the regulation of biological pathways with pro- or antiviral effects [25].
lncRNAs, sequences of RNA longer than 200 nucleotides that are not translated into proteins [26], can sponge miRNAs to moderate their regulatory effect on mRNAs; the association between lncRNAs and miRNAs is essential for gene regulation [27]. Various lncRNAs are involved in modulating mammalian spermatogenesis [28]. Changes in miRNA and lncRNA expression profiles in patients with non-obstructive azoospermia supported their role in male infertility [20]. In line with this scenario, miRNAs and lncRNAs could also have potential therapeutic applications. Nevertheless, the molecular mechanism of miRNAs and lncRNAs on the course of male infertility associated with COVID-19 remains poorly understood.
In this study, the expression profiles of miRNAs and lncRNAs which regulate ACE2 “the hottest targets of SARS-CoV-2” and TMPRSS2 “S protein priming” were analyzed comprehensively in infertile men by bioinformatics approaches. We proposed potential biomarkers which might help to understand the effects of COVID-19 on male infertility.
2. Materials and Methods
2.1. Predicting the Interactions miRNA–ACE2 and miRNA–TMPRSS2
The miRNAs that target ACE2 and TMPRSS2 were predicted by mirDIP database Version 4.1.0.3 [29]. This integrated database covers 30 datasets including TargetScan, RNAhybrid, mirTar, mirbase, DIANA, etc. [29]. The score class was limited to high and very high interactions.
2.2. Screening the miRNAs Associated with Male Infertility
In order to filter the retrieved miRNAs associated with male infertility, differentially expressed miRNAs (DE-miRNAs) in human testes from infertile men were obtained from the study of Abu et al. [30]. |Log FC| > 3 and p value < 0.05 were considered statistically significant to identify differentially expressed miRNAs.
2.3. Predicting the Interactions lncRNAs–miRNAs
The identified DE-miRNAs were submitted to miRWalk2 [31] to predict the association between miRNA and lncRNA.
2.4. Filtering the lncRNAs Associated with Male Infertility
The predicted lncRNAs in the last step were screened according to lncRNAs associated with male infertility that have been identified by Joshi and Lu [32,33]. |Log FC| > 3 and p value < 0.05 were considered statistically significant to identify differentially expressed lncRNAs. The computational procedure of our study is represented in Figure 1. miRNAs able to regulate ACE2 and TMPRSS2 and associated with male infertility were identified through mirDIP and the study of Abu et al. [30].
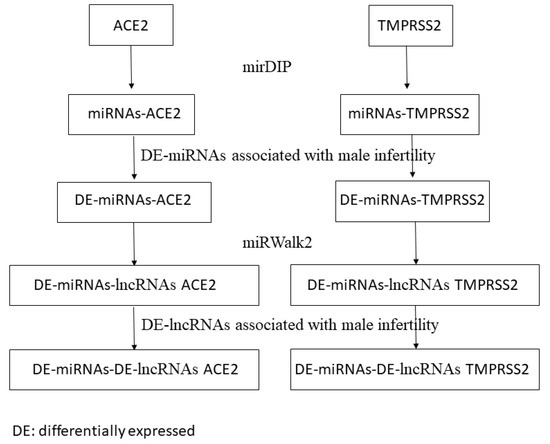
Figure 1.
Workflow of the proposed computational approach. DE: differentially expressed in infertile men; DE: differentially expressed.
In addition, we identified the interactions between these miRNAs and differentially expressed lncRNAs in infertile men identified by Joshi and Lu [32,33].
2.5. Gene Ontology Analysis
Gene Ontology (GO) and panther version 16.0 were used to perform functional classification of identified lncRNAs in categories such as molecular function, biological process and cellular component [34].
2.6. Testis-Specific lncRNAs
The Gini index was used to explore testis-specific lncRNAs. The Gini index was calculated to evaluate the specificity of expression of lncRNAs compared to different healthy tissues. We used the public database GTEX that contains the expression levels in 30 human normal tissues: adipose tissue, adrenal gland, bladder, blood, blood vessel, brain, breast, cervix uteri, colon, esophagus, fallopian tube, heart, kidney, liver, lung, muscle, nerve, ovary, pancreas, pituitary, prostate, salivary gland, skin, small intestine, spleen, stomach, testis, thyroid, uterus, and vagina [35]. We analyzed the Gini index for each normal tissue and quantified the specificity of lncRNAs for testis. It can assume a value from 0 to 1. We defined a lncRNA specific for testis if the Gini index is <= 0.15. We considered 10167 lncRNAs in GTEX data as reported from the lncRNome database [36].
3. Results
3.1. miRNAs Associated with Male Infertility Regulate ACE2 and TMPRSS2
From our bioinformatics analysis we predicted that 80 miRNAs and 92 miRNAs can regulate ACE2 and TMPRSS2, respectively. Ten miRNAs (miR-1208, miR-141-3p, miR-182-5p, miR-300, miR-331-3p, miR-362-5p, miR-381-3p, miR-4308, miR-582-5p and miR-587) were found in common between ACE2 and TMPRSS2.
Furthermore, we explored the miRNAs that regulate ACE2 and TMPRSS2 and are associated with male infertility according to the study of Abu et al. [30]. Four miRNAs (miR-125a-5p, miR-125b-5p, miR-574-5p, and miR-936) were associated with male infertility and were also predicted to modulate ACE2 expression. miR-204-5p, with an effective role in male infertility, was presented as a possible regulator of TMPRSS2. Supplementary File 1 shows the list of all predicted miRNAs as regulators of ACE2 and TMPRSS2, the miRNAs associated with male infertility, and the extracted miRNAs from the predicted ones which were also associated with male infertility.
3.2. lncRNAs–miRNAs Associated with Male Infertility
A total of 5612 lncRNAs were predicted to interact with miR-125a-5p and miR-125b-5p, and 3416 lncRNAs were predicted to interact with miR-936. miR-204-5p was proposed to have interactions with 6569 lncRNAs.
We extracted 349 unique lncRNAs which were associated with male infertility. A total of 155 lncRNAs have interactions with miR-125a-5p and miR-125b-5p, 122 lncRNAs with miR-936, and 187 lncRNAs with miR-204-5p. Supplementary File 2 shows the list of 349 lncRNAs.
3.3. Functional Annotations
The biological role of 349 lncRNAs was evaluated through a functional analysis. From the 349 unique lncRNAs, 323 lncRNAs were annotated by PANTHER and functional analysis of the lncRNAs showed that the top molecular functions (Figure 2a) were “bindings” (40.5% of lncRNAs) and “catalytic activity” (28.6% of lncRNAs); the top biological processes (Figure 2b) were “cellular process” (31% of lncRNAs) and “biological regulation” (16.9% of lncRNAs); and the top cellular components (Figure 2c) were “cellular anatomical entity” (50% of lncRNAs) and “intracellular” (39.2% of lncRNAs).
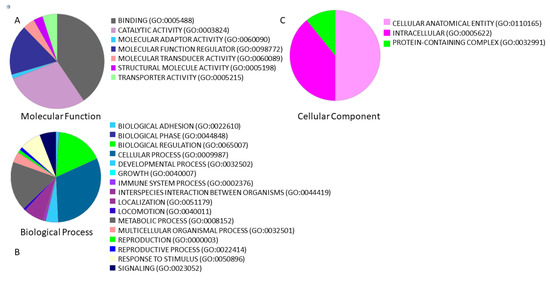
Figure 2.
Functional classification using PANTHER of 349 lncRNAs associated with male infertility and interacting with miR-125a-5p, miR-125b-5p, miR-936 and miR-204-5p. Overall, we found 349 unique lncRNAs. The percentage of lncRNAs involved in (A) molecular function, (B) biological processes, and (C) cellular component is represented by a pie chart.
Biological process analysis demonstrated that the identified lncRNAs were involved in the immune system and reproductive process.
3.4. Possible Roles of lncRNAs in Male Reproductive Disorder Associated with COVID-19
In order to investigate lncRNAs which could play a regulatory role in COVID-19, and especially its male reproductive system consequences, we performed an analysis to select testis-specific lncRNAs. We found that the expression levels of 9 of 349 lncRNAs (GRM7-AS3, ARHGAP26-AS1, BSN-AS1, KRBOX1-AS1, CACNA1C-IT3, AC012361.1, FGF14-IT1, AC012494.1, and GS1-24F4.2) were specific for testis tissue, demonstrating their possible crucial role in the male reproductive system.
GRM7-AS3, ARHGAP26-AS1, BSN-AS1, and KRBOX1-AS1 interact with miR-936, and CACNA1C-IT3, AC012361.1, FGF14-IT1, AC012494.1, and GS1-24F4.2 interact with miR-204-5p.
4. Discussion
In this study, we analyzed the potential association of the dysregulated miRNAs and lncRNAs in male infertility and their association with ACE2 and TMPRSS2. First, we studied miRNAs that could regulate two crucial genes for viral proliferation in the infected host, ACE2 and TMPRSS2. Furthermore, we selected those miRNAs that regulate ACE2 and TMPRSS2 and are associated with male infertility. We identified four miRNAs, miR-125a-5p, miR-125b-5p, miR-574–5p, and miR-936, that regulate ACE2 and are differentially expressed in infertile men. TMPRSS2 is regulated by miR-204-5p, a differentially expressed miRNA in infertile men.
A previous study demonstrated a potential role of miR-125b-5p in hepatitis B virus (HBV) and COVID-19. Indeed, miR-125b-5p and HBV DNA levels were positively associated, demonstrating its role in HBV infection [37]. Regarding COVID-19, miR-125b-5p could modify the risk of SARS-CoV-2 infection in lung cancer patients [38].
miR-125a-5p and miR-125b-5p were found to be over-expressed in Sertoli cells, and in the epididymis of fertile men [39,40]. Salas et al. analyzed miRNA profiles of patients with teratozoospermia and oligozoospermia and miR-125a-3p was found to be downregulated in both the conditions [41]. In addition, previous studies reported a potential interaction between ACE2 and histone deacetylase, HDAC2, suggesting a regulatory network that involves miR-125a–ACE2–HDAC2 [42]. Although the role of HDAC2 in spermatogenesis is not completely understood, previous studies demonstrated that genes regulated by HDAC2 are involved in spermatogonial stem cells [43].
In our study, miR-574–5p was found to be down-expressed in Sertoli cells of infertile men. We can hypothesize that an increase in ACE2 expression reported in infertile men with COVID-19 can be due to the downregulation of miR-574–5p [44]. In addition, miR-574–5p was also proposed as an agent with antiviral activity in HBV, downregulating the expression of HBV polymerase mRNA [45]. A possible mechanism that reinforces its possible role in COVID-19 treatment is reported by a recent study that showed that the upregulation of miR-574–5p inhibits TLR4/ NF-kB signaling and downregulates the production of proinflammatory cytokines in patients with acute respiratory distress syndrome [46]. In line with this scenario, miR-574–5p could reduce the cytokine storm, one of the most common causes of death in patients with COVID-19 [46]. Proinflammatory cytokines are important regulators of testis development and male fertility, suggesting a complex regulatory network of miR-574–5p–ACE2–TLR4/NF-kB–cytokines.
miR-936 is indicated as a regulator of ACE2 in placentas [47]. In addition, a previous study showed that a fibroblast growth factor, FGF2, is a direct target of miR-936 [48]. In a previous study on Zika virus, the inhibition of FGF2 affected viral replication through the inhibition of the MAPK pathway, which is associated with normal FGF/FGFR activity [49]. FGF2 seems to also play a crucial role in male reproductive tissues [50]. In addition, several studies demonstrated the positive correlation between FGF2 and angiotensin, suggesting a small regulatory circuit that involves miR-936–ACE2–FGF2 in COVID-19 and male infertility [51].
In our study, we found that TMPRSS2 is regulated by miR-204-5p. miR-204 was found to be over-expressed in prostate cancer cell lines compared to human prostate tissue. In prostate cancer, different genomic rearrangements can occur, such as the most common fusion of androgen receptor TMPRSS2 with ERG. miR-204 is a TMPRSS2/ERG oncofusion negative regulator and can act as a tumor suppressor or oncomiR, regulating the genes under androgen receptor control [52]. In addition, miR−204b−5p was found to be abundant in the spermatozoa of the epididymis, suggesting its crucial role in the male reproductive system [53].
Furthermore, we explored the interactions between differentially expressed lncRNAs in infertile men and the miRNAs reported above. From this analysis, we identified 349 lncRNAs as potential biomarkers explaining the potential effects of COVID-19 on the male reproductive system. The role of 349 lncRNAs was analyzed with a pathway analysis and we found that they are involved mainly in regulation and binding.
In order to define a selected number of lncRNAs, we evaluated the testis-specific lncRNAs in the 349 lncRNAs. We found that 9 out of 349 lncRNAs were testis specific: GRM7-AS3, ARHGAP26-AS1, BSN-AS1, and KRBOX1-AS1 interacting with miR-936, and CACNA1C-IT3, AC012361.1, FGF14-IT1, AC012494.1, and GS1-24F4.2 interacting with miR-204-5p. Little is known about the role of these lncRNAs. GRM7-AS3 and KRBOX1-AS1 are more known and studied in the literature.
GRM7-AS3 is complementary to a functional RNA, GRM7. Currently, there are no studies that reported an association between GRM7-AS3, COVID-19 and the male reproductive system. A previous study reported that GRM7 plays a role in neurologic diseases such as depression, epilepsy and bipolar disorder, regulating synaptic activity [54].
KRBOX1-AS1 has also been correlated with programmed cell death and cell proliferation in rectal cancer [55]. Although KRBOX1 was found to be over-expressed in testis by a previous study, its role has not been investigated [56].
As these lncRNAs were found to be differentially expressed in infertile men and their expression in healthy men was testis-specific, we proposed these lncRNAs as biomarkers that could explain the association between COVID-19 and male reproductive disorder. miRNAs interacting with these lncRNAs (miR-936 and miR-204-5p) could also play an important role in this molecular mechanism.
Overall, this study predicted the interactions of DE-miRNAs and DE-lncRNAs from infertile men with ACE2 and TMPRSS2 using bioinformatics approaches. We assumed the proposed miRNAs and lncRNAs can be potential biomarkers to examine the effect of SARS-CoV-2 on testis and spermatogenesis damages. However, further experimental study is required to confirm the current findings through comparing the infertile men and normal cases after COVID-19 infection.
5. Conclusions
miRNAs and lncRNAs are involved in various mechanisms of COVID-19 infection and male infertility, but their roles are not fully understood. The present study proposed the miRNAs and lncRNAs as possible diagnostic tools regarding the pathogenic role of SARS-CoV-2 in male infertility.
The miRNAs, including miR-125a-5p, miR-125b-5p, miR-574–5p, miR-936 and miR-204-5p, and the associated lncRNAs, including GRM7-AS3, ARHGAP26-AS1, BSN-AS1, KRBOX1-AS1, CACNA1C-IT3, AC012361.1, FGF14-IT1, AC012494.1, and GS1-24F4.2, could shed a light on possible diagnostic applications in male infertility after SARS-CoV-2 infection. Further studies are encouraged to validate the current findings by comparing normal and infertile men after infection with COVID-19.
Supplementary Materials
The supplementary files are available online at https://www.mdpi.com/article/10.3390/cells10061480/s1. Supplementary File 1 shows the list of gene targets of ACE2 and TMPRSS2. Supplementary File 2 shows the list of 349 lncRNAs associated with male infertility and interacting with miRNAs associated with COVID-19 and male infertility.
Author Contributions
Conceptualization, S.S. and C.C.; methodology, S.S. and C.C.; software, S.S. and C.C.; validation, S.S. and C.C.; resources, I.C.; writing—original draft preparation, S.S., C.C., P.M., and B.N.J.; supervision, I.C.; funding acquisition, I.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The Medical Ethics Committee of Shiraz University of Medical Sciences approved the study protocol (Permission number: IR.SUMS.REC.1399.032).
Data Availability Statement
Data supporting reported results can be found in publicly archived datasets: https://gtexportal.org/home/.
Acknowledgments
This study was conducted with support from Shiraz University of Medical Sciences, Vice-Chancellor of Research and Technology with the grant number of 98-01-106-22068. We would like to thank the project Grant SysBioNet, Italian Roadmap Research Infrastructures 2012 for the financial support.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Cava, C.; Bertoli, G.; Castiglioni, I. A protein interaction map identifies existing drugs targeting SARS-CoV-2. BMC Pharmacol. Toxicol. 2020, 21, 65. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://covid19.who.int/ (accessed on 25 April 2021).
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhou, J.; Fu, S.; Fu, J.; Zhou, B.; Chen, H.; Fu, J.; Wei, C. Prostate adenocarcinoma and COVID-19: The possible impacts of TMPRSS2 expressions in susceptibility to SARS-CoV-2. J. Cell. Mol. Med. 2021, 25, 4157–4165. [Google Scholar] [CrossRef]
- Bwire, G.M. Coronavirus: Why Men are More Vulnerable to Covid-19 Than Women? SN Compr. Clin. Med. 2020, 2, 874–876. [Google Scholar] [CrossRef]
- Ruan, Y.; Hu, B.; Liu, Z.; Liu, K.; Jiang, H.; Li, H.; Li, R.; Luan, Y.; Liu, X.; Yu, G.; et al. No detection of SARS-CoV-2 from urine, expressed prostatic secretions, and semen in 74 recovered COVID-19 male patients: A perspective and urogenital evaluation. Andrology 2021, 9, 99–106. [Google Scholar] [CrossRef]
- Pan, F.; Xiao, X.; Guo, J.; Song, Y.; Li, H.; Patel, D.P.; Spivak, A.M.; Alukal, J.; Zhang, X.; Xiong, C.; et al. No evidence of severe acute respiratory syndrome–coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil. Steril. 2020, 113, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 2020, 27, 876–889.e12. [Google Scholar] [CrossRef]
- Salonia, A.; Pontillo, M.; Capogrosso, P.; Gregori, S.; Tassara, M.; Boeri, L.; Carenzi, C.; Abbate, C.; Cignoli, D.; Ferrara, A.M.; et al. Severely low testosterone in males with COVID-19: A case-control study. Andrology 2021. [Google Scholar] [CrossRef]
- Li, D.; Jin, M.; Bao, P.; Zhao, W.; Zhang, S. Clinical Characteristics and Results of Semen Tests Among Men with Coronavirus Disease 2019. JAMA Netw. Open 2020, 3, e208292. [Google Scholar] [CrossRef]
- Ma, X.; Guan, C.; Chen, R.; Wang, Y.; Feng, S.; Wang, R.; Qu, G.; Zhao, S.; Wang, F.; Wang, X.; et al. Pathological and molecular examinations of postmortem testis biopsies reveal SARS-CoV-2 infection in the testis and spermatogenesis damage in COVID-19 patients. Cell. Mol. Immunol. 2021, 18, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xie, W.; Li, D.; Shi, L.; Ye, G.; Mao, Y.; Xiong, Y.; Sun, H.; Zheng, F.; Chen, Z.; et al. Evaluation of sex-related hormones and semen characteristics in reproductive-aged male COVID-19 patients. J. Med. Virol. 2021, 93, 456–462. [Google Scholar] [CrossRef]
- Shen, Q.; Xiao, X.; Aierken, A.; Yue, W.; Wu, X.; Liao, M.; Hua, J. The ACE2 expression in Sertoli cells and germ cells may cause male reproductive disorder after SARS-CoV-2 infection. J. Cell. Mol. Med. 2020, 24, 9472–9477. [Google Scholar] [CrossRef]
- Vashisht, A.; Gahlay, G.K. Using miRNAs as diagnostic biomarkers for male infertility: Opportunities and challenges. Mol. Hum. Reprod. 2020, 26, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long Non-coding RNAs and MicroRNAs Interplay in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Sabetian, S.; Zarei, M.; Jahromi, B.N.; Morowvat, M.H.; Tabei, S.M.B.; Cava, C. Exploring the dysregulated mRNAs–miRNAs–lncRNAs interactions associated to idiopathic non-obstructive azoospermia. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Qin, Y.; Li, Z.; Dong, J.; Dai, J.; Lu, C.; Guo, X.; Zhao, Y.; Zhu, Y.; Zhang, W.; et al. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: Significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum. Reprod. 2013, 28, 1827–1836. [Google Scholar] [CrossRef]
- Singaravelu, R.; O’Hara, S.; Jones, D.M.; Chen, R.; Taylor, N.G.; Srinivasan, P.; Quan, C.; Roy, D.; Steenbergen, R.H.; Kumar, A.; et al. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol. 2015, 11, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Delshad, E.; Shamsabadi, F.T.; Bahramian, S.; Mehravar, F.; Maghsoudi, H.; Shafiee, M. In silico identification of novel lncRNAs with a potential role in diagnosis of gastric cancer. J. Biomol. Struct. Dyn. 2019, 38, 1954–1962. [Google Scholar] [CrossRef]
- Sahlu, B.W.; Zhao, S.; Wang, X.; Umer, S.; Zou, H.; Huang, J.; Zhu, H. Long noncoding RNAs: New insights in modulating mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Backes, C.; Leidinger, P.; Keller, A.; Lubbad, A.M.; Hammadeh, M.; Meese, E. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil. Steril. 2014, 101, 78–86.e2. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk–Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef]
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 2020, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, D.; Wang, P.; Sun, W.; Xue, X.; Hu, Y.; Xie, C.; Ma, Y. RNA-Sequencing and Bioinformatics Analysis of Long Noncoding RNAs and mRNAs in the asthenozoospermia. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Cava, C.; Bertoli, G.; Castiglioni, I. Portrait of Tissue-Specific Coexpression Networks of Noncoding RNAs (miRNA and lncRNA) and mRNAs in Normal Tissues. Comput. Math. Methods Med. 2019, 2019, 9029351. [Google Scholar] [CrossRef]
- Bhartiya, D.; Pal, K.; Ghosh, S.; Kapoor, S.; Jalali, S.; Panwar, B.; Jain, S.; Sati, S.; Sengupta, S.; Sachidanandan, C.; et al. lncRNome: A comprehensive knowledgebase of human long noncoding RNAs. Database 2013, 2013, bat034. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, X.; Ma, Z.; Lin, Y.; Lu, M. MicroRNA-125b-5p mediates post-transcriptional regulation of hepatitis B virus replication via the LIN28B/let-7 axis. RNA Biol. 2017, 14, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Wu, J.; Deng, C.; Tan, J.; Liu, H.; Zhong, L. Lung adenocarcinoma patients have higher risk of SARS-CoV-2 infection. Aging 2021, 13, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Q.; Zhang, W.; Li, J.; Li, Z.; Tang, Z.; Li, Y.; Han, C.; Hall, S.H.; Zhang, Y. Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim. Biophys. Sin. 2010, 42, 145–153. [Google Scholar] [CrossRef]
- Hayashi, K.; Lopes, S.M.C.D.S.; Kaneda, M.; Tang, F.; Hajkova, P.; Lao, K.; O’Carroll, D.; Das, P.P.; Tarakhovsky, A.; Miska, E.A.; et al. MicroRNA Biogenesis Is Required for Mouse Primordial Germ Cell Development and Spermatogenesis. PLoS ONE 2008, 3, e1738. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Godo, A.; Grossmann, M.; Pons, M.C.; F-Fernández, S.; Garrido, N.; Anton, E. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil. Steril. 2015, 104, 591–601. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; I Nakaya, H. ACE2 Expression Is Increased in the Lungs of Patients with Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Kofman, A.E.; Huszar, J.M.; Payne, C.J. Transcriptional Analysis of Histone Deacetylase Family Members Reveal Similarities Between Differentiating and Aging Spermatogonial Stem Cells. Stem Cell Rev. Rep. 2013, 9, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Bozgeyik, I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene 2021, 27, 100831. [Google Scholar] [CrossRef]
- Wu, W.; Wu, D.; Yan, W.; Wang, Y.; You, J.; Wan, X.; Xi, D.; Luo, X.; Han, M.; Ning, Q. Interferon-Induced Macrophage-Derived Exosomes Mediate Antiviral Activity Against Hepatitis B Virus Through miR-574-5p. J. Infect. Dis. 2021, 223, 686–698. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhou, W.; Rui, Y.; Liu, L.; Chen, B.; Su, X. MicroRNA-574-5p Attenuates Acute Respiratory Distress Syndrome by Targeting HMGB1. Am. J. Respir. Cell Mol. Biol. 2021, 64, 196–207. [Google Scholar] [CrossRef]
- Wang, Y.; Lumbers, E.R.; Arthurs, A.; De Meaultsart, C.C.; Mathe, A.; Avery-Kiejda, K.A.; Roberts, C.; Pipkin, F.B.; Marques, F.Z.; Morris, B.J.; et al. Regulation of the human placental (pro)renin receptor-prorenin-angiotensin system by microRNAs. Mol. Hum. Reprod. 2018, 24, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, S.; Wu, S.; Ni, Y.; Pan, Z. MicroRNA-936 targets FGF2 to inhibit epithelial ovarian cancer aggressiveness by deactivating the PI3K/Akt pathway. OncoTargets Ther. 2019, 12, 5311–5322. [Google Scholar] [CrossRef] [PubMed]
- Limonta, D.; Jovel, J.; Kumar, A.; Lu, J.; Hou, S.; Airo, A.M.; Lopez-Orozco, J.; Wong, C.P.; Saito, L.; Branton, W.; et al. Fibroblast Growth Factor 2 Enhances Zika Virus Infection in Human Fetal Brain. J. Infect. Dis. 2019, 220, 1377–1387. [Google Scholar] [CrossRef]
- Saucedo, L.; Sobarzo, C.; Brukman, N.G.; Guidobaldi, H.A.; Lustig, L.; Giojalas, L.C.; Buffone, M.G.; Vazquez-Levin, M.H.; Marín-Briggiler, C. Involvement of fibroblast growth factor 2 (FGF2) and its receptors in the regulation of mouse sperm physiology. Reproduction 2018, 156, 163–172. [Google Scholar] [CrossRef]
- Tamura, M.; Matsuzuka, T. Angiotensin II Signaling. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Todorova, K.; Metodiev, M.V.; Metodieva, G.; Mincheff, M.; Fernández, N.; Hayrabedyan, S. Micro-RNA-204 Participates in TMPRSS2/ERG Regulation and Androgen Receptor Reprogramming in Prostate Cancer. Horm. Cancer 2017, 8, 28–48. [Google Scholar] [CrossRef]
- Belleannee, C.; Calvo, É.; Caballero, J.; Sullivan, R. Epididymosomes Convey Different Repertoires of MicroRNAs Throughout the Bovine Epididymis. Biol. Reprod. 2013, 89, 30. [Google Scholar] [CrossRef] [PubMed]
- Nho, K.; Ramanan, V.K.; Horgusluoglu, E.; Kim, S.; Inlow, M.H.; Risacher, S.L.; McDonald, B.C.; Farlow, M.R.; Foroud, T.M.; Gao, S.; et al. Comprehensive Gene- and Pathway-Based Analysis of Depressive Symptoms in Older Adults. J. Alzheimer’s Dis. 2015, 45, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Nevalainen, J.; Skarp, S.; Savolainen, E.-R.; Ryynänen, M.; Järvenpää, J. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J. Périnat. Med. 2017, 45, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Deng, H.; Chen, Q.; Wu, Q.; Li, X.; Jiang, S.; Wang, F.; Ye, F.; Ou, L.; Gao, H. Comprehensive Analysis of Differential Immunocyte Infiltration and Potential ceRNA Networks Involved in the Development of Atrial Fibrillation. BioMed Res. Int. 2020, 2020, 8021208. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).