In Silico Identification of miRNA–lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Predicting the Interactions miRNA–ACE2 and miRNA–TMPRSS2
2.2. Screening the miRNAs Associated with Male Infertility
2.3. Predicting the Interactions lncRNAs–miRNAs
2.4. Filtering the lncRNAs Associated with Male Infertility
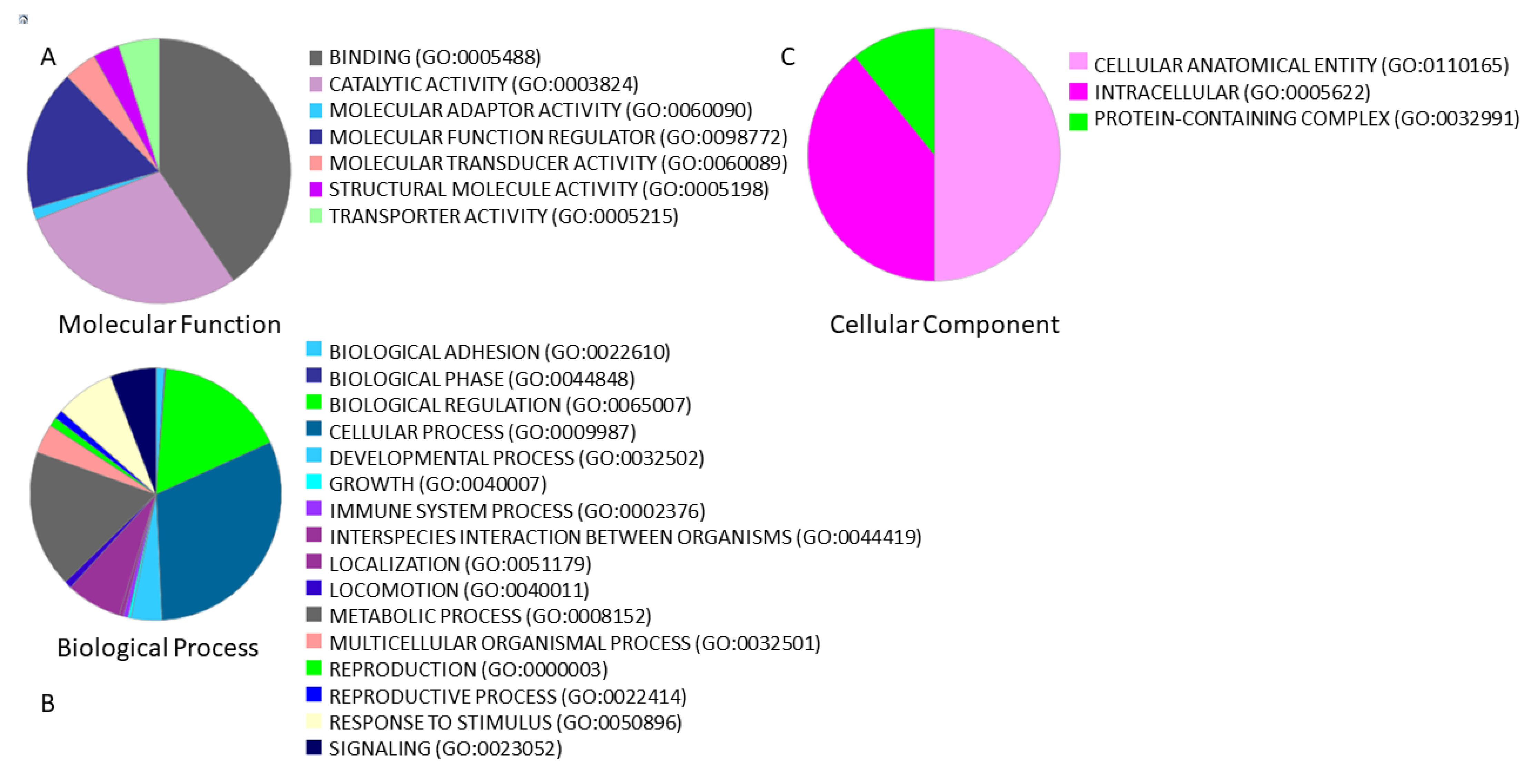
2.5. Gene Ontology Analysis
2.6. Testis-Specific lncRNAs
3. Results
3.1. miRNAs Associated with Male Infertility Regulate ACE2 and TMPRSS2
3.2. lncRNAs–miRNAs Associated with Male Infertility
3.3. Functional Annotations
3.4. Possible Roles of lncRNAs in Male Reproductive Disorder Associated with COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Cava, C.; Bertoli, G.; Castiglioni, I. A protein interaction map identifies existing drugs targeting SARS-CoV-2. BMC Pharmacol. Toxicol. 2020, 21, 65. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://covid19.who.int/ (accessed on 25 April 2021).
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Zhou, J.; Fu, S.; Fu, J.; Zhou, B.; Chen, H.; Fu, J.; Wei, C. Prostate adenocarcinoma and COVID-19: The possible impacts of TMPRSS2 expressions in susceptibility to SARS-CoV-2. J. Cell. Mol. Med. 2021, 25, 4157–4165. [Google Scholar] [CrossRef]
- Bwire, G.M. Coronavirus: Why Men are More Vulnerable to Covid-19 Than Women? SN Compr. Clin. Med. 2020, 2, 874–876. [Google Scholar] [CrossRef]
- Ruan, Y.; Hu, B.; Liu, Z.; Liu, K.; Jiang, H.; Li, H.; Li, R.; Luan, Y.; Liu, X.; Yu, G.; et al. No detection of SARS-CoV-2 from urine, expressed prostatic secretions, and semen in 74 recovered COVID-19 male patients: A perspective and urogenital evaluation. Andrology 2021, 9, 99–106. [Google Scholar] [CrossRef]
- Pan, F.; Xiao, X.; Guo, J.; Song, Y.; Li, H.; Patel, D.P.; Spivak, A.M.; Alukal, J.; Zhang, X.; Xiong, C.; et al. No evidence of severe acute respiratory syndrome–coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil. Steril. 2020, 113, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 2020, 27, 876–889.e12. [Google Scholar] [CrossRef]
- Salonia, A.; Pontillo, M.; Capogrosso, P.; Gregori, S.; Tassara, M.; Boeri, L.; Carenzi, C.; Abbate, C.; Cignoli, D.; Ferrara, A.M.; et al. Severely low testosterone in males with COVID-19: A case-control study. Andrology 2021. [Google Scholar] [CrossRef]
- Li, D.; Jin, M.; Bao, P.; Zhao, W.; Zhang, S. Clinical Characteristics and Results of Semen Tests Among Men with Coronavirus Disease 2019. JAMA Netw. Open 2020, 3, e208292. [Google Scholar] [CrossRef]
- Ma, X.; Guan, C.; Chen, R.; Wang, Y.; Feng, S.; Wang, R.; Qu, G.; Zhao, S.; Wang, F.; Wang, X.; et al. Pathological and molecular examinations of postmortem testis biopsies reveal SARS-CoV-2 infection in the testis and spermatogenesis damage in COVID-19 patients. Cell. Mol. Immunol. 2021, 18, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xie, W.; Li, D.; Shi, L.; Ye, G.; Mao, Y.; Xiong, Y.; Sun, H.; Zheng, F.; Chen, Z.; et al. Evaluation of sex-related hormones and semen characteristics in reproductive-aged male COVID-19 patients. J. Med. Virol. 2021, 93, 456–462. [Google Scholar] [CrossRef]
- Shen, Q.; Xiao, X.; Aierken, A.; Yue, W.; Wu, X.; Liao, M.; Hua, J. The ACE2 expression in Sertoli cells and germ cells may cause male reproductive disorder after SARS-CoV-2 infection. J. Cell. Mol. Med. 2020, 24, 9472–9477. [Google Scholar] [CrossRef]
- Vashisht, A.; Gahlay, G.K. Using miRNAs as diagnostic biomarkers for male infertility: Opportunities and challenges. Mol. Hum. Reprod. 2020, 26, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long Non-coding RNAs and MicroRNAs Interplay in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabetian, S.; Zarei, M.; Jahromi, B.N.; Morowvat, M.H.; Tabei, S.M.B.; Cava, C. Exploring the dysregulated mRNAs–miRNAs–lncRNAs interactions associated to idiopathic non-obstructive azoospermia. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Qin, Y.; Li, Z.; Dong, J.; Dai, J.; Lu, C.; Guo, X.; Zhao, Y.; Zhu, Y.; Zhang, W.; et al. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: Significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum. Reprod. 2013, 28, 1827–1836. [Google Scholar] [CrossRef]
- Singaravelu, R.; O’Hara, S.; Jones, D.M.; Chen, R.; Taylor, N.G.; Srinivasan, P.; Quan, C.; Roy, D.; Steenbergen, R.H.; Kumar, A.; et al. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol. 2015, 11, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Delshad, E.; Shamsabadi, F.T.; Bahramian, S.; Mehravar, F.; Maghsoudi, H.; Shafiee, M. In silico identification of novel lncRNAs with a potential role in diagnosis of gastric cancer. J. Biomol. Struct. Dyn. 2019, 38, 1954–1962. [Google Scholar] [CrossRef]
- Sahlu, B.W.; Zhao, S.; Wang, X.; Umer, S.; Zou, H.; Huang, J.; Zhu, H. Long noncoding RNAs: New insights in modulating mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Backes, C.; Leidinger, P.; Keller, A.; Lubbad, A.M.; Hammadeh, M.; Meese, E. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil. Steril. 2014, 101, 78–86.e2. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk–Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [Green Version]
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 2020, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, D.; Wang, P.; Sun, W.; Xue, X.; Hu, Y.; Xie, C.; Ma, Y. RNA-Sequencing and Bioinformatics Analysis of Long Noncoding RNAs and mRNAs in the asthenozoospermia. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Cava, C.; Bertoli, G.; Castiglioni, I. Portrait of Tissue-Specific Coexpression Networks of Noncoding RNAs (miRNA and lncRNA) and mRNAs in Normal Tissues. Comput. Math. Methods Med. 2019, 2019, 9029351. [Google Scholar] [CrossRef]
- Bhartiya, D.; Pal, K.; Ghosh, S.; Kapoor, S.; Jalali, S.; Panwar, B.; Jain, S.; Sati, S.; Sengupta, S.; Sachidanandan, C.; et al. lncRNome: A comprehensive knowledgebase of human long noncoding RNAs. Database 2013, 2013, bat034. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Zhang, X.; Ma, Z.; Lin, Y.; Lu, M. MicroRNA-125b-5p mediates post-transcriptional regulation of hepatitis B virus replication via the LIN28B/let-7 axis. RNA Biol. 2017, 14, 1389–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Liu, Y.; Wu, J.; Deng, C.; Tan, J.; Liu, H.; Zhong, L. Lung adenocarcinoma patients have higher risk of SARS-CoV-2 infection. Aging 2021, 13, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Q.; Zhang, W.; Li, J.; Li, Z.; Tang, Z.; Li, Y.; Han, C.; Hall, S.H.; Zhang, Y. Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim. Biophys. Sin. 2010, 42, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Lopes, S.M.C.D.S.; Kaneda, M.; Tang, F.; Hajkova, P.; Lao, K.; O’Carroll, D.; Das, P.P.; Tarakhovsky, A.; Miska, E.A.; et al. MicroRNA Biogenesis Is Required for Mouse Primordial Germ Cell Development and Spermatogenesis. PLoS ONE 2008, 3, e1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Godo, A.; Grossmann, M.; Pons, M.C.; F-Fernández, S.; Garrido, N.; Anton, E. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil. Steril. 2015, 104, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; I Nakaya, H. ACE2 Expression Is Increased in the Lungs of Patients with Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Kofman, A.E.; Huszar, J.M.; Payne, C.J. Transcriptional Analysis of Histone Deacetylase Family Members Reveal Similarities Between Differentiating and Aging Spermatogonial Stem Cells. Stem Cell Rev. Rep. 2013, 9, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozgeyik, I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene 2021, 27, 100831. [Google Scholar] [CrossRef]
- Wu, W.; Wu, D.; Yan, W.; Wang, Y.; You, J.; Wan, X.; Xi, D.; Luo, X.; Han, M.; Ning, Q. Interferon-Induced Macrophage-Derived Exosomes Mediate Antiviral Activity Against Hepatitis B Virus Through miR-574-5p. J. Infect. Dis. 2021, 223, 686–698. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhou, W.; Rui, Y.; Liu, L.; Chen, B.; Su, X. MicroRNA-574-5p Attenuates Acute Respiratory Distress Syndrome by Targeting HMGB1. Am. J. Respir. Cell Mol. Biol. 2021, 64, 196–207. [Google Scholar] [CrossRef]
- Wang, Y.; Lumbers, E.R.; Arthurs, A.; De Meaultsart, C.C.; Mathe, A.; Avery-Kiejda, K.A.; Roberts, C.; Pipkin, F.B.; Marques, F.Z.; Morris, B.J.; et al. Regulation of the human placental (pro)renin receptor-prorenin-angiotensin system by microRNAs. Mol. Hum. Reprod. 2018, 24, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, S.; Wu, S.; Ni, Y.; Pan, Z. MicroRNA-936 targets FGF2 to inhibit epithelial ovarian cancer aggressiveness by deactivating the PI3K/Akt pathway. OncoTargets Ther. 2019, 12, 5311–5322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limonta, D.; Jovel, J.; Kumar, A.; Lu, J.; Hou, S.; Airo, A.M.; Lopez-Orozco, J.; Wong, C.P.; Saito, L.; Branton, W.; et al. Fibroblast Growth Factor 2 Enhances Zika Virus Infection in Human Fetal Brain. J. Infect. Dis. 2019, 220, 1377–1387. [Google Scholar] [CrossRef]
- Saucedo, L.; Sobarzo, C.; Brukman, N.G.; Guidobaldi, H.A.; Lustig, L.; Giojalas, L.C.; Buffone, M.G.; Vazquez-Levin, M.H.; Marín-Briggiler, C. Involvement of fibroblast growth factor 2 (FGF2) and its receptors in the regulation of mouse sperm physiology. Reproduction 2018, 156, 163–172. [Google Scholar] [CrossRef]
- Tamura, M.; Matsuzuka, T. Angiotensin II Signaling. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Todorova, K.; Metodiev, M.V.; Metodieva, G.; Mincheff, M.; Fernández, N.; Hayrabedyan, S. Micro-RNA-204 Participates in TMPRSS2/ERG Regulation and Androgen Receptor Reprogramming in Prostate Cancer. Horm. Cancer 2017, 8, 28–48. [Google Scholar] [CrossRef]
- Belleannee, C.; Calvo, É.; Caballero, J.; Sullivan, R. Epididymosomes Convey Different Repertoires of MicroRNAs Throughout the Bovine Epididymis. Biol. Reprod. 2013, 89, 30. [Google Scholar] [CrossRef] [PubMed]
- Nho, K.; Ramanan, V.K.; Horgusluoglu, E.; Kim, S.; Inlow, M.H.; Risacher, S.L.; McDonald, B.C.; Farlow, M.R.; Foroud, T.M.; Gao, S.; et al. Comprehensive Gene- and Pathway-Based Analysis of Depressive Symptoms in Older Adults. J. Alzheimer’s Dis. 2015, 45, 1197–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevalainen, J.; Skarp, S.; Savolainen, E.-R.; Ryynänen, M.; Järvenpää, J. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J. Périnat. Med. 2017, 45, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Deng, H.; Chen, Q.; Wu, Q.; Li, X.; Jiang, S.; Wang, F.; Ye, F.; Ou, L.; Gao, H. Comprehensive Analysis of Differential Immunocyte Infiltration and Potential ceRNA Networks Involved in the Development of Atrial Fibrillation. BioMed Res. Int. 2020, 2020, 8021208. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabetian, S.; Castiglioni, I.; Jahromi, B.N.; Mousavi, P.; Cava, C. In Silico Identification of miRNA–lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection. Cells 2021, 10, 1480. https://doi.org/10.3390/cells10061480
Sabetian S, Castiglioni I, Jahromi BN, Mousavi P, Cava C. In Silico Identification of miRNA–lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection. Cells. 2021; 10(6):1480. https://doi.org/10.3390/cells10061480
Chicago/Turabian StyleSabetian, Soudabeh, Isabella Castiglioni, Bahia Namavar Jahromi, Pegah Mousavi, and Claudia Cava. 2021. "In Silico Identification of miRNA–lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection" Cells 10, no. 6: 1480. https://doi.org/10.3390/cells10061480
APA StyleSabetian, S., Castiglioni, I., Jahromi, B. N., Mousavi, P., & Cava, C. (2021). In Silico Identification of miRNA–lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection. Cells, 10(6), 1480. https://doi.org/10.3390/cells10061480





