Chamber-Specific Protein Expression during Direct Cardiac Reprogramming
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Cardiac Reprogramming
2.2. Immunocytochemistry of Reprogrammed Cells In Vitro
2.3. High-Content Imaging and Analysis
2.4. Quantitative Real Time PCR (qPCR)
2.5. In Vivo Cardiac Reprogramming
2.6. Immunohistochemistry on Heart Sections
2.7. Isolation of Cardiomyocytes
2.8. Statistical Analyses
3. Results
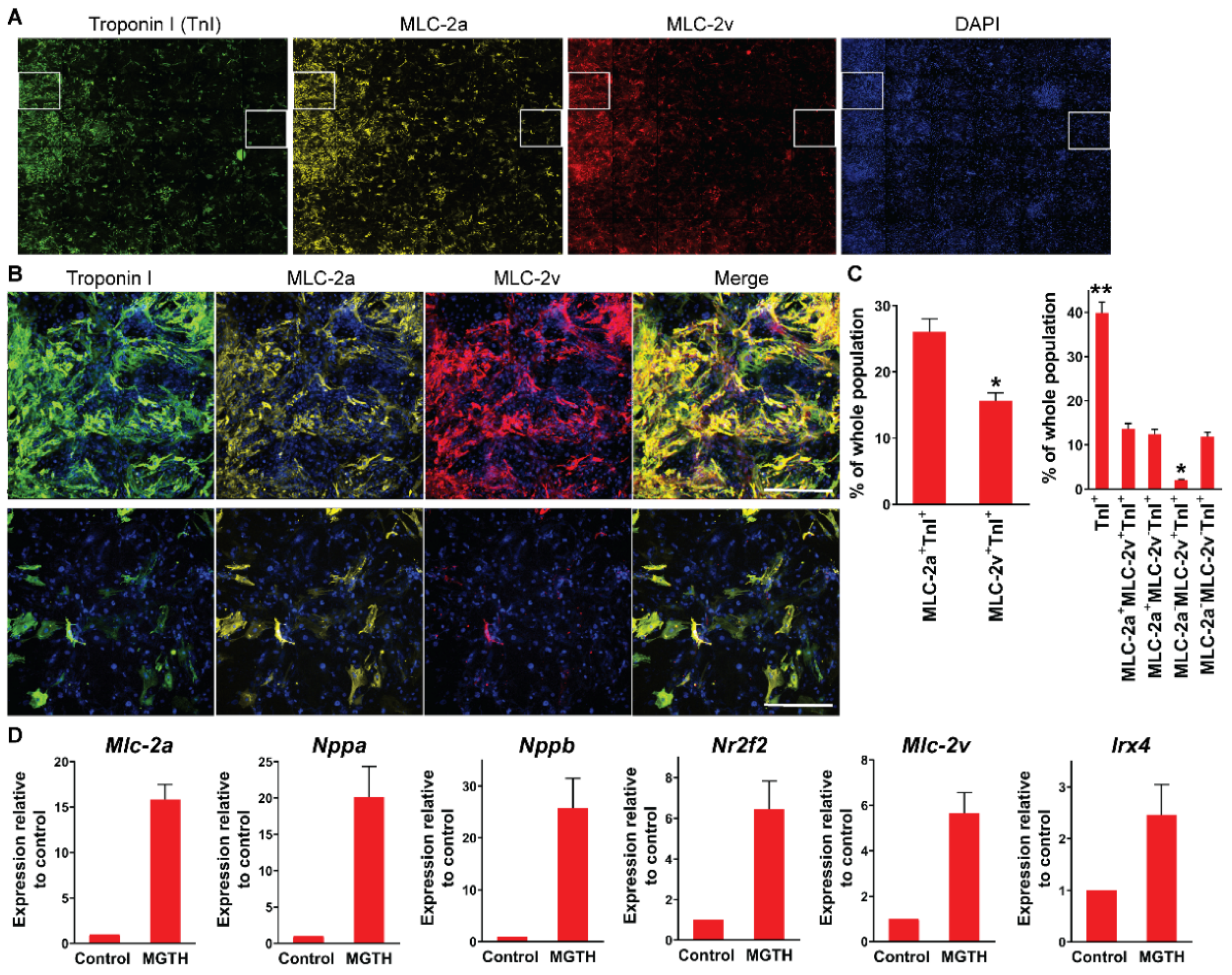
3.1. Chamber Protein Expression during In Vitro Cardiac Reprogramming
3.2. Chamber Protein Expression during In Vivo Cardiac Reprogramming Post-MI
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Wong, C.K.; Tsang, S.Y. Differential gene expressions in atrial and ventricular myocytes: Insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am. J. Physiol. Cell Physiol. 2010, 299, C1234–C1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Nam, Y.J. Assessing Cardiac Reprogramming using High Content Imaging Analysis. J. Vis. Exp. 2020, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, W.; Nam, Y.J. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Sci. Rep. 2019, 9, 14970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, A.D.; Kim, L.J.; Nam, Y.J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019, 9, 6362. [Google Scholar] [CrossRef] [PubMed]
- Gao, E.; Lei, Y.H.; Shang, X.; Huang, Z.M.; Zuo, L.; Boucher, M.; Fan, Q.; Chuprun, J.K.; Ma, X.L.; Koch, W.J. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ. Res. 2010, 107, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Nam, Y.J. Generation of MLC-2v-tdTomato knock-in reporter mouse line. Genesis 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.J.; Lubczyk, C.; Bhakta, M.; Zang, T.; Fernandez-Perez, A.; McAnally, J.; Bassel-Duby, R.; Olson, E.N.; Munshi, N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 2014, 141, 4267–4278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103.e105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, A.; Baek, S.T.; Banfi, S.; Eskiocak, B.; Tallquist, M.D. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 2011, 49, 870–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2016, 135, 978–995. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, L.; Yin, C.; Liu, J.; Qian, L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc. Res. 2015, 108, 217–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoffels, V.M.; Habets, P.E.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000, 223, 266–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, D.; Lamers, W.H.; Moorman, A.F. Patterns of expression in the developing myocardium: Towards a morphologically integrated transcriptional model. Cardiovasc. Res. 1998, 38, 25–53. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Villalpando, J.; Zhang, W.; Nam, Y.-J. Chamber-Specific Protein Expression during Direct Cardiac Reprogramming. Cells 2021, 10, 1513. https://doi.org/10.3390/cells10061513
Zhang Z, Villalpando J, Zhang W, Nam Y-J. Chamber-Specific Protein Expression during Direct Cardiac Reprogramming. Cells. 2021; 10(6):1513. https://doi.org/10.3390/cells10061513
Chicago/Turabian StyleZhang, Zhentao, Jesse Villalpando, Wenhui Zhang, and Young-Jae Nam. 2021. "Chamber-Specific Protein Expression during Direct Cardiac Reprogramming" Cells 10, no. 6: 1513. https://doi.org/10.3390/cells10061513




