DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy
Abstract
:1. Introduction
1.1. Cell Cycle
1.2. RNA Helicases
2. The Role of RNA Helicases in Regulation of Cell Cycle Progression
2.1. RNA Helicases Regulate G1-S Phase Transition
2.2. RNA Helicases Regulate S Phase Progression
2.3. RNA Helicases Regulate G2/M Phase Transition
2.4. RNA Helicases Regulate Cytokinesis
2.5. RNA Helicases Target CDK Inhibitor p21
3. Development of RNA Helicase Inhibitors for Clinical Treatment
3.1. Different Targeting Strategies Are Used to Develop Compounds against RNA Helicases
3.1.1. Compounds That Inhibit the ATPase Activity of RNA Helicase
3.1.2. Compounds That Inhibit the Helicase Activity of RNA Helicases
3.1.3. Compounds That Stabilize RNA Helicases onto RNA, Resulting in the Inhibition of Translation Initiation
3.1.4. Compounds That Regulate Both ATPase and RNA Helicase Activities
3.2. The Therapeutic Potential of RNA Helicase Inhibitors in the Treatment of Various Disorders
4. RNA Helicases and Phase-Separated Organelles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Panagopoulos, A.; Altmeyer, M. The Hammer and the Dance of Cell Cycle Control. Trends Biochem. Sci. 2021, 46, 301–314. [Google Scholar] [CrossRef]
- Guiducci, G.; Stojic, L. Long Noncoding RNAs at the Crossroads of Cell Cycle and Genome Integrity. Trends Genet. 2021, 37, 528–546. [Google Scholar] [CrossRef]
- Jongsma, M.L.; Berlin, I.; Neefjes, J. On the move: Organelle dynamics during mitosis. Trends Cell Biol. 2015, 25, 112–124. [Google Scholar] [CrossRef]
- Lemmens, B.; Lindqvist, A. DNA replication and mitotic entry: A brake model for cell cycle progression. J. Cell Biol. 2019, 218, 3892–3902. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alonso, D.; Malumbres, M. Mammalian cell cycle cyclins. Semin. Cell Dev. Biol. 2020, 107, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zieske, J.D.; Francesconi, C.M.; Guo, X. Cell cycle regulators at the ocular surface. Exp. Eye Res. 2004, 78, 447–456. [Google Scholar] [CrossRef]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef]
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215. [Google Scholar] [CrossRef] [PubMed]
- Jarmoskaite, I.; Russell, R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA 2011, 2, 135–152. [Google Scholar] [CrossRef] [Green Version]
- Cordin, O.; Hahn, D.; Beggs, J.D. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012, 24, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Owttrim, G.W. RNA helicases: Diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013, 10, 96–110. [Google Scholar] [CrossRef] [Green Version]
- Soultanas, P.; Wigley, D.B. Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci. 2001, 26, 47–54. [Google Scholar] [CrossRef]
- Bourgeois, C.F.; Mortreux, F.; Auboeuf, D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef]
- Wang, Y.; Li, G.; Deng, M.; Liu, X.; Huang, W.; Zhang, Y.; Liu, M.; Chen, Y. The multifaceted functions of RNA helicases in the adaptive cellular response to hypoxia: From mechanisms to therapeutics. Pharmacol. Ther. 2020, 221, 107783. [Google Scholar] [CrossRef]
- van Voss, M.R.H.; van Diest, P.J.; Raman, V. Targeting RNA helicases in cancer: The translation trap. Biochim. Biophys. Acta. Rev. Cancer 2017, 1868, 510–520. [Google Scholar] [CrossRef]
- Putra, V.; Hulme, A.J.; Tee, A.E.; Sun, J.Q.; Atmadibrata, B.; Ho, N.; Chen, J.; Gao, J.; Norris, M.D.; Haber, M.; et al. The RNA-helicase DDX21 upregulates CEP55 expression and promotes neuroblastoma. Mol. Oncol. 2021, 15, 1162–1179. [Google Scholar] [CrossRef]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, L.X.; Zhou, J.X.; Harris, P.C.; Calvet, J.P.; Li, X. RNA helicase p68 inhibits the transcription and post-transcription of Pkd1 in ADPKD. Theranostics 2020, 10, 8281–8297. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Wisskirchen, C.; Ludersdorfer, T.H.; Müller, D.A.; Moritz, E.; Pavlovic, J. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 2011, 85, 8646–8655. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Thakur, M.K. Interaction of estrogen receptor-alpha transactivation domain with nuclear proteins of mouse brain: p68 RNA helicase shows age- and sex-specific change. J. Neurosci. Res. 2009, 87, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Lennox, A.L.; Hoye, M.L.; Jiang, R.; Johnson-Kerner, B.L.; Suit, L.A.; Venkataramanan, S.; Sheehan, C.J.; Alsina, F.C.; Fregeau, B.; Aldinger, K.A.; et al. Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development. Neuron 2020, 106, 404–420.e8. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, S.; Okawa, S.; Hillje, A.L.; González-Cano, L.; Del Sol, A.; Schwamborn, J.C. The RNA helicase DDX6 regulates cell-fate specification in neural stem cells via miRNAs. Nucleic Acids Res. 2015, 43, 2638–2654. [Google Scholar] [CrossRef] [Green Version]
- Nyamao, R.M.; Wu, J.; Yu, L.; Xiao, X.; Zhang, F.M. Roles of DDX5 in the tumorigenesis, proliferation, differentiation, metastasis and pathway regulation of human malignancies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Luo, Z.; Zhou, L.; Li, X.; Jiang, T.; Fu, E. DDX5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating beta-catenin signaling pathway. Cancer Sci. 2015, 106, 1303–1312. [Google Scholar] [CrossRef]
- Wang, R.; Jiao, Z.; Li, R.; Yue, H.; Chen, L. p68 RNA helicase promotes glioma cell proliferation in vitro and in vivo via direct regulation of NF-kappaB transcription factor p50. Neuro. Oncol. 2012, 14, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Guturi, K.K.; Sarkar, M.; Bhowmik, A.; Das, N.; Ghosh, M.K. DEAD-box protein p68 is regulated by beta-catenin/transcription factor 4 to maintain a positive feedback loop in control of breast cancer progression. Breast Cancer Res. 2014, 16, 496. [Google Scholar] [CrossRef] [Green Version]
- Sha, M.; Lin, M.; Wang, J.; Ye, J.; Xu, J.; Xu, N. Long non-coding RNA MIAT promotes gastric cancer growth and metastasis through regulation of miR-141/DDX5 pathway. J. Exp. Clin. Cancer Res. 2018, 37, 58. [Google Scholar] [CrossRef] [Green Version]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef] [Green Version]
- Cannizzaro, E.; Bannister, A.J.; Han, N.; Alendar, A.; Kouzarides, T. DDX3X RNA helicase affects breast cancer cell cycle progression by regulating expression of KLF4. FEBS Lett. 2018, 592, 2308–2322. [Google Scholar] [CrossRef]
- Kong, T.; Xue, Y.; Cencic, R.; Zhu, X.; Monast, A.; Fu, Z. eIF4A Inhibitors Suppress Cell-Cycle Feedback Response and Acquired Resistance to CDK4/6 Inhibition in Cancer. Mol. Cancer Ther. 2019, 18, 2158–2170. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.H.; Yu, H.I.; Cho, W.C.; Tarn, W.Y. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene 2015, 34, 2790–2800. [Google Scholar] [CrossRef]
- Nicol, S.M.; Bray, S.E.; Black, H.D.; Lorimore, S.A.; Wright, E.G.; Lane, D.P. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 2013, 32, 3461–3469. [Google Scholar] [CrossRef] [Green Version]
- Hume, S.; Dianov, G.L.; Ramadan, K. A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 2020, 48, 12483–12501. [Google Scholar] [CrossRef]
- Lavoie, J.N.; L’Allemain, G.; Brunet, A.; Muller, R.; Pouyssegur, J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 1996, 271, 20608–20616. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, H.C.; Clurman, B.E. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005, 24, 2776–2786. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Stewart, Z.A.; Pietenpol, J.A. p53 Signaling and cell cycle checkpoints. Chem. Res. Toxicol. 2001, 14, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.H.; Chen, C.M.; Cheng, P.L.; Shih, J.W.; Tsou, A.P.; Lee, Y.H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006, 66, 6579–6588. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.W.; Liu, W.S.; Wang, J.; Chen, C.Y.; Cheng, Y.W.; Lee, H. Reduced p21(WAF1/CIP1) via alteration of p53-DDX3 pathway is associated with poor relapse-free survival in early-stage human papillomavirus-associated lung cancer. Clin. Cancer Res. 2011, 17, 1895–1905. [Google Scholar] [CrossRef] [Green Version]
- Heerma van Voss, M.R.; Kammers, K.; Vesuna, F.; Brilliant, J.; Bergman, Y.; Tantravedi, S. Global Effects of DDX3 Inhibition on Cell Cycle Regulation Identified by a Combined Phosphoproteomics and Single Cell Tracking Approach. Transl. Oncol. 2018, 11, 755–763. [Google Scholar] [CrossRef]
- Tantravedi, S.; Vesuna, F.; Winnard, P.T., Jr.; Martin, A.; Lim, M.; Eberhart, C.G.; Berlinicke, C.; Raabe, E.; van Diest, P.J.; Raman, V. Targeting DDX3 in Medulloblastoma Using the Small Molecule Inhibitor RK-33. Transl. Oncol. 2019, 12, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Verma, D.; Burton, A.; Swaminathan, S. Cellular RNA Helicase DHX9 Interacts with the Essential Epstein-Barr Virus (EBV) Protein SM and Restricts EBV Lytic Replication. J. Virol. 2019, 93, e01244-18. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Vesuna, F.; Tantravedi, S.; Bol, G.M.; Heerma van Voss, M.R.; Nugent, K.; Malek, R.; Gabrielson, K.; van Diest, P.J.; Tran, P.T.; et al. RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3. Cancer Res. 2016, 76, 6340–6350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015, 7, 648–669. [Google Scholar] [CrossRef]
- Heerma van Voss, M.R.; Vesuna, F.; Trumpi, K.; Brilliant, J.; Berlinicke, C.; de Leng, W.; Kranenburg, O.; Offerhaus, G.J.; Burger, H.; van der Wall, E.; et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 2015, 6, 28312–28326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Vesuna, F.; Botlagunta, M.; Bol, G.M.; Irving, A.; Bergman, Y.; Hosmane, R.S.; Kato, Y.; Winnard, P.T., Jr.; Raman, V. NZ51, a ring-expanded nucleoside analog, inhibits motility and viability of breast cancer cells by targeting the RNA helicase DDX3. Oncotarget 2015, 6, 29901–29913. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.C.; Chang, W.C.; Shieh, S.Y.; Tarn, W.Y. DDX3 regulates cell growth through translational control of cyclin E1. Mol. Cell Biol. 2010, 30, 5444–5453. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Feng, W.; Yuan, Z.; Weber, J.D.; Zhang, Y. DHX33 Interacts with AP-2beta To Regulate Bcl-2 Gene Expression and Promote Cancer Cell Survival. Mol. Cell Biol. 2019, 39, e00017-19. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Liu, Y.; Wang, X.; Yuan, B.; Zhang, Y. Role of DHX33 in c-Myc-induced cancers. Carcinogenesis 2017, 38, 649–660. [Google Scholar] [CrossRef]
- Yuan, B.; Wang, X.; Fan, C.; You, J.; Liu, Y.; Weber, J.D.; Zhong, H.; Zhang, Y. DHX33 Transcriptionally Controls Genes Involved in the Cell Cycle. Mol. Cell Biol. 2016, 36, 2903–2917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yu, J.; Wang, X.; Zhang, Y. The RNA helicase DHX33 is required for cancer cell proliferation in human glioblastoma and confers resistance to PI3K/mTOR inhibition. Cell Signal. 2019, 54, 170–178. [Google Scholar] [CrossRef]
- Wang, X.; Feng, W.; Peng, C.; Chen, S.; Ji, H.; Zhong, H.; Ge, W.; Zhang, Y. Targeting RNA helicase DHX33 blocks Ras-driven lung tumorigenesis in vivo. Cancer Sci. 2020, 111, 3564–3575. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jia, X.; Wang, C.; Xu, J.; Gao, S.J.; Lu, C. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ. 2019, 26, 1750–1765. [Google Scholar] [CrossRef]
- Yuan, D.; Chen, Y.; Yang, Z.; Li, G.; Wu, M.; Jiang, J.; Li, D.; Yu, Q. SPOP attenuates migration and invasion of choriocarcinoma cells by promoting DHX9 degradation. Am. J. Cancer Res. 2020, 10, 2428–2445. [Google Scholar] [PubMed]
- Hong, H.; An, O.; Chan, T.H.M.; Ng, V.H.E.; Kwok, H.S.; Lin, J.S.; Qi, L.; Han, J.; Tay, D.J.T.; Tang, S.J.; et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res. 2018, 46, 7953–7969. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Ray, P.; Liu, J.; Kuan, C.T.; Xu, J.; Hsu, D.; Sullenger, B.A.; White, R.R.; Clary, B.M. In Vivo Selection Against Human Colorectal Cancer Xenografts Identifies an Aptamer That Targets RNA Helicase Protein DHX9. Mol. Ther. Nucleic Acids 2016, 5, e315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.; Di Paola, D.; Malina, A.; Mills, J.R.; Kreps, A.; Grosse, F.; Tang, H.; Zannis-Hadjopoulos, M.; Larsson, O.; Pelletier, J. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J. Biol. Chem. 2014, 289, 22798–22814. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.L.; Liu, J.Y.; Yang, J.E.; Yu, X.M.; Chen, Z.L.; Chen, Y.J.; Kuang, M.; Zhu, Y.; Zhuang, S.M. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology 2019, 70, 259–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thacker, U.; Pauzaite, T.; Tollitt, J.; Twardowska, M.; Harrison, C.; Dowle, A.; Coverley, D.; Copeland, N.A. Identification of DHX9 as a cell cycle regulated nucleolar recruitment factor for CIZ1. Sci. Rep. 2020, 10, 18103. [Google Scholar] [CrossRef] [PubMed]
- Guenard, F.; Labrie, Y.; Ouellette, G.; Beauparlant, C.J.; Durocher, F.; Inherit BRCAs. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J. Hum. Genet. 2009, 54, 152–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Li, Z.; Hu, X.; Xie, F.; Kuang, S.; Zhan, B.; Gao, W.; Chen, X.; Gao, S.; Li, Y.; et al. Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation. Adv. Sci. 2020, 7, 2000532. [Google Scholar] [CrossRef]
- Zhang, Y.; Baysac, K.C.; Yee, L.F.; Saporita, A.J.; Weber, J.D. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014, 16, 449. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.; Lee, S.; Choi, H.S.; Kim, S.N.; Lee, E.; Shin, Y.; Seo, J.; Kim, B.; Jung, Y.; Kim, W.K.; et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. 2011, 17, 700–709. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, Z.; Qin, W.; Sun, T.; Lu, S.; Li, Y.; Wang, Y.; Hu, X.; Xu, D.; Wu, Y.; et al. Long non-coding RNA ZFAS1 promotes colorectal cancer tumorigenesis and development through DDX21-POLR1B regulatory axis. Aging 2020, 12, 22656–22687. [Google Scholar] [CrossRef]
- Santoriello, C.; Sporrij, A.; Yang, S.; Flynn, R.A.; Henriques, T.; Dorjsuren, B.; Custo Greig, E.; McCall, W.; Stanhope, M.E.; Fazio, M.; et al. RNA helicase DDX21 mediates nucleotide stress responses in neural crest and melanoma cells. Nat. Cell Biol. 2020, 22, 372–379. [Google Scholar] [CrossRef]
- Holmstrom, T.H.; Mialon, A.; Kallio, M.; Nymalm, Y.; Mannermaa, L.; Holm, T.; Johansson, H.; Black, E.; Gillespie, D.; Salminen, T.A.; et al. c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J. Biol. Chem. 2008, 283, 7046–7053. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Camacho, C.V.; Nagari, A.; Malladi, V.S.; Challa, S.; Kraus, W.L. Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol. Cell 2019, 75, 1270–1285. [Google Scholar] [CrossRef]
- Merrick, W.C. eIF4F: A retrospective. J. Biol. Chem. 2015, 290, 24091–24099.e14. [Google Scholar] [CrossRef] [Green Version]
- Raza, F.; Waldron, J.A.; Quesne, J.L. Translational dysregulation in cancer: eIF4A isoforms and sequence determinants of eIF4A dependence. Biochem. Soc. Trans. 2015, 43, 1227–1233. [Google Scholar] [CrossRef]
- Rubio, C.A.; Weisburd, B.; Holderfield, M.; Arias, C.; Fang, E.; DeRisi, J.L.; Fanidi, A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome. Biol. 2014, 15, 476. [Google Scholar] [CrossRef]
- Busetto, V.; Barbosa, I.; Basquin, J.; Marquenet, E.; Hocq, R.; Hennion, M.; Paternina, J.A.; Namane, A.; Conti, E.; Bensaude, O.; et al. Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes. Nucleic Acids Res. 2020, 48, 5670–5683. [Google Scholar] [CrossRef]
- Ryu, I.; Won, Y.S.; Ha, H.; Kim, E.; Park, Y.; Kim, M.K.; Kwon, D.H.; Choe, J.; Song, H.K.; Jung, H.; et al. eIF4A3 Phosphorylation by CDKs Affects NMD during the Cell Cycle. Cell Rep. 2019, 26, 2126–2139.e9. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Gao, X.; Wang, M.; Qiao, Y.; Xu, Y.; Yang, J.; Dong, N.; He, J.; Sun, Q.; Lv, G.; et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 2016, 7, 22159–22173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.W.; Cheng, S.C. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 2015, 29, 81–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Ma, Y.; Huang, P.; Du, A.; Yang, X.; Zhang, S.; Xing, C.; Liu, F.; Cao, J. Lentiviral DDX46 knockdown inhibits growth and induces apoptosis in human colorectal cancer cells. Gene 2015, 560, 237–244. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.M.; He, W.T.; Chen, H.; Zhu, H.W.; Liu, T.; Zhang, J.H.; Song, T.N.; Zhou, Y.L. Knockdown of DDX46 inhibits proliferation and induces apoptosis in esophageal squamous cell carcinoma cells. Oncol. Rep. 2016, 36, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Xu, M.; Zhong, W.; Hu, Y.; Wang, G. Knockdown of DDX46 suppresses the proliferation and invasion of gastric cancer through inactivating Akt/GSK-3beta/beta-catenin pathway. Exp. Cell Res. 2021, 399, 112448. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, D.; Li, G.; Wang, X. Knockdown of DDX46 Inhibits the Invasion and Tumorigenesis in Osteosarcoma Cells. Oncol. Res. 2017, 25, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005, 30, 630–641. [Google Scholar] [CrossRef]
- Rohaly, G.; Chemnitz, J.; Dehde, S.; Nunez, A.M.; Heukeshoven, J.; Deppert, W.; Dornreiter, I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell 2005, 122, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zonta, E.; Bittencourt, D.; Samaan, S.; Germann, S.; Dutertre, M.; Auboeuf, D. The RNA helicase DDX5/p68 is a key factor promoting c-fos expression at different levels from transcription to mRNA export. Nucleic Acids Res. 2013, 41, 554–564. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, L.; Lapik, Y.R.; Wang, M.; Pestov, D.G. Mammalian DEAD box protein Ddx51 acts in 3’ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol. Cell Biol. 2010, 30, 2947–2956. [Google Scholar] [CrossRef] [Green Version]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, H.; Zhao, C.; Li, W.; Xu, H.; Chen, Y. The DEAD-box RNA helicase 51 controls non-small cell lung cancer proliferation by regulating cell cycle progression via multiple pathways. Sci. Rep. 2016, 6, 26108. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Rohe, A.; Platzer, C.; Najjar, A.; Erdmann, F.; Sippl, W. Regulation of G2/M Transition by Inhibition of WEE1 and PKMYT1 Kinases. Molecules 2017, 22, 2045. [Google Scholar] [CrossRef] [Green Version]
- Mueller, P.R.; Leise, W.F., 3rd. Measurement of Wee kinase activity. Methods Mol. Biol. 2005, 296, 299–328. [Google Scholar]
- Taylor, W.R.; Stark, G.R. Regulation of the G2/M transition by p53. Oncogene 2001, 20, 1803–1815. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, J.; Ye, M.; Liu, F.; Wu, S.; Huang, J.; Shi, G. Ddx56 maintains proliferation of mouse embryonic stem cells via ribosome assembly and interaction with the Oct4/Sox2 complex. Stem. Cell Res. Ther. 2020, 11, 314. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, X.; Kourkoumelis, N.; Shen, Y.; Huang, W. Integrated Analysis of DEAD-Box Helicase 56: A Potential Oncogene in Osteosarcoma. Front. Bioeng. Biotechnol. 2020, 8, 588. [Google Scholar] [CrossRef] [PubMed]
- Pryszlak, M.; Wiggans, M.; Chen, X.; Jaramillo, J.E.; Burns, S.E.; Richards, L.M.; Pugh, T.J.; Kaplan, D.R.; Huang, X.; Dirks, P.B.; et al. The DEAD-box helicase DDX56 is a conserved stemness regulator in normal and cancer stem cells. Cell Rep. 2021, 34, 108903. [Google Scholar] [CrossRef]
- Kouyama, Y.; Masuda, T.; Fujii, A.; Ogawa, Y.; Sato, K.; Tobo, T.; Wakiyama, H.; Yoshikawa, Y.; Noda, M.; Tsuruda, Y.; et al. Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sc. 2019, 110, 3132–3144. [Google Scholar] [CrossRef] [Green Version]
- Mazloomian, A.; Araki, S.; Ohori, M.; El-Naggar, A.M.; Yap, D.; Bashashati, A.; Nakao, S.; Sorensen, P.H.; Nakanishi, A.; Shah, S.; et al. Pharmacological systems analysis defines EIF4A3 functions in cell-cycle and RNA stress granule formation. Commun. Biol. 2019, 2, 165. [Google Scholar] [CrossRef]
- Mierzwa, B.; Gerlich, D.W. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell 2014, 31, 525–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollard, T.D.; O’Shaughnessy, B. Molecular Mechanism of Cytokinesis. Annu. Rev. Biochem. 2019, 88, 661–689. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, T.; Araki, S.; Ohori, M.; El-Naggar, A.M.; Yap, D.; Bashashati, A.; Nakao, S.; Sorensen, P.H.; Nakanishi, A.; Shah, S.; et al. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010, 21, 2953–2965. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.I.; Yamazaki, T.; Harada, K.; Seno, S.; Matsuda, H.; Masuda, S. URH49 exports mRNA by remodeling complex formation and mediating the NXF1-dependent pathway. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194480. [Google Scholar] [CrossRef]
- Alessi, A.F.; Khivansara, V.; Han, T.; Freeberg, M.A.; Moresco, J.J.; Tu, P.G.; Montoye, E.; Yates, J.R., 3rd; Karp, X.; Kim, J.K. Casein kinase II promotes target silencing by miRISC through direct phosphorylation of the DEAD-box RNA helicase CGH-1. Proc. Natl. Acad. Sci. USA 2015, 112, E7213–E7222. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, K.; Iwatsuki, A.; Sugito, N.; Shinohara, H.; Kuranaga, Y.; Oshikawa, Y.; Tajirika, T.; Futamura, M.; Yoshida, K.; Uchiyama, K.; et al. Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 2018, 57, 579–589. [Google Scholar] [CrossRef]
- Audhya, A.; Hyndman, F.; McLeod, I.X.; Maddox, A.S.; Yates, J.R., 3rd; Desai, A.; Oegema, K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 2005, 171, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Peters, D.; Radine, C.; Reese, A.; Budach, W.; Sohn, D.; Janicke, R.U. The DEAD-box RNA helicase DDX41 is a novel repressor of p21(WAF1/CIP1) mRNA translation. J. Biol. Chem. 2017, 292, 8331–8341. [Google Scholar] [CrossRef] [Green Version]
- Dai, L.; Liu, Y.; Liu, J.; Wen, X.; Xu, Z.; Wang, Z.; Sun, H.; Tang, S.; Maguire, A.R.; Quan, J.; et al. A novel cyclinE/cyclinA-CDK inhibitor targets p27(Kip1) degradation, cell cycle progression and cell survival: Implications in cancer therapy. Cancer Lett. 2013, 333, 103–112. [Google Scholar] [CrossRef]
- Bali, A.; O’Brien, P.M.; Edwards, L.S.; Sutherland, R.L.; Hacker, N.F.; Henshall, S.M. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin. Cancer Res. 2004, 10, 5168–5177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juan, G.; Ardelt, B.; Li, X.; Mikulski, S.M.; Shogen, K.; Ardelt, W.; Mittelman, A.; Darzynkiewicz, Z. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia 1998, 12, 1241–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T.; Kawabe, T.; Ohara, H.; Ducommun, B.; Itoh, M.; Okamoto, T. Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J. Biol. Chem. 2001, 276, 42971–42977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef] [Green Version]
- Polprasert, C.; Schulze, I.; Sekeres, M.A.; Makishima, H.; Przychodzen, B.; Hosono, N.; Singh, J.; Padgett, R.A.; Gu, X.; Phillips, J.G.; et al. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell 2015, 27, 658–670. [Google Scholar] [CrossRef] [Green Version]
- Weinreb, J.T.; Gupta, V.; Sharvit, E.; Weil, R.; Bowman, T.V. Ddx41 inhibition of DNA damage signaling permits erythroid progenitor expansion in zebrafish. Haematologica 2021. [Google Scholar] [CrossRef]
- Iyer, R.S.; Nicol, S.M.; Quinlan, P.R.; Thompson, A.M.; Meek, D.W.; Fuller-Pace, F.V. The RNA helicase/transcriptional co-regulator, p68 (DDX5), stimulates expression of oncogenic protein kinase, Polo-like kinase-1 (PLK1), and is associated with elevated PLK1 levels in human breast cancers. Cell Cycle 2014, 13, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Rossow, K.L.; Grande, J.P.; Janknecht, R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007, 67, 7572–7578. [Google Scholar] [CrossRef] [Green Version]
- Li, L.X.; Zhou, J.X.; Wang, X.; Zhang, H.; Harris, P.C.; Calvet, J.P.; Li, X. Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis. Sci. Adv. 2020, 6, eabb3154. [Google Scholar] [CrossRef]
- Sithole, N.; Williams, C.A.; Abbink, T.E.M.; Lever, A.M.L. DDX5 potentiates HIV-1 transcription as a co-factor of Tat. Retrovirology 2020, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Kost, G.C.; Yang, M.Y.; Li, L.; Zhang, Y.; Liu, C.Y.; Kim, D.J.; Ahn, C.H.; Lee, Y.B.; Liu, Z.R. A Novel Anti-Cancer Agent, 1-(3,5-Dimethoxyphenyl)-4-[(6-Fluoro-2-Methoxyquinoxalin-3-yl)Aminocarbonyl] Piperazine (RX-5902), Interferes With beta-Catenin Function Through Y593 Phospho-p68 RNA Helicase. J. Cell Biochem. 2015, 116, 1595–1601. [Google Scholar] [CrossRef]
- Ito, M.; Iwatani, M.; Kamada, Y.; Sogabe, S.; Nakao, S.; Tanaka, T.; Kawamoto, T.; Aparicio, S.; Nakanishi, A.; Imaeda, Y. Discovery of selective ATP-competitive eIF4A3 inhibitors. Bioorg. Med. Chem. 2017, 25, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.C.; Sikirzhytski, V.; Aksenova, M.; Lucius, M.D.; Levon, G.H.; Mack, Z.T.; Pollack, C.; Odhiambo, D.; Broude, E.; Lizarraga, S.B.; et al. Pharmacological inhibition of DEAD-Box RNA Helicase 3 attenuates stress granule assembly. Biochem. Pharmacol. 2020, 182, 114280. [Google Scholar] [CrossRef] [PubMed]
- Heerma van Voss, M.R.; Vesuna, F.; Bol, G.M.; Afzal, J.; Tantravedi, S.; Bergman, Y.; Kammers, K.; Lehar, M.; Malek, R.; Ballew, M.; et al. Targeting mitochondrial translation by inhibiting DDX3: A novel radiosensitization strategy for cancer treatment. Oncogene 2018, 37, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Steinberger, J.; Shen, L.; Kiniry, S.J.; Naineni, S.K.; Cencic, R.; Amiri, M.; Aboushawareb, S.A.E.; Chu, J.; Maiga, R.I.; Yachnin, B.J.; et al. Identification and characterization of hippuristanol-resistant mutants reveals eIF4A1 dependencies within mRNA 5’ leader regions. Nucleic Acids Res. 2020, 48, 9521–9537. [Google Scholar] [CrossRef]
- Chu, J.; Pelletier, J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim. Biophys. Acta 2015, 1849, 781–791. [Google Scholar] [CrossRef]
- Bordeleau, M.E.; Matthews, J.; Wojnar, J.M.; Lindqvist, L.; Novac, O.; Jankowsky, E.; Sonenberg, N.; Northcote, P.; Teesdale-Spittle, P.; Pelletier, J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA 2005, 102, 10460–10465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordeleau, M.E.; Cencic, R.; Lindqvist, L.; Oberer, M.; Northcote, P.; Wagner, G.; Pelletier, J. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 2006, 13, 1287–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanghvi, V.R.; Mohan, P.; Singh, K.; Cao, L.; Berishaj, M.; Wolfe, A.L.; Schatz, J.H.; Lailler, N.; de Stanchina, E.; Viale, A.; et al. NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy. Cancers 2021, 13, 639. [Google Scholar] [CrossRef]
- Capasso, A.; Bagby, S.M.; Dailey, K.L.; Currimjee, N.; Yacob, B.W.; Ionkina, A.; Frank, J.G.; Kim, D.J.; George, C.; Lee, Y.B.; et al. First-in-Class Phosphorylated-p68 Inhibitor RX-5902 Inhibits beta-Catenin Signaling and Demonstrates Antitumor Activity in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2019, 18, 1916–1925. [Google Scholar] [CrossRef] [Green Version]
- Botlagunta, M.; Kollapalli, B.; Kakarla, L.; Gajarla, S.P.; Gade, S.P.; Dadi, C.L.; Penumadu, A.; Javeed, S. In vitro anti-cancer activity of doxorubicin against human RNA helicase, DDX3. Bioinformation 2016, 12, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Wilky, B.A.; Kim, C.; McCarty, G.; Montgomery, E.A.; Kammers, K.; DeVine, L.R.; Cole, R.N.; Raman, V.; Loeb, D.M. RNA helicase DDX3: A novel therapeutic target in Ewing sarcoma. Oncogene 2016, 35, 2574–2583. [Google Scholar] [CrossRef]
- Tsumuraya, T.; Ishikawa, C.; Machijima, Y.; Nakachi, S.; Senba, M.; Tanaka, J.; Mori, N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem. Pharmacol. 2011, 81, 713–722. [Google Scholar] [CrossRef]
- Tsai, B.P.; Jimenez, J.; Lim, S.; Fitzgerald, K.D.; Zhang, M.; Chuah, C.T.; Axelrod, H.; Wilson, L.; Ong, S.T.; Semler, B.L.; et al. A novel Bcr-Abl-mTOR-eIF4A axis regulates IRES-mediated translation of LEF-1. Open Biol. 2014, 4, 140180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, C.; Tanaka, J.; Katano, H.; Senba, M.; Mori, N. Hippuristanol reduces the viability of primary effusion lymphoma cells both in vitro and in vivo. Mar. Drugs 2013, 11, 3410–3424. [Google Scholar] [CrossRef]
- Kuznetsov, G.; Xu, Q.; Rudolph-Owen, L.; Tendyke, K.; Liu, J.; Towle, M.; Zhao, N.; Marsh, J.; Agoulnik, S.; Twine, N.; et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol. Cancer Ther. 2009, 8, 1250–1260. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Pelletier, J. Selective targeting of the DEAD-box RNA helicase eukaryotic initiation factor (eIF) 4A by natural products. Nat. Prod. Rep. 2020, 37, 609–616. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Liu, Z.R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 2006, 127, 139–155. [Google Scholar] [CrossRef] [Green Version]
- Tentler, J.J.; Lang, J.; Capasso, A.; Kim, D.J.; Benaim, E.; Lee, Y.B.; Eisen, A.; Bagby, S.M.; Hartman, S.J.; Yacob, B.W.; et al. RX-5902, a novel beta-catenin modulator, potentiates the efficacy of immune checkpoint inhibitors in preclinical models of triple-negative breast Cancer. BMC Cancer 2020, 20, 1063. [Google Scholar] [CrossRef]
- Diamond, J.; Eckhardt, G.; Gluck, L.; Gutierrez, M.; Peterson, C.; Pila, R.; Benaim, E. Phase 1 study of RX-5902, a novel orally bioavailable inhibitor of phosphorylated P68, which prevents β-catenin translocation in advanced solid tumors. Ann. Oncol. 2017, 28, v83. [Google Scholar] [CrossRef]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Zwicker, D.; Decker, M.; Jaensch, S.; Hyman, A.A.; Julicher, F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. USA 2014, 111, E2636–E2645. [Google Scholar] [CrossRef] [Green Version]
- Jao, L.E.; Akef, A.; Wente, S.R. A role for Gle1, a regulator of DEAD-box RNA helicases, at centrosomes and basal bodies. Mol. Biol. Cell 2017, 28, 120–127. [Google Scholar] [CrossRef]
- Iserman, C.; Desroches Altamirano, C.; Jegers, C.; Friedrich, U.; Zarin, T.; Fritsch, A.W.; Mittasch, M.; Domingues, A.; Hersemann, L.; Jahnel, M.; et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 2020, 181, 818–831.e19. [Google Scholar] [CrossRef] [PubMed]
- Begovich, K.; Wilhelm, J.E. An In Vitro Assembly System Identifies Roles for RNA Nucleation and ATP in Yeast Stress Granule Formation. Mol. Cell 2020, 79, 991–1007.e4. [Google Scholar] [CrossRef] [PubMed]
- Mugler, C.F.; Hondele, M.; Heinrich, S.; Sachdev, R.; Vallotton, P.; Koek, A.Y.; Chan, L.Y.; Weis, K. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 2016, 5, e18746. [Google Scholar] [CrossRef]
- Sachdev, R.; Hondele, M.; Linsenmeier, M.; Vallotton, P.; Mugler, C.F.; Arosio, P.; Weis, K. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. eLife 2019, 8, e41415. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Luo, E.C.; Haggerty, C.; Aigner, S.; Charlton, J.; Brumbaugh, J.; Ji, F.; Rabano Jimenez, I.; Clowers, K.J.; Huebner, A.J.; et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem. Cell 2019, 25, 622–638.e13. [Google Scholar]
- Chen, W.; Hu, Y.; Lang, C.F.; Brown, J.S.; Schwabach, S.; Song, X.; Zhang, Y.; Munro, E.; Bennett, K.; Zhang, D.; et al. The Dynamics of P Granule Liquid Droplets Are Regulated by the Caenorhabditis elegans Germline RNA Helicase GLH-1 via Its ATP Hydrolysis Cycle. Genetics 2020, 215, 421–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voronina, E.; Paix, A.; Seydoux, G. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 2012, 139, 3732–3740. [Google Scholar] [CrossRef] [Green Version]
- Marnik, E.A.; Updike, D.L. Membraneless organelles: P granules in Caenorhabditis elegans. Traffic 2019, 20, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Klosin, A.; Oltsch, F.; Harmon, T.; Honigmann, A.; Julicher, F.; Hyman, A.A.; Zechner, C. Phase separation provides a mechanism to reduce noise in cells. Science 2020, 367, 464–468. [Google Scholar] [CrossRef]
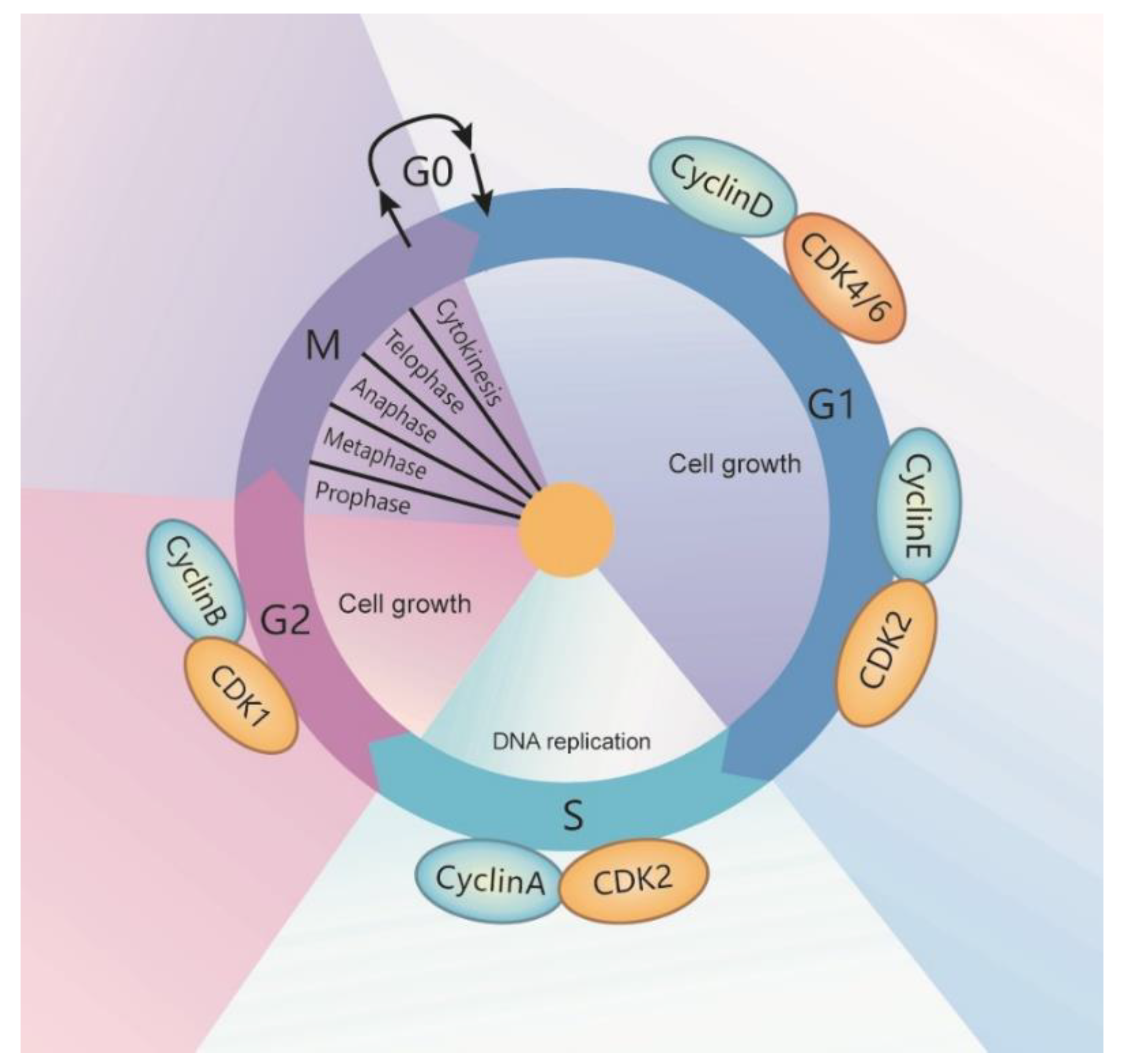
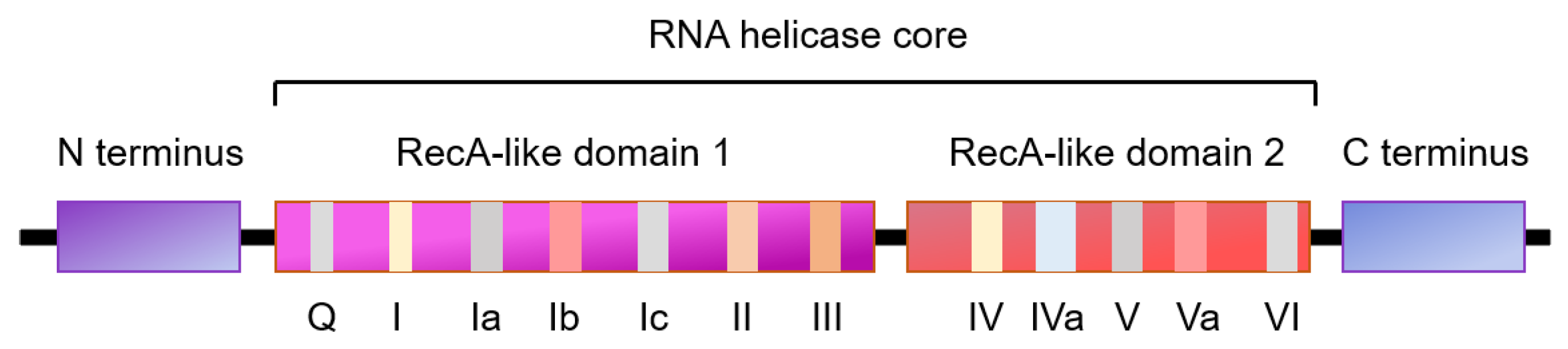
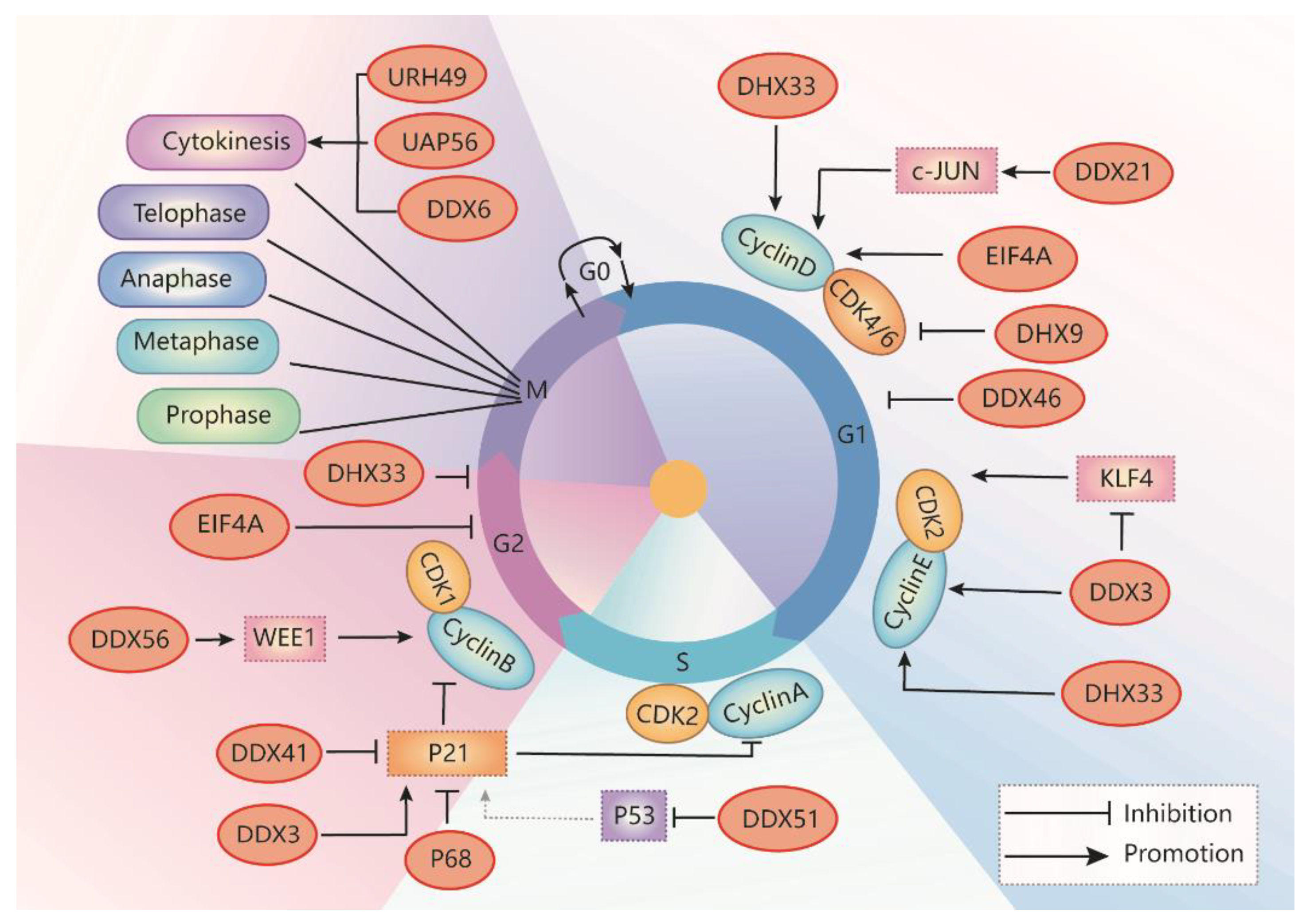
| Cell Cycle Stage | RNA Helicases | Intracellular Location | Expression |
|---|---|---|---|
| G1-S phase transition | Ded1/DDX3 | Nuclear speckles and cytoplasm | Upregulation in medulloblastoma, colorectal, breast, prostate, and lung cancer |
| DHX33 | Nucleus and nucleoli | Upregulation in lung cancers, hepatocellular carcinoma, lymphoma, colon cancer, and glioblastoma | |
| DHX9 | Nucleus | Upregulation in cervical cancer, breast cancer, prostate cancer, colorectal cancer, hepatocellular carcinoma, and Ewing sarcoma | |
| DDX21 | Nucleus and cytoplasm | Dysregulation in colon cancer, lymphomas, neuroblastoma, and breast cancers | |
| eIF4A | Nucleus and cytoplasm | Dysregulation in pancreatic cancer, breast cancer, prostate cancer | |
| DDX46 | Focal nuclear | Upregulated in colorectal carcinoma, esophageal squamous cell carcinoma, gastric cancer, and osteosarcoma cells | |
| S phase progression | DDX51 | Predominantly in nuclear | Dysregulation in NSCLC |
| G2-M phase transition | DDX56 | Nucleolus | Upregulation of DDX56 in various cancer, including osteosarcoma, colorectal cancer, and relates to a poor prognosis |
| DHX33 | Nucleus and nucleoli | See above | |
| Mitosis | UAP56 | Nucleus and cytoplasm | Not clear |
| URH49 | Nucleus and cytoplasm | Not clear | |
| Cytokinesis | UAP56 | Nucleus and cytoplasm | Not clear |
| URH49 | Nucleus and cytoplasm | Not clear | |
| DDX6 | Nucleus and cytoplasm | colorectal cancer and hepatocellular carcinoma | |
| Regulate the expression of p21(WAF1/CIP1) | DDX41 | Nucleus and cytoplasm | DDX41 mutant leads to anemia and acute myeloid leukemia. DDX41 increased in cervical cancer. |
| DDX5 | Mostly in nucleus, cytoplasmic levels of DDX5 increased in the G2/M phase | p68 increased in a range of cancers except for hepatocellular carcinoma | |
| DDX3 | Predominantly in nuclear speckles and at low levels in cytoplasm | See above |
| RNA Helicases | Inhibitors | Chemical Structure | Mechanisms of Action | Diseases | Model | Toxicity or Tissue-Specific |
|---|---|---|---|---|---|---|
| DDX3 | RK-33 | 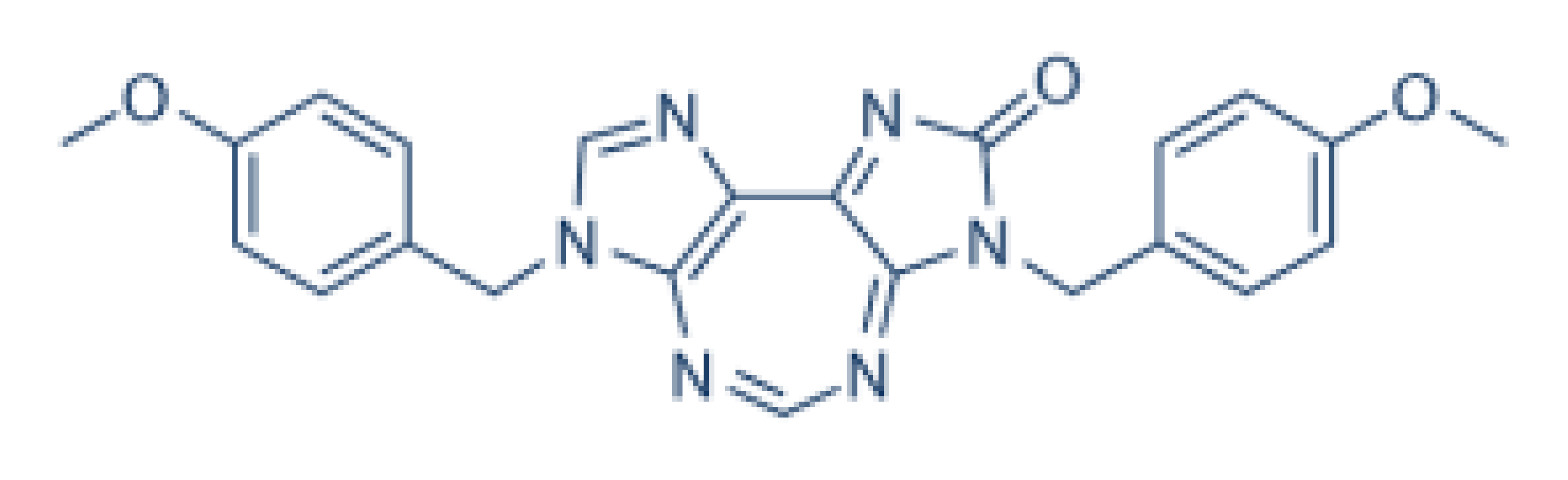 | Inhibition of helicase activity | Lung cancer, medulloblastoma, prostate cancer, Ewing sarcoma, and colorectal cancer | In vitro, and animal models | No discernable toxicity in animal models |
| NZ51 | 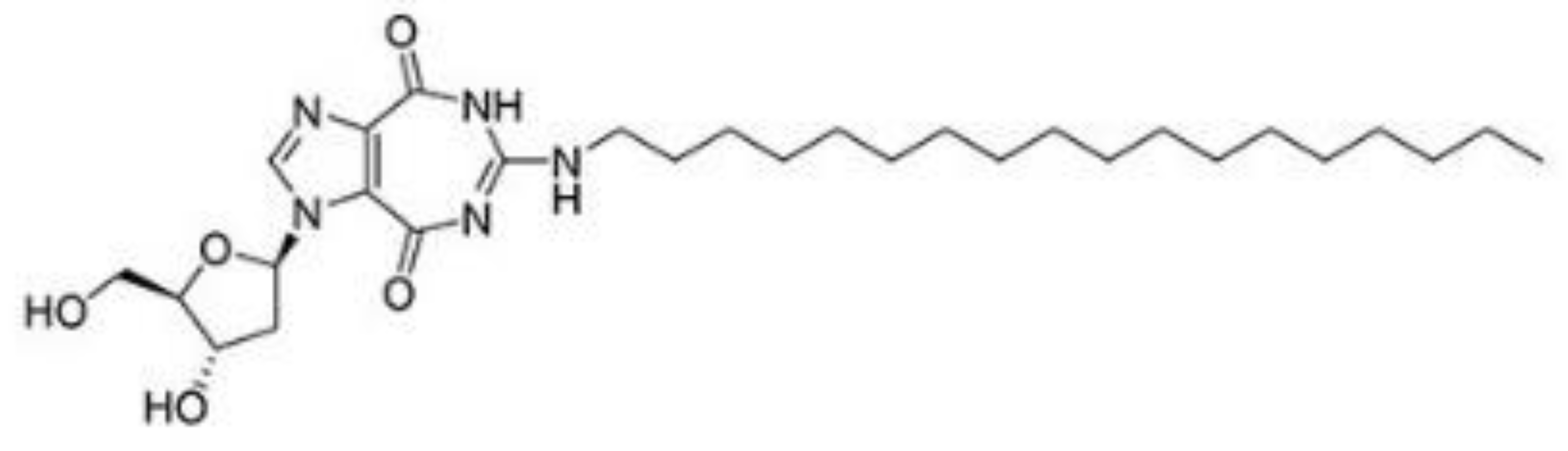 | Inhibition of helicase activity | Breast cancer | In vitro | Not clear | |
| Doxorubicin | 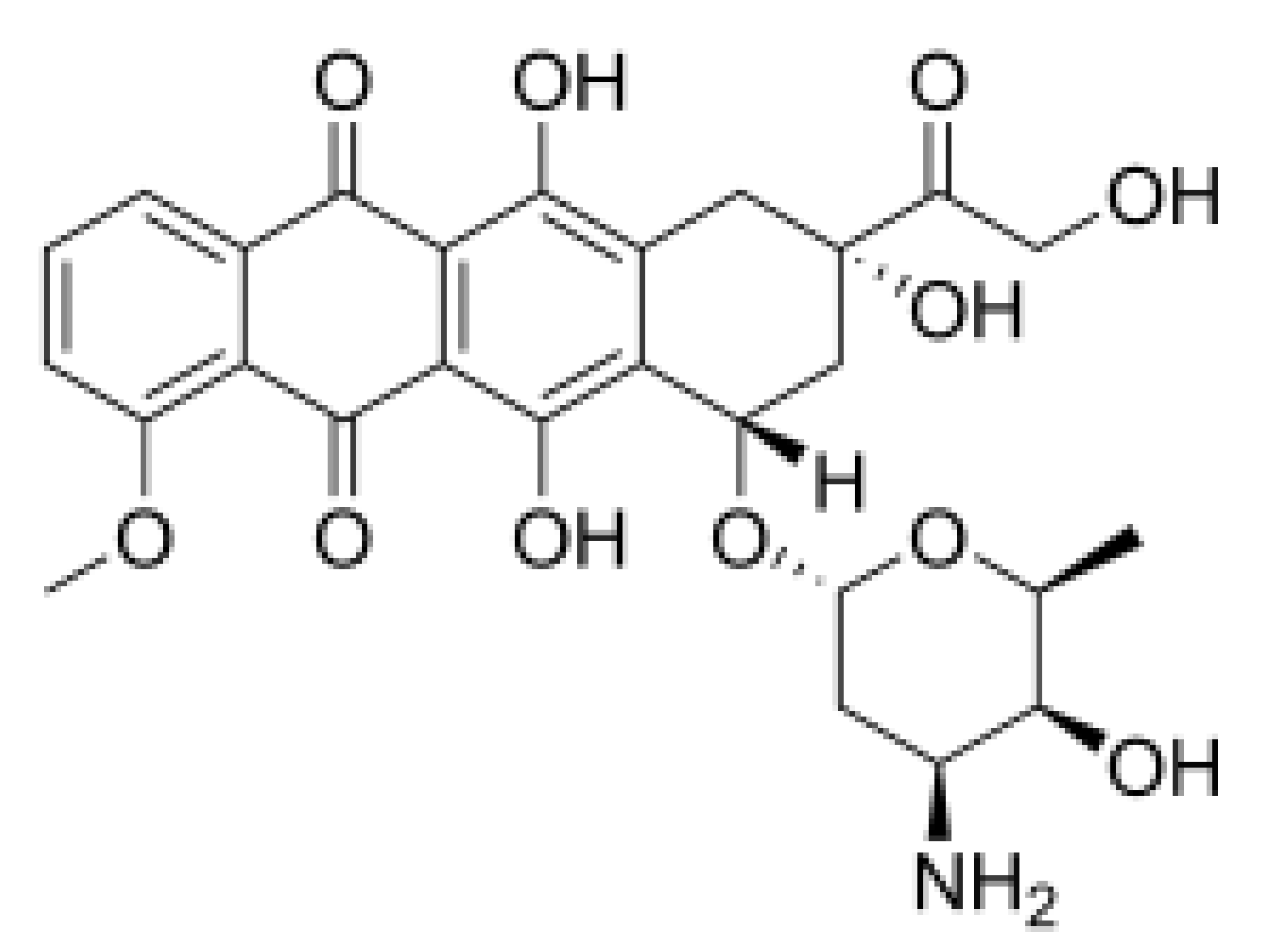 | Inhibition of ATPase activity | Oral squamous cell carcinoma | In vitro | Cardiotoxicity | |
| eIF4A | Compound 18 |  | Inhibition of ATPase activity | Exon junction complex | NA | IC50: 0.97 μmol/L |
| Silvestrol | 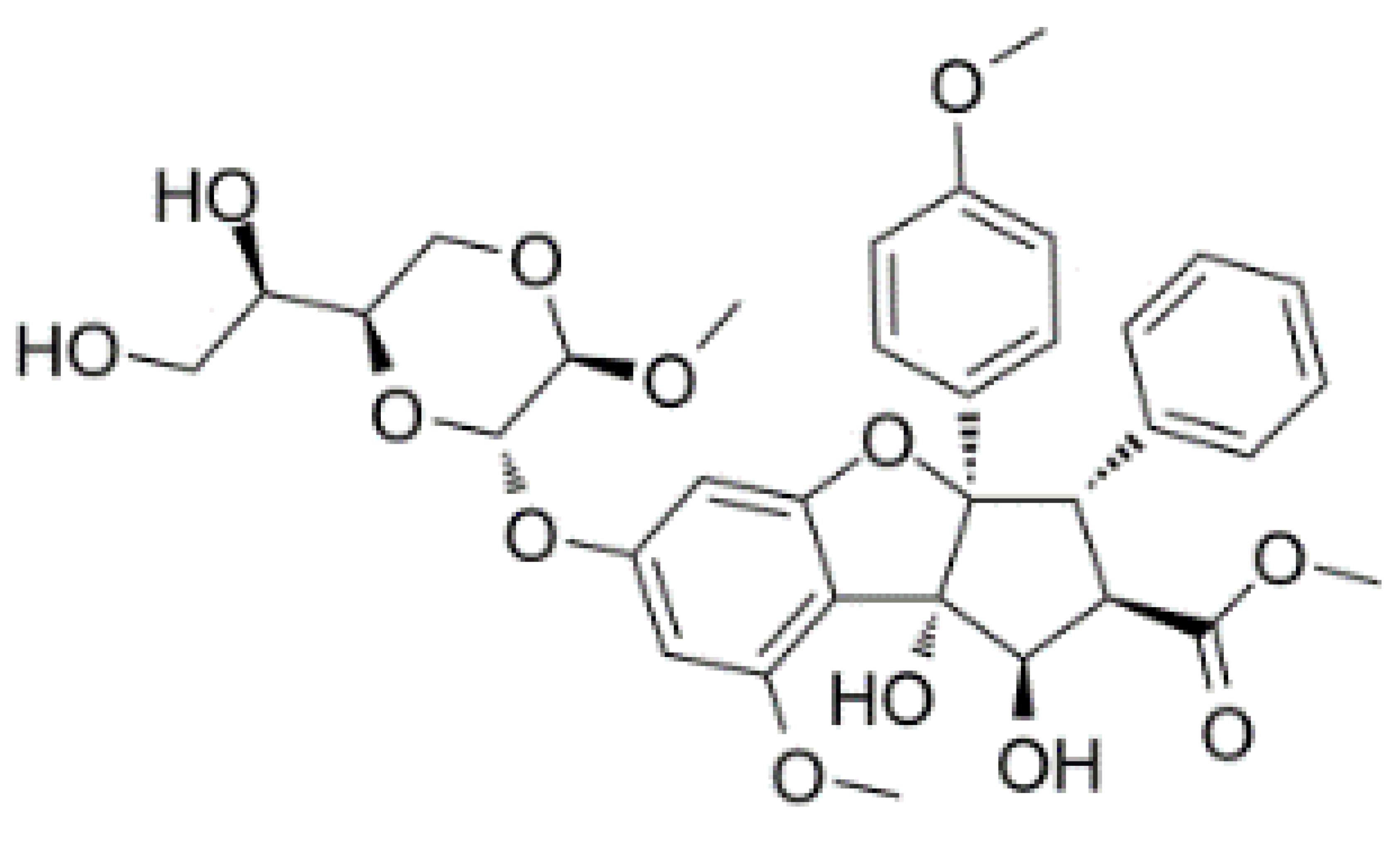 | Stabilization of RNA helicase onto RNA | Breast cancer | In vitro | Not toxic in vitro and in vivo at concentrations of effective activity | |
| Hippuristanol | 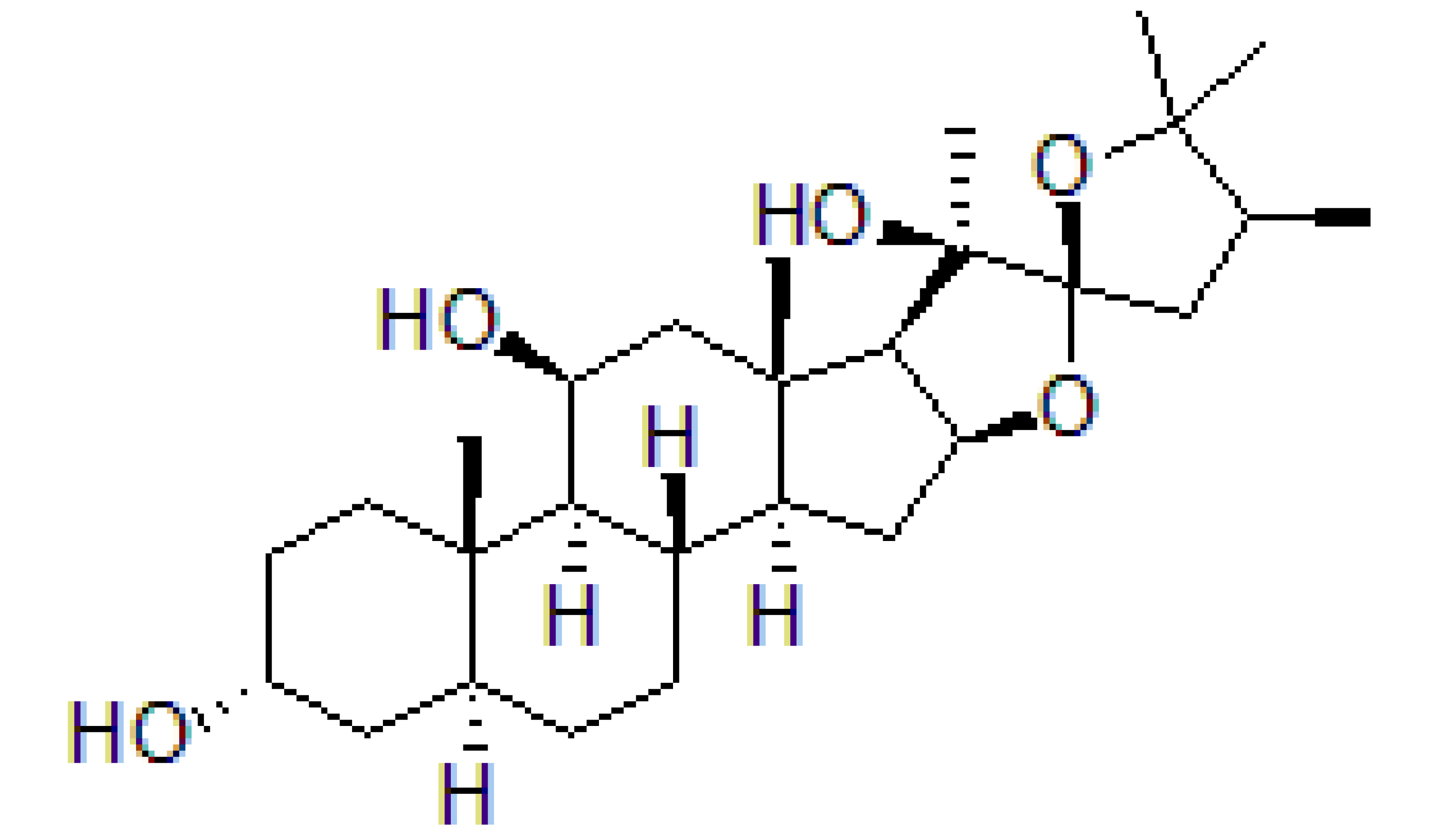 | Inhibition of helicase activity | Leukemia | In vitro | Not clear | |
| CR-1-31-B | 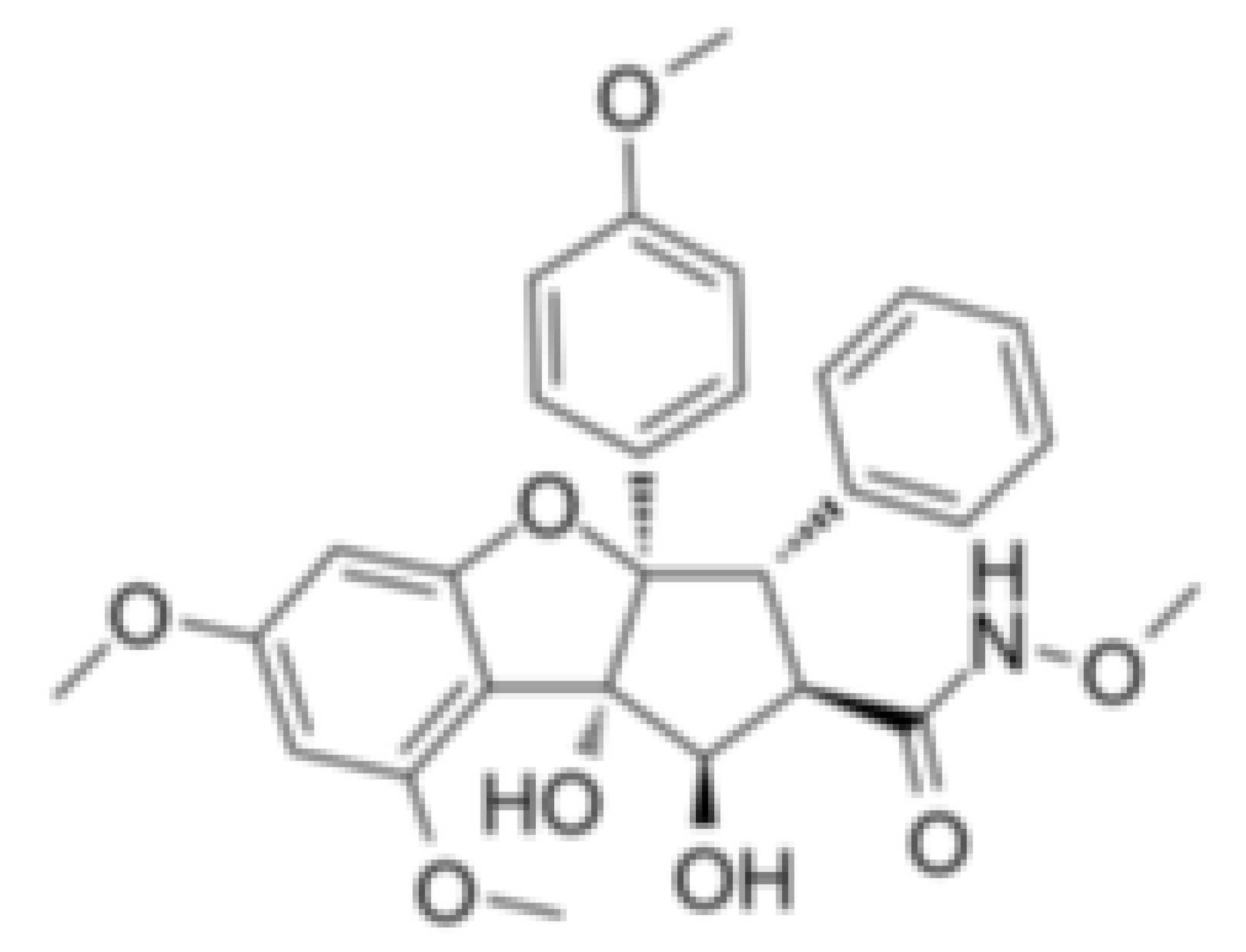 | Stabilization of RNA helicase onto RNA | Breast cancer | In vitro | Not clear | |
| Pateamine A | 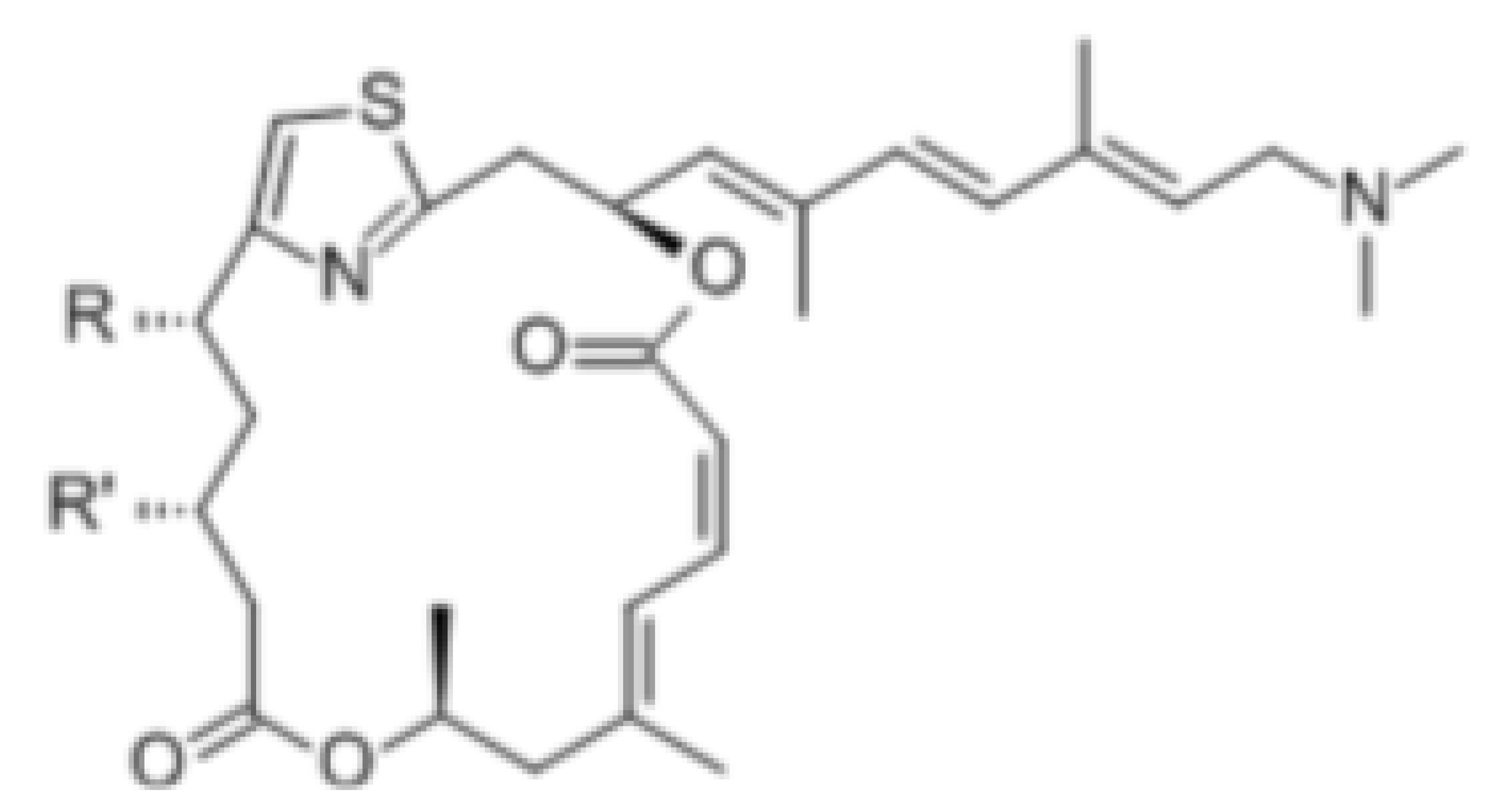 | Regulation of ATPase and RNA helicase activity | Melanoma | In vitro, and animal models | Low toxicity to quiescent cells | |
| DDX6 | RX-5902 |  | Inhibition of ATPase activity | TNBC | In vitro, preclinical models of TNBC, phase I study | Not clear |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Li, X. DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy. Cells 2021, 10, 1540. https://doi.org/10.3390/cells10061540
Zhang L, Li X. DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy. Cells. 2021; 10(6):1540. https://doi.org/10.3390/cells10061540
Chicago/Turabian StyleZhang, Lu, and Xiaogang Li. 2021. "DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy" Cells 10, no. 6: 1540. https://doi.org/10.3390/cells10061540
APA StyleZhang, L., & Li, X. (2021). DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy. Cells, 10(6), 1540. https://doi.org/10.3390/cells10061540





