Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom
Abstract
1. Introduction
2. Rho GTPase Family in Dictyostelium discoideum
3. A Phylogeny of the Rho GTPase Family in Amoebozoa
4. Comparative Analysis of the Rho Signalling in Dictyostelium and Mammalian Cells
4.1. Rac1 GTPases
4.1.1. WASP Family Proteins as Rac1 Effectors
4.1.2. Formins as Rac1 Effectors
4.1.3. Coronins as Rac1 Interactors
4.1.4. PAK Kinases as Rac1 Effectors
4.1.5. IQGAP-Related Proteins as Rac1 Effectors
4.1.6. Filamins as Rac1 Effectors
4.2. RacF1, RacF2, and RacB
4.3. RacA
4.4. RacC
4.5. RacH, RacD and RacP
4.6. RacE
4.7. Other Rho GTPases (G, L, I, J, M, N, O, and Q)
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABD | actin-binding domain |
| ABI | ABL interactor |
| Abp1 | actin-binding protein 1 |
| ACA | adenylyl cyclase A |
| AID | autoinhibitory domain |
| Arp2/3 | actin related protein 2/3 |
| BTB | broad-complex, tamtrack, bric à brac |
| CCP | clathrin-coated pit |
| CCV | clathrin-coated vesicle |
| CHD | calponin homology domain |
| CI | cortexillin I |
| CII | cortexillin II |
| CLIP-170 | cytoplasmic linker protein-170 |
| CME | clathrin-mediated endocytosis |
| CRIB | Cdc42/Rac interactive binding |
| CYRI | CYFIP-related Rac interactor |
| DAD | Diaphanous-autoregulatory domain |
| DD | dimerization domain |
| DID | Diaphanous-inhibitory domain |
| DRF | Diaphanous-related formin |
| DUF1394 | domain of unknown function |
| EGFR | epidermal growth factor receptor |
| ERK | extracellular signal-regulated kinase |
| FcγR | Fcγ receptor |
| FH1 | formin homology 1 |
| FH2 | formin homology 2 |
| FH3 | formin homology 3 |
| FRET | fluorescence resonance energy transfer |
| GAP | GTPase activating protein |
| GEF | guanine–nucleotide exchange factor |
| GBD | GTPase binding domain |
| GPCR | G protein-coupled receptor |
| GRD | GAP-related domain |
| HSPC300 | hematopoietic stem/progenitor cell protein 300 |
| JMY | junction-mediating and -regulatory protein |
| MEF | mouse embryonic fibroblast |
| MHCK | myosin II heavy chain kinase |
| MIHCK | myosin I heavy chain kinase |
| MLC | myosin II light chain |
| MLCK | MLC kinase |
| NAP1 | Nck-associated protein 1 |
| NPF | nucleation promoting factor |
| N-WASP | neural WASP |
| PAK | p21-activated kinase |
| PBD | p21 binding domain |
| PH | pleckstrin homology |
| PI(3,4)P2 | phosphatidylinositol (3,4)-bisphosphate |
| PI3K | phosphatidylinositol 3-kinase |
| PIP3 | phosphatidylinositol (3,4,5)-trisphosphate |
| PIR121 | 121F-specific p53 inducible RNA |
| PKB | protein kinase B |
| PRD | proline rich domain |
| REM | Rho effector homology |
| RGCT | RasGAP C-terminus |
| RhoGDI | Rho GDP-dissociation inhibitor |
| Rif | Rho in filopodia |
| RKH | ROK-kinectin homology |
| SRA1 | specifically RAC1-associated protein 1 |
| VEGF | vascular endothelial growth factor |
| VEGFR2 | VEGF receptor 2 |
| WASH | WASP and SCAR homologue |
| WASP | Wiskott–Aldrich syndrome protein |
| WAVE/SCAR | WASP family verprolin-homologous protein/suppressor of cAMP receptor |
| WCA | WASP-homology-2, also known as verprolin-homology, cofilin-homology, or connecting or central, acidic |
| WHAMM | WASP homolog associated with actin, membranes, and microtubules |
| WRC | WAVE regulatory complex |
| Y2H | yeast-two-hybrid |
References
- Ridley, A.J.; Hall, A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992, 70, 389–399. [Google Scholar] [CrossRef]
- Ridley, A.J.; Paterson, H.F.; Johnston, C.L.; Diekmann, D.; Hall, A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992, 70, 401–410. [Google Scholar] [CrossRef]
- Ridley, A.J. Historical Overview of Rho GTPases. Methods Mol. Biol. 2012, 827, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Boureux, A.; Vignal, E.; Faure, S.; Fort, P. Evolution of the Rho Family of Ras-Like GTPases in Eukaryotes. Mol. Biol. Evol. 2007, 24, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Eme, L.; Sharpe, S.C.; Brown, M.W.; Roger, A.J. On the Age of Eukaryotes: Evaluating Evidence from Fossils and Molecular Clocks. Cold Spring Harb. Perspect. Biol. 2014, 6, a016139. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Tice, A.K.; Spiegel, F.W.; Silberman, J.D.; Pánek, T.; Čepička, I.; Kostka, M.; Kosakyan, A.; Alcântara, D.M.C.; Roger, A.J.; et al. Between a Pod and a Hard Test: The Deep Evolution of Amoebae. Mol. Biol. Evol. 2017, 34, 2258–2270. [Google Scholar] [CrossRef]
- Artemenko, Y.; Lampert, T.J.; Devreotes, P.N. Moving towards a paradigm: Common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci. 2014, 71, 3711–3747. [Google Scholar] [CrossRef]
- Bozzaro, S.; Bucci, C.; Steinert, M. Phagocytosis and Host-Pathogen Interactions in Dictyostelium with a Look at Macrophages. Int. Rev. Cell Mol. Biol. 2008, 271, 253–300. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.D.; Bosmani, C.; Barisch, C.; Raykov, L.; Lefrancois, L.H.; Muñoz, E.C.; López-Jiménez, A.T.; Soldati, T. Eat Prey, Live: Dictyostelium discoideum as a Model for Cell-Autonomous Defenses. Front. Immunol. 2018, 8, 1906. [Google Scholar] [CrossRef]
- Dickinson, D.J.; Nelson, W.J.; Weis, W.I. Studying Epithelial Morphogenesis in Dictyostelium. Methods Mol. Biol. 2015, 1189, 267–281. [Google Scholar] [CrossRef]
- Loomis, W.F. Genetic control of morphogenesis in Dictyostelium. Dev. Biol. 2015, 402, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Weijer, C.J. Dictyostelium morphogenesis. Curr. Opin. Genet. Dev. 2004, 14, 392–398. [Google Scholar] [CrossRef]
- Alexandrova, A.Y.; Chikina, A.S.; Svitkina, T.M. Actin Cytoskeleton in Mesenchymal-to-Amoeboid Transition of Cancer Cells. Int. Rev. Cell Mol. Biol. 2020, 356, 197–256. [Google Scholar] [CrossRef] [PubMed]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef]
- King, J.S.; Kay, R.R. The origins and evolution of macropinocytosis. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180158. [Google Scholar] [CrossRef]
- Palm, W.; Thompson, C.B. Nutrient acquisition strategies of mammalian cells. Nature 2017, 546, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Stuelten, C.; Parent, C.A.; Montell, D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.D.; Paschke, P.I.; Kay, R.R. Function of small GTPases in Dictyostelium macropinocytosis. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180150. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Le Berre, M.; Lautenschläger, F.; Maiuri, P.; Callan-Jones, A.; Heuzé, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Bhattacharya, S.; Edwards, M.; Cai, H.; Inoue, T.; Iglesias, P.A.; Devreotes, P.N. Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nat. Cell Biol. 2017, 19, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Medalia, O.; Beck, M.; Ecke, M.; Weber, I.; Neujahr, R.; Baumeister, W.; Gerisch, G. Organization of Actin Networks in Intact Filopodia. Curr. Biol. 2007, 17, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Schirenbeck, A.; Bretschneider, T.; Arasada, R.; Schleicher, M.; Faix, J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 2005, 7, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Zatulovskiy, E.; Tyson, R.; Bretschneider, T.; Kay, R.R. Bleb-driven chemotaxis of Dictyostelium cells. J. Cell Biol. 2014, 204, 1027–1044. [Google Scholar] [CrossRef] [PubMed]
- Weber, I. Is there a pilot in a pseudopod? Eur. J. Cell Biol. 2006, 85, 915–924. [Google Scholar] [CrossRef]
- Cardelli, J. Phagocytosis and Macropinocytosis in Dictyostelium: Phosphoinositide-Based Processes, Biochemically Distinct. Traffic 2001, 2, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Rivero, F. Endocytosis and the Actin Cytoskeleton in Dictyostelium discoideum. Int. Rev. Cell Mol. Biol. 2008, 267, 343–397. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Pang, K.-M.; Knecht, D. The regulation of actin polymerization and cross-linking in Dictyostelium. Biochim. Biophys. Acta (BBA) Gen. Subj. 2001, 1525, 217–227. [Google Scholar] [CrossRef]
- Noegel, A.; Schleicher, M. The actin cytoskeleton of Dictyostelium: A story told by mutants. J. Cell Sci. 2000, 113 Pt 5, 759–766. [Google Scholar] [CrossRef]
- Rosenblum, E.B.; Parent, C.E.; Brandt, E.E. The Molecular Basis of Phenotypic Convergence. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 203–226. [Google Scholar] [CrossRef]
- Speed, M.P.; Arbuckle, K. Quantification provides a conceptual basis for convergent evolution. Biol. Rev. 2016, 92, 815–829. [Google Scholar] [CrossRef]
- Fort, P. Rho signaling: An historical and evolutionary perspective. In Rho Signaling: Molecular Biology in Health and Disease; Fort, P., Blangy, A., Eds.; World Scientific: Singapore, 2018; pp. 3–18. [Google Scholar]
- Eliáš, M.; Klimes, V. Rho GTPases: Deciphering the Evolutionary History of a Complex Protein Family. Methods Mol. Biol. 2012, 827, 13–34. [Google Scholar] [CrossRef]
- Beljan, S.; Bosnar, M.H.; Ćetković, H. Rho Family of Ras-Like GTPases in Early-Branching Animals. Cells 2020, 9, 2279. [Google Scholar] [CrossRef] [PubMed]
- Vlahou, G.; Rivero, F. Rho GTPase signaling in Dictyostelium discoideum: Insights from the genome. Eur. J. Cell Biol. 2006, 85, 947–959. [Google Scholar] [CrossRef]
- Le, Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.R. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef]
- Dever, T.E.; Glynias, M.J.; Merrick, W.C. GTP-binding domain: Three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. USA 1987, 84, 1814–1818. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Milburn, M.V.; Tong, L.; Devos, A.M.; Brunger, A.; Yamaizumi, Z.; Nishimura, S.; Kim, S.H. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science 1990, 247, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P. Fast-cycling Rho GTPases. Small GTPases 2020, 11, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Colicelli, J. Human RAS Superfamily Proteins and Related GTPases. Sci. STKE 2004, 2004, re13. [Google Scholar] [CrossRef] [PubMed]
- Dvorský, R.; Ahmadian, M.R. Always look on the bright site of Rho: Structural implications for a conserved intermolecular interface. EMBO Rep. 2004, 5, 1130–1136. [Google Scholar] [CrossRef]
- Schaefer, A.; Reinhard, N.R.; Hordijk, P.L. Toward understanding RhoGTPase specificity: Structure, function and local activation. Small GTPases 2014, 5, e968004. [Google Scholar] [CrossRef]
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef]
- Garcia-Mata, R.; Boulter, E.; Burridge, K. The ‘invisible hand’: Regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 2011, 12, 493–504. [Google Scholar] [CrossRef]
- Mott, H.; Owen, D. Structures of Ras superfamily effector complexes: What have we learnt in two decades? Crit. Rev. Biochem. Mol. Biol. 2015, 50, 85–133. [Google Scholar] [CrossRef]
- Bishop, A.L.; Hall, A. Rho GTPases and Their Effector Proteins. Biochem. J. 2000, 348 Pt 2, 241–255. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Drechsel, D.; Hall, A. A Conserved Binding Motif Defines Numerous Candidate Target Proteins for Both Cdc42 and Rac GTPases. J. Biol. Chem. 1995, 270, 29071–29074. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.D.; Ridley, A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2018, 217, 447–457. [Google Scholar] [CrossRef]
- Phuyal, S.; Farhan, H. Multifaceted Rho GTPase Signaling at the Endomembranes. Front. Cell Dev. Biol. 2019, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Sit, S.-T.; Manser, E. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Fey, P.; Jimenez-Morales, D.; Dodson, R.J.; Chisholm, R.L. dictyBase 2015: Expanding data and annotations in a new software environment. Genesis 2015, 53, 523–534. [Google Scholar] [CrossRef]
- Rivero, F.; Somesh, B.P. Signal transduction pathways regulated by Rho GTPases in Dictyostelium. J. Muscle Res. Cell Motil. 2002, 23, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Rivero, F.; Xiong, H. Rho Signaling in Dictyostelium discoideum. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Elsevier Inc.: Oxford, UK, 2016; Volume 322, pp. 61–181. [Google Scholar]
- Franco-Barraza, J.; Zamudio-Meza, H.; Franco, E.; Domínguez-Robles, M.D.C.; Villegas-Sepúlveda, N.; Meza, I. Rho signaling in Entamoeba histolytica modulates actomyosin-dependent activities stimulated during invasive behavior. Cell Motil. Cytoskelet. 2006, 63, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Lohia, A.; Samuelson, J. Heterogeneity of Entamoeba histolytica rac genes encoding p21rac homologues. Gene 1996, 173, 205–208. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Fiore-Donno, A.M.; Chao, E.; Kudryavtsev, A.; Berney, C.; Snell, E.A.; Lewis, R. Multigene phylogeny resolves deep branching of Amoebozoa. Mol. Phylogenet. Evol. 2015, 83, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Schilde, C.; Schaap, P. The Amoebozoa. In Dictyostelium Discoideum Protocols, 2nd ed.; Eichinger, L., Rivero, F., Eds.; Methods in Molecular Biology Series 983; Humana Press: Totowa, NJ, USA, 2013; pp. 1–15. [Google Scholar]
- Song, J.; Xu, Q.; Olsen, R.; Loomis, W.F.; Shaulsky, G.; Kuspa, A.; Sucgang, R. Comparing the Dictyostelium and Entamoeba Genomes Reveals an Ancient Split in the Conosa Lineage. PLoS Comput. Biol. 2005, 1, e71. [Google Scholar] [CrossRef]
- Godbold, G.D.; Corbett, K.D.; Mann, B.J. A Rho-like small GTPase of Entamoeba histolytica contains an unusual amino acid residue in a conserved GDP-stabilization region and is not a substrate for C3 exoenzyme. Exp. Parasitol. 2002, 101, 107–110. [Google Scholar] [CrossRef]
- Bosch, D.E.; Wittchen, E.S.; Qiu, C.; Burridge, K.; Siderovski, D. Unique Structural and Nucleotide Exchange Features of the Rho1 GTPase of Entamoeba histolytica. J. Biol. Chem. 2011, 286, 39236–39246. [Google Scholar] [CrossRef]
- Larochelle, D.A.; Vithalani, K.K.; De Lozanne, A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J. Cell Biol. 1996, 133, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jung, D.; Cao, Z.; Chung, C.Y. Adenylyl cyclase localization to the uropod of aggregating Dictyostelium cells requires RacC. Biochem. Biophys. Res. Commun. 2015, 465, 613–619. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Bush, J.; Franek, K.; Cardelli, J. Cloning and characterization of seven novel Dictyostelium discoideum rac-related genes belonging to the rho family of GTPases. Gene 1993, 136, 61–68. [Google Scholar] [CrossRef]
- Santhanam, B.; Cai, H.; Devreotes, P.N.; Shaulsky, G.; Katoh-Kurasawa, M. The GATA transcription factor GtaC regulates early developmental gene expression dynamics in Dictyostelium. Nat. Commun. 2015, 6, 7551. [Google Scholar] [CrossRef] [PubMed]
- Stajdohar, M.; Jeran, L.; Kokosar, J.; Blenkus, D.; Janez, T.; Kuspa, A.; Shaulsky, G.; Zupan, B. DictyExpress: Visual Analytics of NGS Gene Expression in Dictyostelium. 2015. Available online: https://www.dictyexpress.org (accessed on 4 February 2021).
- Muramoto, T.; Urushihara, H. Small GTPase RacF2 affects sexual cell fusion and asexual development in Dictyostelium discoideum through the regulation of cell adhesion. Dev. Growth Differ. 2006, 48, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Dumontier, M.; Hocht, P.; Mintert, U.; Faix, J. Rac1 GTPases control filopodia formation, cell motility, endocytosis, cytokinesis and development in Dictyostelium. J. Cell Sci. 2000, 113 Pt 12, 2253–2265. [Google Scholar] [CrossRef]
- Palmieri, S.J.; Nebl, T.; Pope, R.K.; Seastone, D.J.; Lee, E.; Hinchcliffe, E.H.; Sluder, G.; Knecht, D.; Cardelli, J.; Luna, E.J. Mutant Rac1B expression in Dictyostelium: Effects on morphology, growth, endocytosis, development, and the actin cytoskeleton. Cell Motil. Cytoskelet. 2000, 46, 285–304. [Google Scholar] [CrossRef]
- Wang, Y.; Senoo, H.; Sesaki, H.; Iijima, M. Rho GTPases orient directional sensing in chemotaxis. Proc. Natl. Acad. Sci. USA 2013, 110, E4723–E4732. [Google Scholar] [CrossRef]
- Cotteret, S.; Chernoff, J. The evolutionary history of effectors downstream of Cdc42 and Rac. Genome Biol. 2002, 3, REVIEWS0002. [Google Scholar] [CrossRef]
- Chung, C.Y.; Feoktistov, A.; Hollingsworth, R.J.; Rivero, F.; Mandel, N.S. An attenuating role of a WASP-related protein, WASP-B, in the regulation of F-actin polymerization and pseudopod formation via the regulation of RacC during Dictyostelium chemotaxis. Biochem. Biophys. Res. Commun. 2013, 436, 719–724. [Google Scholar] [CrossRef] [PubMed]
- De La Roche, M.; Mahasneh, A.; Lee, S.-F.; Rivero, F.; Côté, G.P. Cellular Distribution and Functions of Wild-Type and Constitutively Activated Dictyostelium PakB. Mol. Biol. Cell 2005, 16, 238–247. [Google Scholar] [CrossRef][Green Version]
- Han, J.W.; Leeper, L.; Rivero, F.; Chung, C.Y. Role of RacC for the Regulation of WASP and Phosphatidylinositol 3-Kinase during Chemotaxis of Dictyostelium. J. Biol. Chem. 2006, 281, 35224–35234. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Rivero, F.; Park, K.C.; Huang, E.; Funamoto, S.; Firtel, R.A. Dictyostelium PAKc Is Required for Proper Chemotaxis. Mol. Biol. Cell 2004, 15, 5456–5469. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Rivero, F.; Meili, R.; Lee, S.; Apone, F.; Firtel, R.A. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004, 23, 4177–4189. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Müller-Taubenberger, A.; Faix, J.; Rivero, F.; Noegel, A.A. A Cdc42- and Rac-interactive binding (CRIB) domain mediates functions of coronin. Proc. Natl. Acad. Sci. USA 2014, 111, E25–E33. [Google Scholar] [CrossRef] [PubMed]
- Mullins, R.D.; Heuser, J.A.; Pollard, T.D. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 1998, 95, 6181–6186. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Borisy, G.G. Cellular Motility Driven by Assembly and Disassembly of Actin Filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Burianek, L.E.; Soderling, S.H. Under lock and key: Spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol. 2013, 24, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.S.; Kakalis, L.T.; Abdul-Manan, N.; Liu, G.A.; Rosen, M.K. Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature 2000, 404, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Sasaki, T.; Takai, Y.; Takenawa, T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 1998, 391, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Ma, L.; Miki, H.; Lopez, M.; Kirchhausen, T.; Takenawa, T.; Kirschner, M.W. The Interaction between N-WASP and the Arp2/3 Complex Links Cdc42-Dependent Signals to Actin Assembly. Cell 1999, 97, 221–231. [Google Scholar] [CrossRef]
- Rotty, J.D.; Wu, C.; Bear, J.E. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 2012, 14, 7–12. [Google Scholar] [CrossRef]
- Tomasevic, N.; Jia, Z.; Russell, A.; Fujii, T.; Hartman, J.J.; Clancy, S.; Wang, M.; Beraud, C.; Wood, K.W.; Sakowicz, R. Differential Regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry 2007, 46, 3494–3502. [Google Scholar] [CrossRef]
- Bear, J.E.; Rawls, J.; Saxe, C.L. SCAR, a WASP-related Protein, Isolated as a Suppressor of Receptor Defects in Late Dictyostelium Development. J. Cell Biol. 1998, 142, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Suetsugu, S.; Takenawa, T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998, 17, 6932–6941. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Borek, D.; Padrick, S.; Gomez, T.S.; Metlagel, Z.; Ismail, A.M.; Umetani, J.; Billadeau, D.D.; Otwinowski, Z.; Rosen, M.K. Structure and control of the actin regulatory WAVE complex. Nature 2010, 468, 533–538. [Google Scholar] [CrossRef]
- Eden, S.; Rohatgi, R.; Podtelejnikov, A.V.; Mann, M.; Kirschner, M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 2002, 418, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Gautreau, A.; Ho, H.-Y.H.; Li, J.; Steen, H.; Gygi, S.P.; Kirschner, M.W. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 2004, 101, 4379–4383. [Google Scholar] [CrossRef] [PubMed]
- Stovold, C.F.; Millard, T.H.; Machesky, L.M. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 2005, 6, 11. [Google Scholar] [CrossRef]
- Innocenti, M.; Zucconi, A.; Disanza, A.; Frittoli, E.; Areces, L.B.; Steffen, A.; Stradal, T.; Di Fiore, P.P.; Carlier, M.-F.; Scita, G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nature 2004, 6, 319–327. [Google Scholar] [CrossRef]
- Kim, Y.; Sung, J.Y.; Ceglia, I.; Lee, K.-W.; Ahn, J.-H.; Halford, J.M.; Kim, A.M.; Kwak, S.P.; Park, J.B.; Ryu, S.H.; et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 2006, 442, 814–817. [Google Scholar] [CrossRef]
- Suetsugu, S.; Kurisu, S.; Oikawa, T.; Yamazaki, D.; Oda, A.; Takenawa, T. Optimization of WAVE2 complex–induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J. Cell Biol. 2006, 173, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Derivery, E.; Lombard, B.; Loew, D.; Gautreau, A. The Wave complex is intrinsically inactive. Cell Motil. Cytoskelet. 2009, 66, 777–790. [Google Scholar] [CrossRef]
- Ismail, A.M.; Padrick, S.; Chen, B.; Umetani, J.; Rosen, M.K. The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 2009, 16, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Lebensohn, A.M.; Kirschner, M.W. Activation of the WAVE Complex by Coincident Signals Controls Actin Assembly. Mol. Cell 2009, 36, 512–524. [Google Scholar] [CrossRef]
- Oikawa, T.; Yamaguchi, H.; Itoh, T.; Kato, M.; Ijuin, T.; Yamazaki, D.; Suetsugu, S.; Takenawa, T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nature 2004, 6, 420–426. [Google Scholar] [CrossRef]
- Myers, S.A.; Han, J.W.; Lee, Y.; Firtel, R.A.; Chung, C.Y. ADictyosteliumHomologue of WASP Is Required for Polarized F-Actin Assembly during Chemotaxis. Mol. Biol. Cell 2005, 16, 2191–2206. [Google Scholar] [CrossRef]
- Veltman, D.M.; Insall, R.H. WASP Family Proteins: Their Evolution and Its Physiological Implications. Mol. Biol. Cell 2010, 21, 2880–2893. [Google Scholar] [CrossRef]
- Carnell, M.; Zech, T.; Calaminus, S.D.; Ura, S.; Hagedorn, M.; Johnston, S.A.; May, R.C.; Soldati, T.; Machesky, L.M.; Insall, R.H. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 2011, 193, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, C.J.; Qualmann, B.; Kessels, M.M.; Almers, W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur. J. Cell Biol. 2004, 83, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Veltman, D.M.; Auciello, G.; Spence, H.J.; Machesky, L.M.; Rappoport, J.Z.; Insall, R.H. Functional analysis of Dictyostelium IBARa reveals a conserved role of the I-BAR domain in endocytosis. Biochem. J. 2011, 436, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Veltman, D.M.; King, J.; Machesky, L.M.; Insall, R.H. SCAR knockouts in Dictyostelium: WASP assumes SCAR’s position and upstream regulators in pseudopods. J. Cell Biol. 2012, 198, 501–508. [Google Scholar] [CrossRef]
- Benesch, S.; Polo, S.; Lai, F.P.L.; Anderson, K.I.; Stradal, T.; Wehland, J.; Rottner, K. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J. Cell Sci. 2005, 118, 3103–3115. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Amato, C.; Thomason, P.A.; Insall, R.H. WASP family proteins and formins compete in pseudopod- and bleb-based migration. J. Cell Biol. 2018, 217, 701–714. [Google Scholar] [CrossRef]
- Amato, C.; Thomason, P.; Davidson, A.J.; Swaminathan, K.; Ismail, S.; Machesky, L.M.; Insall, R.H. WASP Restricts Active Rac to Maintain Cells’ Front-Rear Polarization. Curr. Biol. 2019, 29, 4169–4182.e4. [Google Scholar] [CrossRef] [PubMed]
- Hinze, C.; Boucrot, E. Local actin polymerization during endocytic carrier formation. Biochem. Soc. Trans. 2018, 46, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, M.; Gerboth, S.; Rottner, K.; Lai, F.P.L.; Hertzog, M.; Stradal, T.; Frittoli, E.; Didry, D.; Polo, S.; Disanza, A.; et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005, 7, 969–976. [Google Scholar] [CrossRef]
- Croisé, P.; Estay-Ahumada, C.; Gasman, S.; Ory, S. Rho GTPases, phosphoinositides, and actin: A Tripartite Framework for Efficient Vesicular Trafficking. Small GTPases 2014, 5, e29469. [Google Scholar] [CrossRef]
- Lamaze, C.; Chuang, T.-H.; Terlecky, L.J.; Bokoch, G.M.; Schmid, S.L. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature 1996, 382, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Campellone, K.G.; Welch, M.D. A Nucleator Arms Race: Cellular Control of Actin Assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef]
- Swaminathan, K.; Stumpf, M.; Müller, R.; Horn, A.-C.; Schmidbauer, J.; Eichinger, L.; Müller-Taubenberger, A.; Faix, J.; Noegel, A.A. Coronin7 regulates WASP and SCAR through CRIB mediated interaction with Rac proteins. Sci. Rep. 2015, 5, 14437. [Google Scholar] [CrossRef]
- Filić, V.; Marinović, M.; Faix, J.; Weber, I. A dual role for Rac1 GTPases in the regulation of cell motility. J. Cell Sci. 2012, 125, 387–398. [Google Scholar] [CrossRef]
- Niedergang, F.; Grinstein, S. How to build a phagosome: New concepts for an old process. Curr. Opin. Cell Biol. 2018, 50, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Cox, D. Cdc42 Regulates FcγReceptor-mediated Phagocytosis through the Activation and Phosphorylation of Wiskott-Aldrich Syndrome Protein (WASP) and Neural-WASP. Mol. Biol. Cell 2009, 20, 4500–4508. [Google Scholar] [CrossRef]
- Blagg, S.L.; Stewart, M.; Sambles, C.; Insall, R.H. PIR121 Regulates Pseudopod Dynamics and SCAR Activity in Dictyostelium. Curr. Biol. 2003, 13, 1480–1487. [Google Scholar] [CrossRef]
- Caracino, D.; Jones, C.; Compton, M.; Saxe, C.L. The N-Terminus ofDictyosteliumScar Interacts with Abi and HSPC300 and Is Essential for Proper Regulation and Function. Mol. Biol. Cell 2007, 18, 1609–1620. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ibarra, N.; Blagg, S.L.; Vazquez, F.; Insall, R. Nap1 Regulates Dictyostelium Cell Motility and Adhesion through SCAR-Dependent and -Independent Pathways. Curr. Biol. 2006, 16, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Thomason, P.A.; Lilla, S.; Schaks, M.; Tang, Q.; Goode, B.L.; Machesky, L.M.; Rottner, K.; Insall, R.H. Cell–substrate adhesion drives Scar/WAVE activation and phosphorylation by a Ste20-family kinase, which controls pseudopod lifetime. PLoS Biol. 2020, 18, e3000774. [Google Scholar] [CrossRef]
- Chen, B.; Chou, H.-T.; Brautigam, C.A.; Xing, W.; Yang, S.; Henry, L.; Doolittle, L.K.; Walz, T.; Rosen, M.K. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 2017, 6, 6. [Google Scholar] [CrossRef]
- Schaks, M.; Singh, S.P.; Kage, F.; Thomason, P.; Klünemann, T.; Steffen, A.; Blankenfeldt, W.; Stradal, T.E.; Insall, R.H.; Rottner, K. Distinct Interaction Sites of Rac GTPase with WAVE Regulatory Complex Have Non-Redundant Functions In Vivo. Curr. Biol. 2018, 28, 3674–3684.e6. [Google Scholar] [CrossRef]
- Marinović, M.; Šoštar, M.; Filić, V.; Antolović, V.; Weber, I. Quantitative imaging of Rac1 activity in Dictyostelium cells with a fluorescently labelled GTPase-binding domain from DPAKa kinase. Histochem. Cell Biol. 2016, 146, 267–279. [Google Scholar] [CrossRef]
- Veltman, D.M.; Williams, T.D.; Bloomfield, G.; Chen, B.-C.; Betzig, E.; Insall, R.; Kay, R.R. A plasma membrane template for macropinocytic cups. eLife 2016, 5, e20085. [Google Scholar] [CrossRef] [PubMed]
- King, J.S.; Veltman, D.M.; Georgiou, M.; Baum, B.; Insall, R. SCAR/WAVE is activated at mitosis and drives myosin-independent cytokinesis. J. Cell Sci. 2010, 123, 2246–2255. [Google Scholar] [CrossRef]
- Machesky, L.M.; Mullins, R.D.; Higgs, H.N.; Kaiser, D.A.; Blanchoin, L.; May, R.C.; Hall, M.E.; Pollard, T.D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 1999, 96, 3739–3744. [Google Scholar] [CrossRef]
- Litschko, C.; Linkner, J.; Brühmann, S.; Stradal, T.E.; Reinl, T.; Jänsch, L.; Rottner, K.; Faix, J. Differential functions of WAVE regulatory complex subunits in the regulation of actin-driven processes. Eur. J. Cell Biol. 2017, 96, 715–727. [Google Scholar] [CrossRef]
- Seastone, D.J.; Harris, E.; Temesvari, L.A.; Bear, J.E.; Saxe, C.L.; Cardelli, J. The WASp-like protein Scar regulates macropinocytosis, phagocytosis and endosomal membrane flow in Dictyostelium. J. Cell Sci. 2001, 114, 2673–2683. [Google Scholar] [CrossRef] [PubMed]
- Ura, S.; Pollitt, A.Y.; Veltman, D.M.; Morrice, N.A.; Machesky, L.M.; Insall, R.H. Pseudopod Growth and Evolution during Cell Movement Is Controlled through SCAR/WAVE Dephosphorylation. Curr. Biol. 2012, 22, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Hahne, P.; Sechi, A.; Benesch, S.; Small, J. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 2001, 492, 215–220. [Google Scholar] [CrossRef]
- Suetsugu, S.; Yamazaki, D.; Kurisu, S.; Takenawa, T. Differential Roles of WAVE1 and WAVE2 in Dorsal and Peripheral Ruffle Formation for Fibroblast Cell Migration. Dev. Cell 2003, 5, 595–609. [Google Scholar] [CrossRef]
- Yamazaki, D.; Suetsugu, S.; Miki, H.; Kataoka, Y.; Nishikawa, S.-I.; Fujiwara, T.; Yoshida, N.; Takenawa, T. WAVE2 is required for directed cell migration and cardiovascular development. Nature 2003, 424, 452–456. [Google Scholar] [CrossRef]
- Steffen, A.; Rottner, K.; Ehinger, J.; Innocenti, M.; Scita, G.; Wehland, J.; Stradal, T.E.B. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004, 23, 749–759. [Google Scholar] [CrossRef]
- Yan, C.; Martinez-Quiles, N.; Eden, S.; Shibata, T.; Takeshima, F.; Shinkura, R.; Fujiwara, Y.; Bronson, R.; Snapper, S.B.; Kirschner, M.W.; et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003, 22, 3602–3612. [Google Scholar] [CrossRef]
- Steffen, A.; Ladwein, M.; Dimchev, G.A.; Hein, A.; Schwenkmezger, L.; Arens, S.; Ladwein, K.I.; Holleboom, J.M.; Schur, F.; Small, J.V.; et al. Rac function is critical for cell migration but not required for spreading and focal adhesion formation. J. Cell Sci. 2013, 126, 4572–4588. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Lord, S.J.; Mullins, R.D. WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. J. Cell Biol. 2017, 216, 1673–1688. [Google Scholar] [CrossRef] [PubMed]
- Lommel, S.; Benesch, S.; Rottner, K.; Franz, T.; Wehland, J.; Kühn, R. Actin pedestal formation by enteropathogenicEscherichia coliand intracellular motility ofShigella flexneriare abolished in N-WASP-defective cells. EMBO Rep. 2001, 2, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, C.; Wang, W.; Dovas, A.; Yamaguchi, H.; Sidani, M.; El-Sibai, M.; Desmarais, V.; Holman, H.A.; Kitchen, S.; Backer, J.M.; et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol. 2008, 180, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Snapper, S.B.; Takeshima, F.; Antón, I.; Liu, C.-H.; Thomas, S.M.; Nguyen, D.; Dudley, D.; Fraser, H.; Purich, D.; Lopez-Ilasaca, M.; et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 2001, 3, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Badolato, R.; Sozzani, S.; Malacarne, F.; Bresciani, S.; Fiorini, M.; Borsatti, A.; Albertini, A.; Mantovani, A.; Ugazio, A.G.; Notarangelo, L.D. Monocytes from Wiskott-Aldrich patients display reduced chemotaxis and lack of cell polarization in response to monocyte chemoattractant protein-1 and formyl-methionyl-leucyl-phenylalanine. J. Immunol. 1998, 161, 1026–1033. [Google Scholar] [PubMed]
- Burns, S.; Thrasher, A.J.; Blundell, M.P.; Machesky, L.; Jones, G.E. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood 2001, 98, 1142–1149. [Google Scholar] [CrossRef]
- Ishihara, D.; Dovas, A.; Park, H.; Isaac, B.M.; Cox, D. The Chemotactic Defect in Wiskott-Aldrich Syndrome Macrophages Is Due to the Reduced Persistence of Directional Protrusions. PLoS ONE 2012, 7, e30033. [Google Scholar] [CrossRef]
- Snapper, S.B.; Meelu, P.; Nguyen, D.; Stockton, B.M.; Bozza, P.; Alt, F.W.; Rosen, F.S.; Von Andrian, U.H.; Klein, C. WASP deficiency leads to global defects of directed leukocyte migration in vitro and in vivo. J. Leukoc. Biol. 2005, 77, 993–998. [Google Scholar] [CrossRef]
- Fort, L.; Batista, J.M.; Thomason, P.A.; Spence, H.J.; Whitelaw, J.A.; Tweedy, L.; Greaves, J.; Martin, K.J.; Anderson, K.I.; Brown, P.; et al. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell Biol. 2018, 20, 1159–1171. [Google Scholar] [CrossRef]
- Yagi, S.; Matsuda, M.; Kiyokawa, E. Suppression of Rac1 activity at the apical membrane of MDCK cells is essential for cyst structure maintenance. EMBO Rep. 2012, 13, 237–243. [Google Scholar] [CrossRef]
- Steffen, A.; Faix, J.; Resch, G.P.; Linkner, J.; Wehland, J.; Small, J.V.; Rottner, K.; Stradal, T.E. Filopodia Formation in the Absence of Functional WAVE- and Arp2/3-Complexes. Mol. Biol. Cell 2006, 17, 2581–2591. [Google Scholar] [CrossRef]
- Pruyne, D.; Evangelista, M.; Yang, C.; Bi, E.; Zigmond, S.; Bretscher, A.; Boone, C. Role of Formins in Actin Assembly: Nucleation and Barbed-End Association. Science 2002, 297, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Kovar, D.R.; Harris, E.S.; Mahaffy, R.; Higgs, H.N.; Pollard, T.D. Control of the Assembly of ATP- and ADP-Actin by Formins and Profilin. Cell 2006, 124, 423–435. [Google Scholar] [CrossRef]
- Sagot, I.; Rodal, A.A.; Moseley, J.B.; Goode, B.L.; Pellman, D. An actin nucleation mechanism mediated by Bni1 and Profilin. Nat. Cell Biol. 2002, 4, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Courtemanche, N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018, 10, 1553–1569. [Google Scholar] [CrossRef]
- Faix, J.; Grosse, R. Staying in Shape with Formins. Dev. Cell 2006, 10, 693–706. [Google Scholar] [CrossRef]
- Pruyne, D. Revisiting the Phylogeny of the Animal Formins: Two New Subtypes, Relationships with Multiple Wing Hairs Proteins, and a Lost Human Formin. PLoS ONE 2016, 11, e0164067. [Google Scholar] [CrossRef]
- Kühn, S.; Geyer, M. Formins as effector proteins of Rho GTPases. Small GTPases 2014, 5, e983876. [Google Scholar] [CrossRef] [PubMed]
- Rivero, F.; Muramoto, T.; Meyer, A.-K.; Urushihara, H.; Uyeda, T.Q.P.; Kitayama, C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genom. 2005, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, S.; Schultz, J.; Grosshans, J. Formin’ cellular structures: Physiological Roles of Diaphanous (Dia) in Actin Dynamics. Commun. Integr. Biol. 2013, 6, e27634. [Google Scholar] [CrossRef] [PubMed]
- Gallop, J. Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell Dev. Biol. 2020, 102, 81–89. [Google Scholar] [CrossRef]
- Goh, W.I.; Sudhaharan, T.; Lim, K.B.; Sem, K.P.; Lau, C.L.; Ahmed, S. Rif-mDia1 Interaction Is Involved in Filopodium Formation Independent of Cdc42 and Rac Effectors. J. Biol. Chem. 2011, 286, 13681–13694. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.I.; Lim, K.B.; Sudhaharan, T.; Sem, K.P.; Bu, W.; Chou, A.M.; Ahmed, S. mDia1 and WAVE2 Proteins Interact Directly with IRSp53 in Filopodia and Are Involved in Filopodium Formation. J. Biol. Chem. 2012, 287, 4702–4714. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Czech, L.; Gerboth, S.; Kojima, S.-I.; Scita, G.; Svitkina, T. Novel Roles of Formin mDia2 in Lamellipodia and Filopodia Formation in Motile Cells. PLoS Biol. 2007, 5, e317. [Google Scholar] [CrossRef]
- Mellor, H. The Role of Formins in Filopodia Formation. Biochim. Biophys. Acta 2010, 1803, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Alberts, A.S.; Bouquin, N.; Johnston, L.H.; Treisman, R. Analysis of RhoA-binding Proteins Reveals an Interaction Domain Conserved in Heterotrimeric G Protein β Subunits and the Yeast Response Regulator Protein Skn7. J. Biol. Chem. 1998, 273, 8616–8622. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Wallar, B.J.; Flanders, A.; Swiatek, P.J.; Alberts, A.S. Disruption of the Diaphanous-Related Formin Drf1 Gene Encoding mDia1 Reveals a Role for Drf3 as an Effector for Cdc42. Curr. Biol. 2003, 13, 534–545. [Google Scholar] [CrossRef]
- Ellis, S.; Mellor, H. The novel Rho-family GTPase Rif regulates coordinated actin-based membrane rearrangements. Curr. Biol. 2000, 10, 1387–1390. [Google Scholar] [CrossRef]
- Pellegrin, S.; Mellor, H. The Rho Family GTPase Rif Induces Filopodia through mDia2. Curr. Biol. 2005, 15, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Marshall, T.W.; Uetrecht, A.C.; Schafer, D.A.; Bear, J.E. Coronin 1B Coordinates Arp2/3 Complex and Cofilin Activities at the Leading Edge. Cell 2007, 128, 915–929. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Makhov, A.M.; Schafer, D.A.; Bear, J.E. Coronin 1B Antagonizes Cortactin and Remodels Arp2/3-Containing Actin Branches in Lamellipodia. Cell 2008, 134, 828–842. [Google Scholar] [CrossRef]
- Chan, K.T.; Creed, S.J.; Bear, J.E. Unraveling the enigma: Progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011, 21, 481–488. [Google Scholar] [CrossRef] [PubMed]
- De Hostos, E.; Bradtke, B.; Lottspeich, F.; Guggenheim, R.; Gerisch, G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 1991, 10, 4097–4104. [Google Scholar] [CrossRef]
- Shina, M.C.; Ünal, C.; Eichinger, L.; Müller-Taubenberger, A.; Schleicher, M.; Steinert, M.; Noegel, A.A. A Coronin7 Homolog with Functions in Actin-driven Processes. J. Biol. Chem. 2010, 285, 9249–9261. [Google Scholar] [CrossRef] [PubMed]
- De Hostos, E.L.; Rehfuess, C.; Bradtke, B.; Waddell, D.R.; Albrecht, R.; Murphy, J.; Gerisch, G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 1993, 120, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Maniak, M.; Rauchenberger, R.; Albrecht, R.; Murphy, J.; Gerisch, G. Coronin involved in phagocytosis: Dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell 1995, 83, 915–924. [Google Scholar] [CrossRef]
- Shina, M.C.; Müller-Taubenberger, A.; Unal, C.; Schleicher, M.; Steinert, M.; Eichinger, L.; Müller, R.; Blau-Wasser, R.; Glöckner, G.; Noegel, A.A. Redundant and unique roles of coronin proteins in Dictyostelium. Cell. Mol. Life Sci. 2010, 68, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Vinet, A.F.; Fiedler, T.; Studer, V.; Froquet, R.; Dardel, A.; Cosson, P.; Pieters, J. Initiation of multicellular differentiation in Dictyostelium discoideum is regulated by coronin A. Mol. Biol. Cell 2014, 25, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.-P.; Eichinger, L.; Fernandez, M.P.; Morgan, R.O.; Clemen, C.S. Evolutionary and Functional Diversity of Coronin Proteins. Subcell. Biochem. 2008, 48, 98–109. [Google Scholar] [CrossRef]
- Williamson, R.C.; Cowell, C.A.M.; Hammond, C.L.; Bergen, D.J.M.; Roper, J.A.; Feng, Y.; Rendall, T.; Race, P.R.; Bass, M.D. Coronin-1C and RCC2 guide mesenchymal migration by trafficking Rac1 and controlling GEF exposure. J. Cell Sci. 2014, 127, 4292–4307. [Google Scholar] [CrossRef]
- Ojeda, V.; Castro-Castro, A.; Bustelo, X.R. Coronin1 Proteins Dictate Rac1 Intracellular Dynamics and Cytoskeletal Output. Mol. Cell. Biol. 2014, 34, 3388–3406. [Google Scholar] [CrossRef]
- Castro-Castro, A.; Ojeda, V.; Barreira, M.; Sauzeau, V.; Navarro-Lérida, I.; Muriel, O.; Couceiro, J.R.; Pimentel-Muíños, F.X.; Del Pozo, M.A.; Bustelo, X.R. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011, 30, 3913–3927. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, K.; Swaminathan, K.; Peche, V.S.; Clemen, C.S.; Knyphausen, P.; Lammers, M.; Noegel, A.A.; Rastetter, R.H. Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology. Sci. Rep. 2016, 6, 25411. [Google Scholar] [CrossRef]
- Rybakin, V.; Stumpf, M.; Schulze, A.; Majoul, I.V.; Noegel, A.A.; Hasse, A. Coronin 7, the mammalian POD-1 homologue, localizes to the Golgi apparatus. FEBS Lett. 2004, 573, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Rybakin, V.; Gounko, N.; Späte, K.; Höning, S.; Majoul, I.V.; Duden, R.; Noegel, A.A. Crn7 Interacts with AP-1 and Is Required for the Maintenance of Golgi Morphology and Protein Export from the Golgi. J. Biol. Chem. 2006, 281, 31070–31078. [Google Scholar] [CrossRef]
- Bokoch, G.M. Biology of the p21-Activated Kinases. Annu. Rev. Biochem. 2003, 72, 743–781. [Google Scholar] [CrossRef]
- Kumar, A.; Molli, P.R.; Pakala, S.B.; Nguyen, T.M.B.; Rayala, S.; Kumar, R. PAK thread from amoeba to mammals. J. Cell. Biochem. 2009, 107, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Szczepanowska, J. Involvement of Rac/Cdc42/PAK pathway in cytoskeletal rearrangements. Acta Biochim. Pol. 2009, 56, 225–234. [Google Scholar] [CrossRef]
- Rane, C.K.; Minden, A. P21 activated kinases: Structure, Regulation, and Functions. Small GTPases 2014, 5, e28003. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hannigan, M.; Mo, Z.; Liu, B.; Lu, W.; Wu, Y.; Smrcka, A.V.; Wu, G.; Li, L.; Liu, M.; et al. Directional Sensing Requires G Beta Gamma-Mediated PAK1 and PIX Alpha-Dependent Activation of Cdc42. Cell 2003, 114, 215–227. [Google Scholar] [CrossRef]
- Sells, M.A.; Knaus, U.G.; Bagrodia, S.; Ambrose, D.M.; Bokoch, G.M.; Chernoff, J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 1997, 7, 202–210. [Google Scholar] [CrossRef]
- Goeckeler, Z.M.; Masaracchia, R.A.; Zeng, Q.; Chew, T.-L.; Gallagher, P.; Wysolmerski, R.B. Phosphorylation of Myosin Light Chain Kinase by p21-activated Kinase PAK2. J. Biol. Chem. 2000, 275, 18366–18374. [Google Scholar] [CrossRef] [PubMed]
- Sanders, L.C.; Matsumura, F.; Bokoch, G.M.; De Lanerolle, P. Inhibition of Myosin Light Chain Kinase by p21-Activated Kinase. Science 1999, 283, 2083–2085. [Google Scholar] [CrossRef] [PubMed]
- DerMardirossian, C.; Schnelzer, A.; Bokoch, G.M. Phosphorylation of RhoGDI by Pak1 Mediates Dissociation of Rac GTPase. Mol. Cell 2004, 15, 117–127. [Google Scholar] [CrossRef]
- Shin, E.-Y.; Shim, E.-S.; Lee, C.-S.; Kim, H.K.; Kim, E.-G. Phosphorylation of RhoGDI1 by p21-activated kinase 2 mediates basic fibroblast growth factor-stimulated neurite outgrowth in PC12 cells. Biochem. Biophys. Res. Commun. 2009, 379, 384–389. [Google Scholar] [CrossRef]
- Hashimoto, S.; Tsubouchi, A.; Mazaki, Y.; Sabe, H. Interaction of Paxillin with P21-Activated Kinase (PAK). Association of Paxillin Alpha with the Kinase-Inactive and the Cdc42-Activated Forms of PAK3. J. Biol. Chem. 2001, 276, 6037–6045. [Google Scholar] [CrossRef]
- Abo, A.; Qu, J.; Cammarano, M.S.; Dan, C.; Fritsch, A.; Baud, V.; Belisle, B.; Minden, A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998, 17, 6527–6540. [Google Scholar] [CrossRef]
- Chung, C.Y.; Firtel, R.A. Paka, a Putative Pak Family Member, Is Required for Cytokinesis and the Regulation of the Cytoskeleton in Dictyostelium discoideum Cells during Chemotaxis. J. Cell Biol. 1999, 147, 559–576. [Google Scholar] [CrossRef]
- Tang, M.; Iijima, M.; Kamimura, Y.; Chen, L.; Long, Y.; Devreotes, P. Disruption of PKB signaling restores polarity to cells lacking tumor suppressor PTEN. Mol. Biol. Cell 2011, 22, 437–447. [Google Scholar] [CrossRef]
- Müller-Taubenberger, A.; Bretschneider, T.; Faix, J.; Konzok, A.; Simmeth, E.; Weber, I. Differential localization of the Dictyostelium kinase DPAKa during cytokinesis and cell migration. J. Muscle Res. Cell Motil. 2002, 23, 751–763. [Google Scholar] [CrossRef]
- Li, M.; Quan, C.; Chen, S.; Wang, H.Y. The 14-3-3 protein is an essential component of cyclic AMP signaling for regulation of chemotaxis and development in Dictyostelium. Cell. Signal. 2020, 75, 109739. [Google Scholar] [CrossRef]
- Chung, C.Y.; Potikyan, G.; Firtel, R.A. Control of Cell Polarity and Chemotaxis by Akt/PKB and PI3 Kinase through the Regulation of PAKa. Mol. Cell 2001, 7, 937–947. [Google Scholar] [CrossRef]
- De la Roche, M.A.; Côté, G.P. Regulation of Dictyostelium Myosin I and II. Biochim. Biophys. Acta (BBA) Gen. Subj. 2001, 1525, 245–261. [Google Scholar] [CrossRef]
- Lee, S.-F.; Té, G.P.C. Purification and Characterization of a Dictyostelium Protein Kinase Required for Actin Activation of the Mg2+ATPase Activity of Dictyostelium Myosin ID. J. Biol. Chem. 1995, 270, 11776–11782. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, R.; Von Heyden, Y.; Kistler, C.; Gopaldass, N.A.; Hausherr, S.; Crawley, S.W.; Schwarz, E.C.; Diensthuber, R.P.; Côté, G.P.; Tsiavaliaris, G.; et al. A Myosin IK-Abp1-PakB Circuit Acts as a Switch to Regulate Phagocytosis Efficiency. Mol. Biol. Cell 2010, 21, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; De La Roche, M.; Crawley, S.W.; Li, Z.; Furmaniak-Kazmierczak, E.; Côté, G.P. PakB binds to the SH3 domain of Dictyostelium Abp1 and regulates its effects on cell polarity and early development. Mol. Biol. Cell 2013, 24, 2216–2227. [Google Scholar] [CrossRef]
- Lee, S.-F.; Egelhoff, T.T.; Mahasneh, A.; Côté, G.P. Cloning and Characterization of a Dictyostelium Myosin I Heavy Chain Kinase Activated by Cdc42 and Rac. J. Biol. Chem. 1996, 271, 27044–27048. [Google Scholar] [CrossRef]
- Lee, S.-F.; Mahasneh, A.; de la Roche, M.; Côté, G.P. Regulation of the p21-activated Kinase-relatedDictyostelium Myosin I Heavy Chain Kinase by Autophosphorylation, Acidic Phospholipids, and Ca2+-Calmodulin. J. Biol. Chem. 1998, 273, 27911–27917. [Google Scholar] [CrossRef][Green Version]
- Phillips, J.E.; Gomer, R.H. The p21-Activated Kinase (PAK) Family Member PakD Is Required for Chemorepulsion and Proliferation Inhibition by Autocrine Signals in Dictyostelium discoideum. PLoS ONE 2014, 9, e96633. [Google Scholar] [CrossRef]
- Garcia, M.; Ray, S.; Brown, I.; Irom, J.; Brazill, D. PakD, a Putative p21-Activated Protein Kinase in Dictyostelium discoideum, Regulates Actin. Eukaryot. Cell 2013, 13, 119–126. [Google Scholar] [CrossRef]
- Suess, P.M.; Gomer, R.H. Extracellular Polyphosphate Inhibits Proliferation in an Autocrine Negative Feedback Loop in Dictyostelium discoideum. J. Biol. Chem. 2016, 291, 20260–20269. [Google Scholar] [CrossRef]
- Suess, P.M.; Watson, J.; Chen, W.; Gomer, R.H. Extracellular polyphosphate signals through Ras and Akt to prime Dictyostelium discoideum cells for development. J. Cell Sci. 2017, 130, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Brill, S.; Li, S.; Lyman, C.W.; Church, D.M.; Wasmuth, J.J.; Weissbach, L.; Bernards, A.; Snijders, A.J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996, 16, 4869–4878. [Google Scholar] [CrossRef]
- Hart, M.J.; Callow, M.G.; Souza, B.; Polakis, P. IQGAP1, a Calmodulin-Binding Protein with a RasGAP-Related Domain, Is a Potential Effector for Cdc42Hs. EMBO J. 1996, 15, 2997–3005. [Google Scholar] [CrossRef]
- Kuroda, S.; Fukata, M.; Kobayashi, K.; Nakafuku, M.; Nomura, N.; Iwamatsu, A.; Kaibuchi, K. Identification of IQGAP as a Putative Target for the Small GTPases, Cdc42 and Rac1. J. Biol. Chem. 1996, 271, 23363–23367. [Google Scholar] [CrossRef] [PubMed]
- McCallum, S.J.; Wu, W.J.; Cerione, R.A. Identification of a Putative Effector for Cdc42Hs with High Sequence Similarity to the RasGAP-related Protein IQGAP1 and a Cdc42Hs Binding Partner with Similarity to IQGAP2. J. Biol. Chem. 1996, 271, 21732–21737. [Google Scholar] [CrossRef] [PubMed]
- Weissbach, L.; Settleman, J.; Kalady, M.F.; Snijders, A.J.; Murthy, A.E.; Yan, Y.X.; Bernards, A. Identification of a Human RasGAP-Related Protein Containing Calmodulin-Binding Motifs. J. Biol. Chem. 1994, 269, 20517–20521. [Google Scholar] [CrossRef]
- Wang, S.; Watanabe, T.; Noritake, J.; Fukata, M.; Yoshimura, T.; Itoh, N.; Harada, T.; Nakagawa, M.; Matsuura, Y.; Arimura, N.; et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J. Cell Sci. 2007, 120, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep. 2015, 16, 427–446. [Google Scholar] [CrossRef]
- White, C.D.; Erdemir, H.H.; Sacks, D.B. IQGAP1 and its binding proteins control diverse biological functions. Cell. Signal. 2012, 24, 826–834. [Google Scholar] [CrossRef]
- Bashour, A.-M.; Fullerton, A.T.; Hart, M.J.; Bloom, G.S. IQGAP1, a Rac- and Cdc42-binding Protein, Directly Binds and Cross-links Microfilaments. J. Cell Biol. 1997, 137, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.W.; Cerione, R.A.; Hart, M.J. Identification of an Actin Cytoskeletal Complex That Includes IQGAP and the Cdc42 GTPase. J. Biol. Chem. 1997, 272, 24443–24447. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Kuroda, S.; Fujii, K.; Nakamura, T.; Shoji, I.; Matsuura, Y.; Okawa, K.; Iwamatsu, A.; Kikuchi, A.; Kaibuchi, K. Regulation of Cross-linking of Actin Filament by IQGAP1, a Target for Cdc42. J. Biol. Chem. 1997, 272, 29579–29583. [Google Scholar] [CrossRef]
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Binds ERK2 and Modulates Its Activity. J. Biol. Chem. 2004, 279, 17329–17337. [Google Scholar] [CrossRef]
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Is a Scaffold for Mitogen-Activated Protein Kinase Signaling. Mol. Cell. Biol. 2005, 25, 7940–7952. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.-D.; Joyal, J.L.; Li, Z.; Sacks, D.B. IQGAP1 Integrates Ca2+/Calmodulin and Cdc42 Signaling. J. Biol. Chem. 1999, 274, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Joyal, J.L.; Annan, R.S.; Ho, Y.-D.; Huddleston, M.E.; Carr, S.A.; Hart, M.J.; Sacks, D.B. Calmodulin Modulates the Interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by Nanoelectrospray Tandem Mass Spectrometry. J. Biol. Chem. 1997, 272, 15419–15425. [Google Scholar] [CrossRef]
- Nojima, H.; Adachi, M.; Matsui, T.; Okawa, K.; Tsukita, S.; Tsukita, S. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nat. Cell Biol. 2008, 10, 971–978. [Google Scholar] [CrossRef]
- Swart-Mataraza, J.M.; Li, Z.; Sacks, D.B. IQGAP1 Is a Component of Cdc42 Signaling to the Cytoskeleton. J. Biol. Chem. 2002, 277, 24753–24763. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 Capture Microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef]
- Kuroda, S.; Fukata, M.; Nakagawa, M.; Fujii, K.; Nakamura, T.; Ookubo, T.; Izawa, I.; Nagase, T.; Nomura, N.; Tani, H.; et al. Role of IQGAP1, a Target of the Small GTPases Cdc42 and Rac1, in Regulation of E-Cadherin- Mediated Cell-Cell Adhesion. Science 1998, 281, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Hedman, A.C.; Sacks, D.B. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015, 25, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M.; Kawasaki, A.; Nojima, H.; Nishida, E.; Tsukita, S. Involvement of IQGAP family proteins in the regulation of mammalian cell cytokinesis. Genes Cells 2014, 19, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Brandt, D.T.; Grosse, R. Get to grips: Steering local actin dynamics with IQGAPs. EMBO Rep. 2007, 8, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Noritake, J.; Watanabe, T.; Sato, K.; Wang, S.; Kaibuchi, K. IQGAP1: A key regulator of adhesion and migration. J. Cell Sci. 2005, 118, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Wang, S.; Kaibuchi, K. IQGAPs as key regulators of actin-cytoskeleton dynamics. Cell Struct. Funct. 2015, 40, 69–77. [Google Scholar] [CrossRef]
- Shannon, K.B. IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells. Int. J. Cell Biol. 2012, 2012, 894817. [Google Scholar] [CrossRef]
- Friedberg, F.; Rivero, F. Single and multiple CH (calponin homology) domain containing multidomain proteins in Dictyostelium discoideum: An inventory. Mol. Biol. Rep. 2009, 37, 2853–2862. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Takahashi, Y.; Hasebe, T.; Shirouzu, M.; Yokoyama, S.; Sutoh, K. Dictyostelium IQGAP-related Protein Specifically Involved in the Completion of Cytokinesis. J. Cell Biol. 1997, 137, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Faix, J.; Clougherty, C.; Konzok, A.; Mintert, U.; Murphy, J.; Albrecht, R.; Muhlbauer, B.; Kuhlmann, J. The IQGAP-related protein DGAP1 interacts with Rac and is involved in the modulation of the F-actin cytoskeleton and control of cell motility. J. Cell Sci. 1998, 111 Pt 20, 3059–3071. [Google Scholar] [CrossRef]
- Kurella, V.B.; Richard, J.M.; Parke, C.L.; LeCour, L.F.; Bellamy, H.D.; Worthylake, D.K. Crystal Structure of the GTPase-activating Protein-related Domain from IQGAP1. J. Biol. Chem. 2009, 284, 14857–14865. [Google Scholar] [CrossRef] [PubMed]
- Marinović, M.; Mijanović, L.; Šoštar, M.; Vizovišek, M.; Junemann, A.; Fonović, M.; Turk, B.; Weber, I.; Faix, J.; Filić, V. IQGAP-related protein IqgC suppresses Ras signaling during large-scale endocytosis. Proc. Natl. Acad. Sci. USA 2019, 116, 1289–1298. [Google Scholar] [CrossRef]
- Faix, J.; Weber, I.; Mintert, U.; Köhler, J.; Lottspeich, F.; Marriott, G. Recruitment of cortexillin into the cleavage furrow is controlled by Rac1 and IQGAP-related proteins. EMBO J. 2001, 20, 3705–3715. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Burgute, B.; Rieger, D.; Müller, R.; Rivero, F.; Faix, J.; Schleicher, M.; Noegel, A.A. Regulation of the Actin Cytoskeleton by an Interaction of IQGAP Related Protein GAPA with Filamin and Cortexillin I. PLoS ONE 2010, 5, e15440. [Google Scholar] [CrossRef] [PubMed]
- Faix, J.; Steinmetz, M.; Boves, H.; Kammerer, R.; Lottspeich, F.; Mintert, U.; Murphy, J.; Stock, A.; Aebi, U.; Gerisch, G. Cortexillins, Major Determinants of Cell Shape and Size, Are Actin-Bundling Proteins with a Parallel Coiled-Coil Tail. Cell 1996, 86, 631–642. [Google Scholar] [CrossRef]
- Simson, R.; Wallraff, E.; Faix, J.; Niewöhner, J.; Gerisch, G.; Sackmann, E. Membrane Bending Modulus and Adhesion Energy of Wild-Type and Mutant Cells of Dictyostelium Lacking Talin or Cortexillins. Biophys. J. 1998, 74, 514–522. [Google Scholar] [CrossRef]
- Lee, S.; Shen, Z.; Robinson, D.N.; Briggs, S.; Firtel, R.A. Involvement of the Cytoskeleton in Controlling Leading-Edge Function during Chemotaxis. Mol. Biol. Cell 2010, 21, 1810–1824. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Liu, X.; Kriebel, P.W.; Daniels, M.P.; Korn, E. Actin cross-linking proteins cortexillin I and II are required for cAMP signaling during Dictyostelium chemotaxis and development. Mol. Biol. Cell 2012, 23, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Robinson, D.N.; Nelson, W.J.; Weis, W.I. α-Catenin and IQGAP Regulate Myosin Localization to Control Epithelial Tube Morphogenesis in Dictyostelium. Dev. Cell 2012, 23, 533–546. [Google Scholar] [CrossRef]
- BenseñorL, B.; Kan, H.-M.; Wang, N.; Wallrabe, H.; Davidson, L.A.; Cai, Y.; Schafer, D.A.; Bloom, G.S. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J. Cell Sci. 2007, 120, 658–669. [Google Scholar] [CrossRef]
- Brandt, D.T.; Marion, S.; Griffiths, G.; Watanabe, T.; Kaibuchi, K.; Grosse, R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J. Cell Biol. 2007, 178, 193–200. [Google Scholar] [CrossRef]
- Le Clainche, C.; Schlaepfer, D.; Ferrari, A.; Klingauf, M.; Grohmanova, K.; Veligodskiy, A.; Didry, D.; Le, D.; Egile, C.; Carlier, M.-F.; et al. IQGAP1 Stimulates Actin Assembly through the N-Wasp-Arp2/3 Pathway. J. Biol. Chem. 2007, 282, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Sethi, K.; Cram, E.J.; Zaidel-Bar, R. Stretch-induced actomyosin contraction in epithelial tubes: Mechanotransduction pathways for tubular homeostasis. Semin. Cell Dev. Biol. 2017, 71, 146–152. [Google Scholar] [CrossRef]
- Bañón-Rodríguez, I.; Gálvez-Santisteban, M.; Vergarajauregui, S.; Bosch, M.; Borreguero-Pascual, A.; Martín-Belmonte, F. EGFR controls IQGAP basolateral membrane localization and mitotic spindle orientation during epithelial morphogenesis. EMBO J. 2014, 33, 129–145. [Google Scholar] [CrossRef]
- Nakamura, F.; Stossel, T.P.; Hartwig, J.H. The filamins. Cell Adhes. Migr. 2011, 5, 160–169. [Google Scholar] [CrossRef]
- Stossel, T.P.; Condeelis, J.; Cooley, L.; Hartwig, J.H.; Noegel, A.; Schleicher, M.; Shapiro, S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001, 2, 138–145. [Google Scholar] [CrossRef]
- Popowicz, G.M.; Schleicher, M.; Noegel, A.A.; Holak, T.A. Filamins: Promiscuous Organizers of the Cytoskeleton. Trends Biochem. Sci. 2006, 31, 411–419. [Google Scholar] [CrossRef]
- Cox, D.; Condeelis, J.; Wessels, D.; Soll, D.; Kern, H.; Knecht, D. Targeted disruption of the ABP-120 gene leads to cells with altered motility. J. Cell Biol. 1992, 116, 943–955. [Google Scholar] [CrossRef]
- Cox, D.; Ridsdale, J.A.; Condeelis, J.; Hartwig, J. Genetic deletion of ABP-120 alters the three-dimensional organization of actin filaments in Dictyostelium pseudopods. J. Cell Biol. 1995, 128, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Wessels, D.; Soll, D.R.; Hartwig, J.; Condeelis, J. Re-expression of ABP-120 rescues cytoskeletal, motility, and phagocytosis defects of ABP-120- Dictyostelium mutants. Mol. Biol. Cell 1996, 7, 803–823. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fisher, P.; Noegel, A.; Fechheimer, M.; Rivero, F.; Prassler, J.; Gerisch, G. Photosensory and thermosensory responses in Dictyostelium slugs are specifically impaired by absence of the F-actin cross-linking gelation factor (ABP-120). Curr. Biol. 1997, 7, 889–892. [Google Scholar] [CrossRef]
- Del Valle-Pérez, B.; Martínez, V.G.; Lacasa-Salavert, C.; Figueras, A.; Shapiro, S.S.; Takafuta, T.; Casanovas, O.; Capella, G.; Ventura, F.; Viñals, F. Filamin B Plays a Key Role in Vascular Endothelial Growth Factor-induced Endothelial Cell Motility through Its Interaction with Rac-1 and Vav-2. J. Biol. Chem. 2010, 285, 10748–10760. [Google Scholar] [CrossRef]
- Jacquemet, G.; Morgan, M.R.; Byron, A.; Humphries, J.D.; Choi, C.K.; Chen, C.; Caswell, P.T.; Humphries, M.J. Rac1 is deactivated at integrin activation sites via an IQGAP1/filamin-A/RacGAP1 pathway. J. Cell Sci. 2013, 126, 4121–4135. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Ka, S.H.; Cho, Y.; Choi, E.-J.; Baek, S.H.; Seol, J.H.; Park, D.; et al. Filamin B Serves as a Molecular Scaffold for Type I Interferon-induced c-Jun NH2-terminal Kinase Signaling Pathway. Mol. Biol. Cell 2008, 19, 5116–5130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shifrin, Y.; Pinto, V.I.; Hassanali, A.; Arora, P.D.; McCulloch, C.A. Force-induced apoptosis mediated by the Rac/Pak/p38 signalling pathway is regulated by filamin A. Biochem. J. 2012, 445, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Rivero, F.; Albrecht, R.; Dislich, H.; Bracco, E.; Graciotti, L.; Bozzaro, S.; Noegel, A.A. RacF1, a Novel Member of the Rho Protein Family in Dictyostelium discoideum, Associates Transiently with Cell Contact Areas, Macropinosomes, and Phagosomes. Mol. Biol. Cell 1999, 10, 1205–1219. [Google Scholar] [CrossRef]
- Lee, E.; Seastone, D.J.; Harris, E.; Cardelli, J.A.; Knecht, D.A. RacB Regulates Cytoskeletal Function in Dictyostelium spp. Eukaryot. Cell 2003, 2, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.M.; Dingermann, T.; Knecht, D.A. Regulated expression of myosin II heavy chain and RacB using an inducible tRNA suppressor gene. Gene 2001, 277, 187–197. [Google Scholar] [CrossRef]
- Rivero, F.; Dislich, H.; Glöckner, G.; Noegel, A.A. The Dictyostelium discoideum family of Rho-related proteins. Nucleic Acids Res. 2001, 29, 1068–1079. [Google Scholar] [CrossRef]
- Berthold, J.; Schenková, K.; Rivero, F. Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacol. Sin. 2008, 29, 285–295. [Google Scholar] [CrossRef]
- Ji, W.; Rivero, F. Atypical Rho GTPases of the RhoBTB Subfamily: Roles in Vesicle Trafficking and Tumorigenesis. Cells 2016, 5, 28. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain 1 1Edited by F. Cohen. J. Mol. Biol. 1999, 285, 1353–1361. [Google Scholar] [CrossRef]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.-H.; Vidal, M.; Elledge, S.J.; Harper, J. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Bridges, E.; Sheldon, H.; Kleibeuker, E.; Ramberger, E.; Zois, C.; Barnard, A.; Harjes, U.; Li, J.-L.; Masiero, M.; MacLaren, R.; et al. RHOQ is induced by DLL4 and regulates angiogenesis by determining the intracellular route of the Notch intracellular domain. Angiogenesis 2020, 23, 493–513. [Google Scholar] [CrossRef]
- Hodge, R.G.; Ridley, A.J. Regulation and functions of RhoU and RhoV. Small GTPases 2017, 11, 8–15. [Google Scholar] [CrossRef]
- Leszczynska, K.; Kaur, S.; Wilson, E.; Bicknell, R.; Heath, V.L. The role of RhoJ in endothelial cell biology and angiogenesis. Biochem. Soc. Trans. 2011, 39, 1606–1611. [Google Scholar] [CrossRef]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cdc42—The centre of polarity. J. Cell Sci. 2004, 117, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Pichaud, F.; Walther, R.F.; de Almeida, F.N. Regulation of Cdc42 and its effectors in epithelial morphogenesis. J. Cell Sci. 2019, 132, jcs217869. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.-H.; Lv, L.-C.; Duan, J.; Wu, Y.-M.; He, S.-J.; Hu, Z.-Z.; Xiong, L.-X. Regulating Cdc42 and Its Signaling Pathways in Cancer: Small Molecules and MicroRNA as New Treatment Candidates. Molcules 2018, 23, 787. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Manser, E. Myotonic dystrophy kinase-related Cdc42-binding kinases (MRCK), the ROCK-like effectors of Cdc42 and Rac1. Small GTPases 2015, 6, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Integrin-Mediated Activation of Cdc42 Controls Cell Polarity in Migrating Astrocytes through PKCζ. Cell 2001, 106, 489–498. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Cdc42 regulates GSK-3β and adenomatous polyposis coli to control cell polarity. Nature 2003, 421, 753–756. [Google Scholar] [CrossRef]
- LaRochelle, D.A.; Vithalani, K.K.; De Lozanne, A. Role of Dictyostelium racE in cytokinesis: Mutational analysis and localization studies by use of green fluorescent protein. Mol. Biol. Cell 1997, 8, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Seastone, D.J.; Lee, E.; Bush, J.; Knecht, D.; Cardelli, J. Overexpression of a Novel Rho Family GTPase, RacC, Induces Unusual Actin-based Structures and Positively Affects Phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell 1998, 9, 2891–2904. [Google Scholar] [CrossRef]
- Erickson, J.W.; Zhang, C.-J.; Kahn, R.A.; Evans, T.; Cerione, R.A. Mammalian Cdc42 Is a Brefeldin A-sensitive Component of the Golgi Apparatus. J. Biol. Chem. 1996, 271, 26850–26854. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, L.; Zheng, Y. Gene Targeting of Cdc42 and Cdc42GAP Affirms the Critical Involvement of Cdc42 in Filopodia Induction, Directed Migration, and Proliferation in Primary Mouse Embryonic Fibroblasts. Mol. Biol. Cell 2006, 17, 4675–4685. [Google Scholar] [CrossRef]
- Chernichenko, N.; Omelchenko, T.; Deborde, S.; Bakst, R.L.; He, S.; Chen, C.-H.; Gusain, L.; Vakiani, E.; Katabi, N.; Hall, A.; et al. Cdc42 Mediates Cancer Cell Chemotaxis in Perineural Invasion. Mol. Cancer Res. 2020, 18, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.; Owen, D.; Mott, H.R. Cdc42 in actin dynamics: An ordered pathway governed by complex equilibria and directional effector handover. Small GTPases 2017, 8, 237–244. [Google Scholar] [CrossRef]
- Jeon, P.; Jeon, T.J. WasC, a WASP family protein, is involved in cell adhesion and migration through regulation of F-actin polymerization in Dictyostelium. J. Microbiol. 2020, 58, 696–702. [Google Scholar] [CrossRef]
- Kriebel, P.W.; Barr, V.A.; Parent, C.A. Adenylyl Cyclase Localization Regulates Streaming during Chemotaxis. Cell 2003, 112, 549–560. [Google Scholar] [CrossRef]
- Saran, S.; Meima, M.E.; Alvarez-Curto, E.; Weening, K.E.; Rozen, D.; Schaap, P. cAMP signaling in Dictyostelium. Complexity of CAMP Synthesis, Degradation and Detection. J. Muscle Res. Cell Motil. 2002, 23, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.P.; Tepass, U. Cdc42 and Vesicle Trafficking in Polarized Cells. Traffic 2010, 11, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.; Matas, O.B.; Martínez-Menárguez, J.A.; Mato, E.; Durán, J.M.; Ballesta, J.; Way, M.; Egea, G. Regulation of Protein Transport from the Golgi Complex to the Endoplasmic Reticulum by CDC42 and N-WASP. Mol. Biol. Cell 2002, 13, 866–879. [Google Scholar] [CrossRef]
- Phillips, J.E.; Gomer, R.H. A secreted protein is an endogenous chemorepellant in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 2012, 109, 10990–10995. [Google Scholar] [CrossRef] [PubMed]
- Rijal, R.; Consalvo, K.M.; Lindsey, C.K.; Gomer, R.H. An endogenous chemorepellent directs cell movement by inhibiting pseudopods at one side of cells. Mol. Biol. Cell 2019, 30, 242–255. [Google Scholar] [CrossRef]
- Okuwa, T.; Morio, T.; Saito, T.; Masamune, Y.; Yasukawa, H. Complete Sequences and Expression Kinetics of racG, racH, racI and racJ Genes in Dictyostelium discoideum. Biol. Pharm. Bull. 2001, 24, 84–87. [Google Scholar] [CrossRef]
- Somesh, B.P.; Neffgen, C.; Iijima, M.; Devreotes, P.; Rivero, F. Dictyostelium RacH regulates endocytic vesicular trafficking and is required for localization of vacuolin. Traffic 2006, 7, 1194–1212. [Google Scholar] [CrossRef]
- Balest, A.; Peracino, B.; Bozzaro, S. Legionella pneumophila infection is enhanced in a RacH-null mutant of Dictyostelium. Commun. Integr. Biol. 2011, 4, 194–197. [Google Scholar] [CrossRef]
- Hagedorn, M.; Soldati, T. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell. Microbiol. 2007, 9, 2984. [Google Scholar] [CrossRef][Green Version]
- Hagedorn, M.; Rohde, K.H.; Russell, D.G.; Soldati, T. Infection by Tubercular Mycobacteria Is Spread by Nonlytic Ejection from Their Amoeba Hosts. Science 2009, 323, 1729–1733. [Google Scholar] [CrossRef]
- Humphries, A.C.; Donnelly, S.K.; Way, M. Cdc42 and the RhoGEF Intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J. Cell Sci. 2014, 127, 673–685. [Google Scholar] [CrossRef]
- Gouin, E.; Welch, M.D.; Cossart, P. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 2005, 8, 35–45. [Google Scholar] [CrossRef]
- Gerald, N.; Dai, J.; Ting-Beall, H.P.; De Lozanne, A. A Role for Dictyostelium RacE in Cortical Tension and Cleavage Furrow Progression. J. Cell Biol. 1998, 141, 483–492. [Google Scholar] [CrossRef]
- Robinson, D.N.; Spudich, J.A. Dynacortin, a Genetic Link between Equatorial Contractility and Global Shape Control Discovered by Library Complementation of a Dictyostelium discoideum Cytokinesis Mutant. J. Cell Biol. 2000, 150, 823–838. [Google Scholar] [CrossRef]
- West-Foyle, H.; Kothari, P.; Osborne, J.; Robinson, D.N. 14-3-3 proteins tune non-muscle myosin II assembly. J. Biol. Chem. 2018, 293, 6751–6761. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kee, Y.-S.; Poirier, C.C.; Jelinek, C.; Osborne, J.; Divi, S.; Surcel, A.; Will, M.E.; Eggert, U.S.; Müller-Taubenberger, A.; et al. 14-3-3 Coordinates Microtubules, Rac, and Myosin II to Control Cell Mechanics and Cytokinesis. Curr. Biol. 2010, 20, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Kee, Y.-S.; Ren, Y.; Dorfman, D.; Iijima, M.; Firtel, R.; Iglesias, P.A.; Robinson, D.N. A mechanosensory system governs myosin II accumulation in dividing cells. Mol. Biol. Cell 2012, 23, 1510–1523. [Google Scholar] [CrossRef]
- Ramalingam, N.; Franke, C.; Jaschinski, E.; Winterhoff, M.; Lü, Y.; Brühmann, S.; Junemann, A.; Meier, H.; Noegel, A.A.; Weber, I.; et al. A resilient formin-derived cortical actin meshwork in the rear drives actomyosin-based motility in 2D confinement. Nat. Commun. 2015, 6, 8496. [Google Scholar] [CrossRef] [PubMed]
- Litschko, C.; Brühmann, S.; Csiszár, A.; Stephan, T.; Dimchev, V.; Damiano-Guercio, J.; Junemann, A.; Körber, S.; Winterhoff, M.; Nordholz, B.; et al. Functional integrity of the contractile actin cortex is safeguarded by multiple Diaphanous-related formins. Proc. Natl. Acad. Sci. USA 2019, 116, 3594–3603. [Google Scholar] [CrossRef] [PubMed]
- Senoo, H.; Cai, H.; Wang, Y.; Sesaki, H.; Iijima, M. The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells. Mol. Biol. Cell 2016, 27, 1596–1605. [Google Scholar] [CrossRef]
- Senoo, H.; Kamimura, Y.; Kimura, R.; Nakajima, A.; Sawai, S.; Sesaki, H.; Iijima, M. Phosphorylated Rho–GDP directly activates mTORC2 kinase towards AKT through dimerization with Ras–GTP to regulate cell migration. Nature 2019, 21, 867–878. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2019, 20, 74–88. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Senoo, H.; Wai, M.; Matsubayashi, H.T.; Sesaki, H.; Iijima, M. Hetero-oligomerization of Rho and Ras GTPases Connects GPCR Activation to mTORC2-AKT Signaling. Cell Rep. 2020, 33, 108427. [Google Scholar] [CrossRef]
- Lammers, M.; Meyer, S.; Kühlmann, D.; Wittinghofer, A. Specificity of Interactions between mDia Isoforms and Rho Proteins. J. Biol. Chem. 2008, 283, 35236–35246. [Google Scholar] [CrossRef]
- Maiti, S.; Michelot, A.; Gould, C.; Blanchoin, L.; Sokolova, O.; Goode, B.L. Structure and activity of full-length formin mDia1. Cytoskeleton 2012, 69, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Thumkeo, D.; Ohta, H.; Zhang, Z.; Huang, S.; Kanchanawong, P.; Fuu, T.; Watanabe, S.; Shimada, K.; Fujihara, Y.; et al. mDia1/3 generate cortical F-actin meshwork in Sertoli cells that is continuous with contractile F-actin bundles and indispensable for spermatogenesis and male fertility. PLoS Biol. 2018, 16, e2004874. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Arora, P.D.; Lai, C.C.; Copeland, J.W.; Moraes, T.F.; McCulloch, C.A.; Lavoie, B.D.; Wilde, A. The scaffold-protein IQGAP1 enhances and spatially restricts the actin-nucleating activity of Diaphanous-related formin 1 (DIAPH1). J. Biol. Chem. 2020, 295, 3134–3147. [Google Scholar] [CrossRef]
- Brandwein, D.; Wang, Z. Interaction between Rho GTPases and 14-3-3 Proteins. Int. J. Mol. Sci. 2017, 18, 2148. [Google Scholar] [CrossRef] [PubMed]
- Abdrabou, A.; Brandwein, D.; Liu, C.; Wang, Z. Rac1 S71 Mediates the Interaction between Rac1 and 14-3-3 Proteins. Cells 2019, 8, 1006. [Google Scholar] [CrossRef] [PubMed]
- Obšilová, V.; Kopecka, M.; Kosek, D.; Kacirova, M.; Kylarová, S.; Rezabkova, L.; Obsil, T. Mechanisms of the 14-3-3 protein function: Regulation of protein function through conformational modulation. Physiol. Res. 2014, 63, 155–164. [Google Scholar] [CrossRef]
- Rios, A.; Hernández-Ramírez, V.I.; Moguel, M.; Bahena, A.I.Z.; Rosales-Encina, J.L.; Vargas, M. Ángel; Talamás-Rohana, P. Participation of Rho, ROCK-2, and GAP activities during actin microfilament rearrangements in Entamoeba histolytica induced by fibronectin signaling. Cell Biol. Int. 2008, 32, 984–1000. [Google Scholar] [CrossRef]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Narumiya, S.; Tanji, M.; Ishizaki, T. Rho Signaling, ROCK and MDia1, in Transformation, Metastasis and Invasion. Cancer Metastasis Rev. 2009, 28, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Bosgraaf, L.; van Haastert, P.J. The regulation of myosin II in Dictyostelium. Eur. J. Cell Biol. 2006, 85, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Basant, A.; Glotzer, M. Spatiotemporal Regulation of RhoA during Cytokinesis. Curr. Biol. 2018, 28, R570–R580. [Google Scholar] [CrossRef] [PubMed]
- Maddox, A.S.; Burridge, K. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J. Cell Biol. 2003, 160, 255–265. [Google Scholar] [CrossRef]
- Wheeler, A.P.; Ridley, A.J. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004, 301, 43–49. [Google Scholar] [CrossRef]
- Filić, V.; Marinović, M.; Faix, J.; Weber, I. The IQGAP-related protein DGAP1 mediates signaling to the actin cytoskeleton as an effector and a sequestrator of Rac1 GTPases. Cell. Mol. Life Sci. 2014, 71, 2775–2785. [Google Scholar] [CrossRef]
- Somesh, B.P.; Vlahou, G.; Iijima, M.; Insall, R.H.; Devreotes, P.; Rivero, F. RacG Regulates Morphology, Phagocytosis, and Chemotaxis. Eukaryot. Cell 2006, 5, 1648–1663. [Google Scholar] [CrossRef]
- Amos, L.A.; Löwe, J. Overview of the Diverse Roles of Bacterial and Archaeal Cytoskeletons. Subcell. Biochem. 2017, 84, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, P.R.; Williams, T.A.; Garner, E.; Baum, B. Evolution of polymer formation within the actin superfamily. Mol. Biol. Cell 2017, 28, 2461–2469. [Google Scholar] [CrossRef]
- Wagstaff, J.; Löwe, J. Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol. 2018, 16, 187–201. [Google Scholar] [CrossRef]
- Wickstead, B.; Gull, K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. [Google Scholar] [CrossRef]
- Akıl, C.; Robinson, R.C. Genomes of Asgard archaea encode profilins that regulate actin. Nature 2018, 562, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Akıl, C.; Tran, L.T.; Orhant-Prioux, M.; Baskaran, Y.; Manser, E.; Blanchoin, L.; Robinson, R.C. Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc. Natl. Acad. Sci. USA 2020, 117, 19904–19913. [Google Scholar] [CrossRef] [PubMed]
- Izoré, T.; Kureisaite-Ciziene, D.; McLaughlin, S.H.; Löwe, J. Crenactin forms actin-like double helical filaments regulated by arcadin-2. eLife 2016, 5, e21600. [Google Scholar] [CrossRef] [PubMed]
- Merino, F.; Raunser, S. The mother of all actins? eLife 2016, 5, 161. [Google Scholar] [CrossRef] [PubMed]
- Velle, K.B.; Fritz-Laylin, L.K. Diversity and evolution of actin-dependent phenotypes. Curr. Opin. Genet. Dev. 2019, 58-59, 40–48. [Google Scholar] [CrossRef]
- Velle, K.B.; Fritz-Laylin, L.K. Conserved actin machinery drives microtubule-independent motility and phagocytosis in Naegleria. J. Cell Biol. 2020, 219, 219. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.; Lord, S.J.; Mullins, R.D. Our evolving view of cell motility. Cell Cycle 2017, 16, 1735–1736. [Google Scholar] [CrossRef][Green Version]
- Fritz-Laylin, L.K. The evolution of animal cell motility. Curr. Biol. 2020, 30, R477–R482. [Google Scholar] [CrossRef]
- Kameritsch, P.; Renkawitz, J. Principles of Leukocyte Migration Strategies. Trends Cell Biol. 2020, 30, 818–832. [Google Scholar] [CrossRef] [PubMed]
- Moreau, H.D.; Piel, M.; Voituriez, R.; Lennon-Duménil, A.-M. Integrating Physical and Molecular Insights on Immune Cell Migration. Trends Immunol. 2018, 39, 632–643. [Google Scholar] [CrossRef]
- Spang, A.; Saw, J.H.; Jørgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; Van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Zaremba-Niedzwiedzka, K.; Caceres, E.F.; Saw, J.H.; Bäckström, D.; Juzokaite, L.; Vancaester, E.; Seitz, K.W.; Anantharaman, K.; Starnawski, P.; Kjeldsen, K.U.; et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017, 541, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Dey, G.; Thattai, M.; Baum, B. On the Archaeal Origins of Eukaryotes and the Challenges of Inferring Phenotype from Genotype. Trends Cell Biol. 2016, 26, 476–485. [Google Scholar] [CrossRef]
- Kessin, R.H. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity; Bard, J.B.L., Barlow, P.W., Kirk, D.L., Eds.; Developmental and Cell Biology Series; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Martín-González, J.; Montero-Bullón, J.; Lacal, J. Dictyostelium discoideum as a non-mammalian biomedical model. Microb. Biotechnol. 2021, 14, 111–125. [Google Scholar] [CrossRef]
- Eichinger, L.; Noegel, A.A. Comparative genomics of Dictyostelium discoideum and Entamoeba histolytica. Curr. Opin. Microbiol. 2005, 8, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, R.; Khan, N.A. Biology and pathogenesis of Acanthamoeba. Parasites Vectors 2012, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Hillmann, F.; Forbes, G.; Novohradská, S.; Ferling, I.; Riege, K.; Groth, M.; Westermann, M.; Marz, M.; Spaller, T.; Winckler, T.; et al. Multiple Roots of Fruiting Body Formation in Amoebozoa. Genome Biol. Evol. 2018, 10, 591–606. [Google Scholar] [CrossRef]
- Eichinger, L.; Rivero, F. (Eds.) Dictyostelium Discoideum Protocols, 2nd ed.; Methods in Molecular Biology Series 983; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Manich, M.; Hernandez-Cuevas, N.; Villa, J.D.O.; Syan, S.; Marchat, L.A.; Olivo-Marin, J.-C.; Guillén, N. Morphodynamics of the Actin-Rich Cytoskeleton in Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2018, 8, 179. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.; Prochnik, S.E.; Ginger, M.; Dacks, J.B.; Carpenter, M.; Field, M.C.; Kuo, A.; Paredez, A.; Chapman, J.; Pham, J.; et al. The Genome of Naegleria gruberi Illuminates Early Eukaryotic Versatility. Cell 2010, 140, 631–642. [Google Scholar] [CrossRef]
- Paredez, A.; Assaf, Z.J.; Sept, D.; Timofejeva, L.; Dawson, S.C.; Wang, C.-J.R.; Cande, W.Z. An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 6151–6156. [Google Scholar] [CrossRef] [PubMed]
- Krtková, J.; Xu, J.; Lalle, M.; Steele-Ogus, M.; Alas, G.C.M.; Sept, D.; Paredez, A.R. 14-3-3 Regulates Actin Filament Formation in the Deep-Branching Eukaryote Giardia lamblia. mSphere 2017, 2, e00248-17. [Google Scholar] [CrossRef]
- Fort, P.; Blangy, A. The Evolutionary Landscape of Dbl-Like RhoGEF Families: Adapting Eukaryotic Cells to Environmental Signals. Genome Biol. Evol. 2017, 9, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Guerin, N.A.; Larochelle, D.A. Chimeric analysis of the small GTPase RacE in cytokinesis signaling in Dictyostelium discoideum. Exp. Cell Res. 2004, 295, 226–235. [Google Scholar] [CrossRef]
- Charest, P.G.; Firtel, R.A. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 2006, 401, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Stairs, C.W.; Ettema, T.J. The Archaeal Roots of the Eukaryotic Dynamic Actin Cytoskeleton. Curr. Biol. 2020, 30, R521–R526. [Google Scholar] [CrossRef]
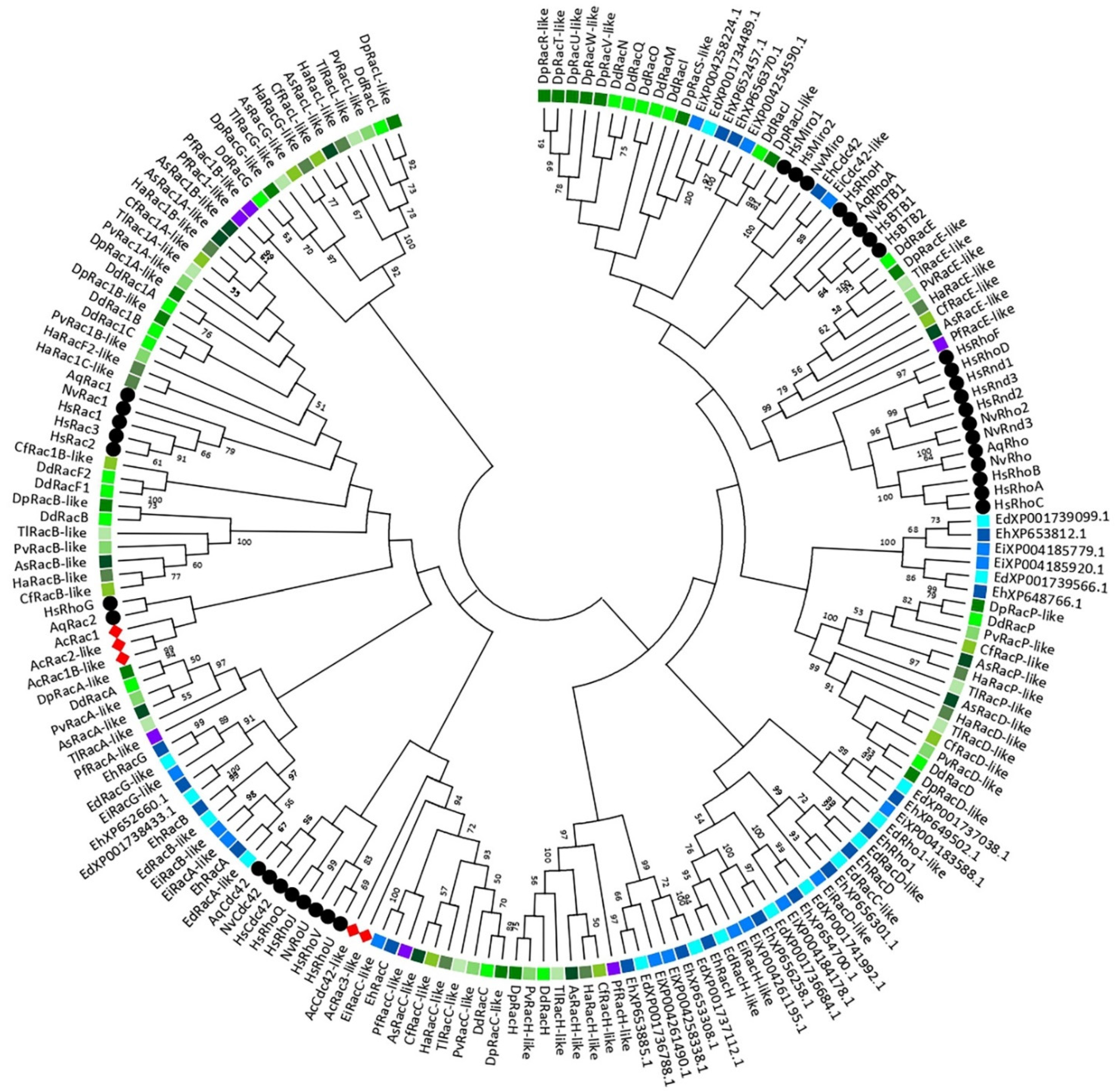
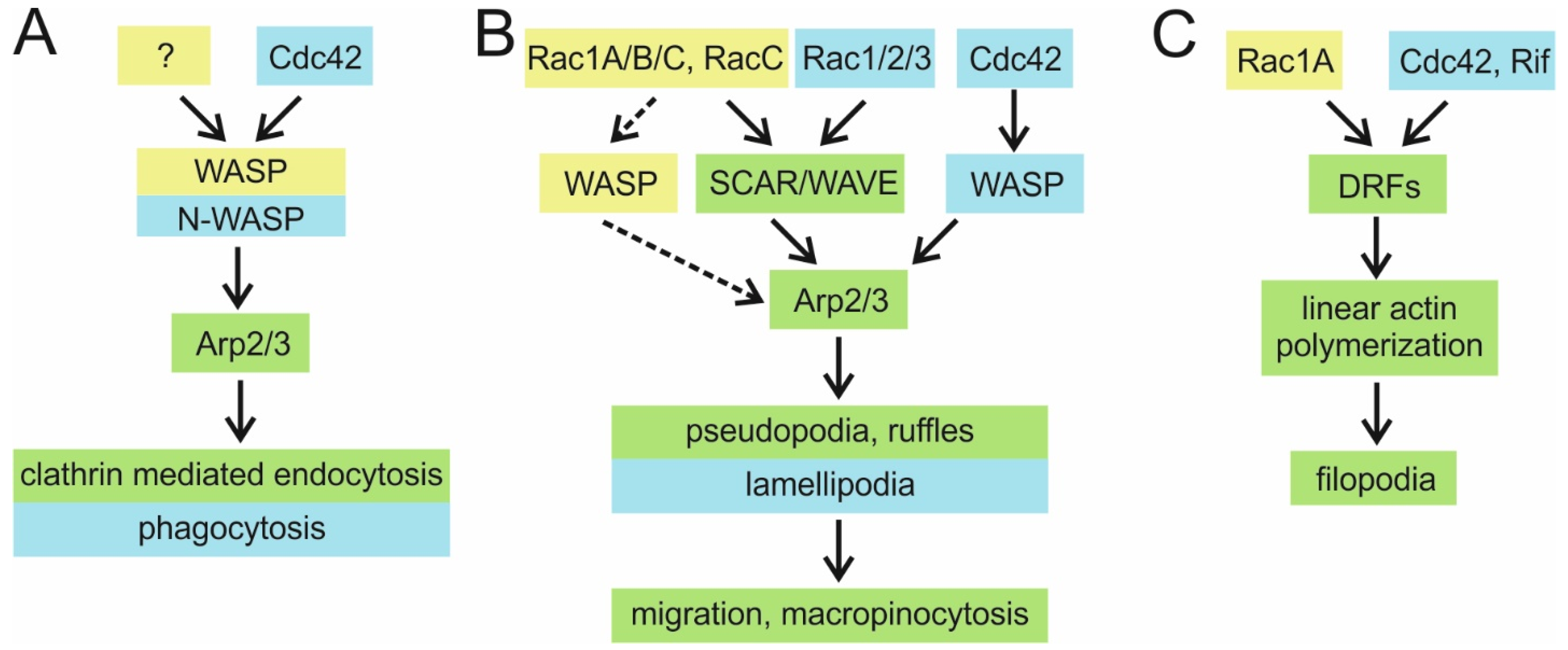
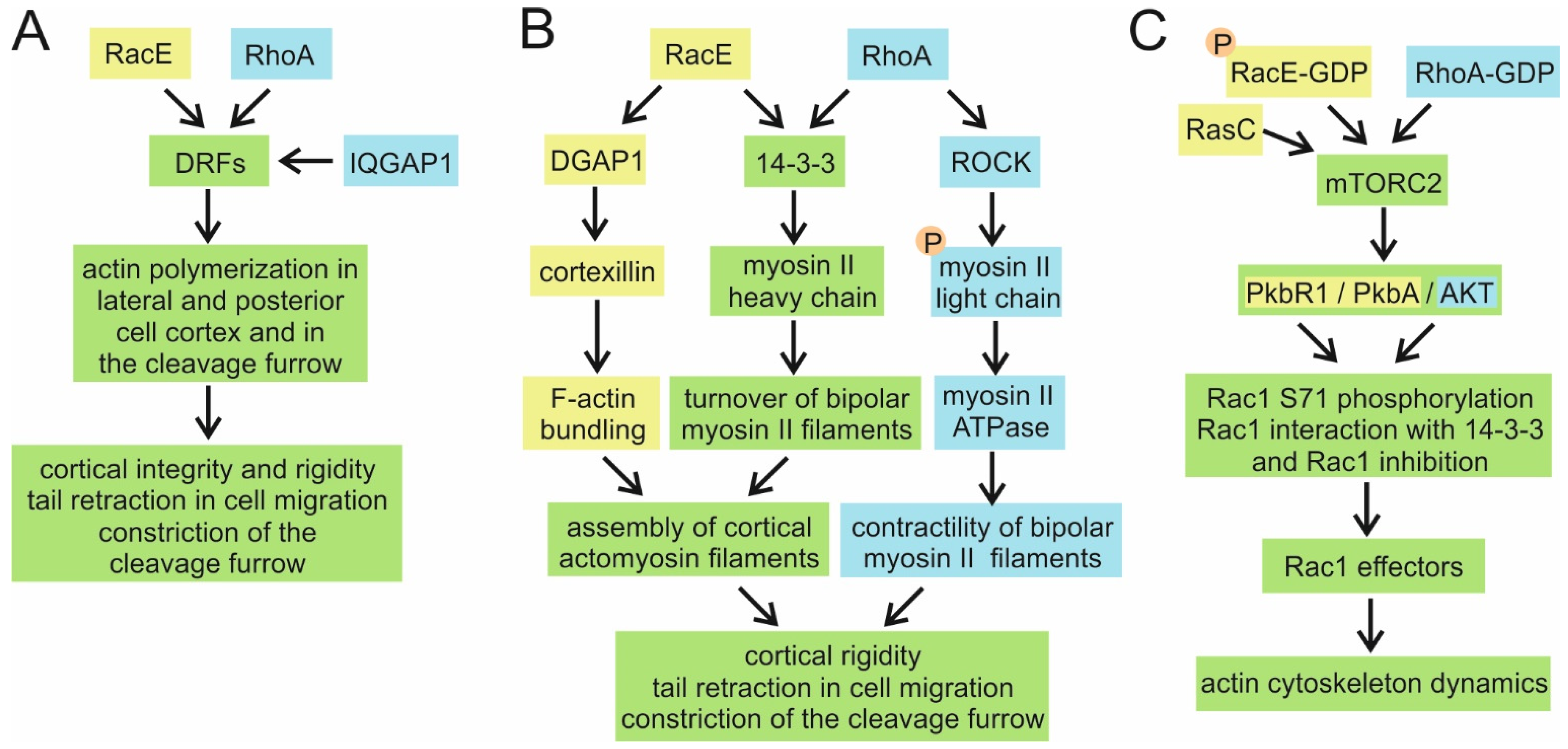
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filić, V.; Mijanović, L.; Putar, D.; Talajić, A.; Ćetković, H.; Weber, I. Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom. Cells 2021, 10, 1592. https://doi.org/10.3390/cells10071592
Filić V, Mijanović L, Putar D, Talajić A, Ćetković H, Weber I. Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom. Cells. 2021; 10(7):1592. https://doi.org/10.3390/cells10071592
Chicago/Turabian StyleFilić, Vedrana, Lucija Mijanović, Darija Putar, Antea Talajić, Helena Ćetković, and Igor Weber. 2021. "Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom" Cells 10, no. 7: 1592. https://doi.org/10.3390/cells10071592
APA StyleFilić, V., Mijanović, L., Putar, D., Talajić, A., Ćetković, H., & Weber, I. (2021). Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom. Cells, 10(7), 1592. https://doi.org/10.3390/cells10071592







