Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development
Abstract
:1. Introduction
2. Types, Origin, and Function of Major Regulatory ncRNAs
3. Evolution, Conservation, Species Specificity, Tissue Specificity, and Genotype- and Stress-Dependent Expression of ncRNAs
4. ncRNAs Mediating Plant Immunity against Attacking Pathogens
5. Deciphering the Molecular Mechanisms of ncRNAs Regulating the Response of Legumes to Water Stress
6. Role of ncRNAs in Plant Adaptation to Salinity Stress
7. Contribution of ncRNAs Attributing Plant Adaptation under Metal Toxicity Stress
8. Molecular Mechanisms of ncRNAs Regulating Nutrient Acquisition and Homeostasis in Legumes
9. Regulatory Role of ncRNAs for Shaping Developmental Processes in Legume Species
10. ncRNAs Orchestrating Nodulation, Symbiosis, and Root Development Processes
11. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, T.J. Legumes of the World. S. Afr. J. Bot. 2007, 73, 272–273. [Google Scholar] [CrossRef] [Green Version]
- Jukanti, A.K.; Gaur, P.M.; Gowda, C.L.; Chibbar, R.N. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): A review. Br. J. Nutr. 2012, 108, S11–S26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koornneef, M.; Alonso-Blanco, C.; Peeters, A.J.M. Genetic approaches in plant physiology. New Phytol. 1997, 137, 1–8. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. 2009, 25, 21–44. [Google Scholar] [CrossRef] [Green Version]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long non-coding RNAs. Mol. Cell. 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Santosh, B.; Varshney, A.; Yadava, P.K. Non-coding RNAs: Biological functions and applications. Cell Biochem. Funct. 2014, 33, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Zhen, Y.; Kudapa, H.; Jagadeeswaran, G.; Hivrale, V.; Varshney, R.K.; Sunkar, R. High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci. 2015, 235, 46–57. [Google Scholar] [CrossRef]
- Li, Z.; Xu, H.; Li, Y.; Wan, X.; Ma, Z.; Cao, J.; Li, Z.; He, F.; Wang, Y.; Wan, L.; et al. Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol. Biol. 2018, 96, 473–492. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Yu, K.; Lü, H.; Zhang, X.; Liu, X.; Sun, C.; Xu, H.; Zhang, J.; He, X.; Zhang, D. Transcriptome-wide identification of novel circular RNAs in soybean in response to low-phosphorus stress. PLoS ONE 2020, 15, e0227243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen, J.; Mysore, K.S.; Zhang, W.H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Expt. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef]
- Liu, D.; Mewalal, R.; Hu, R.; Tuskan, G.A.; Yang, X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic. Res. 2017, 4, 17031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long non-coding RNA in plants in the era of reference sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Yang, L.; Chen, L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Herr, A.J.; Jensen, M.B.; Dalmay, T.; Baulcombe, D.C. RNA polymerase IV directs silencing of endogenous DNA. Science 2005, 308, 118–120. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [Green Version]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.Y.; Poirier, Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 2013, 25, 4166–4182. [Google Scholar] [CrossRef] [Green Version]
- Sousa, C.; Johansson, C.; Charon, C.; Manyani, H.; Sautter, C.; Kondorosi, A.; Crespi, M. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell Biol. 2001, 21, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Nejat, N.; Mantri, N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 2018, 38, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466. [Google Scholar] [CrossRef]
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef]
- De la Rosa, C.; Lozano, L.; Castillo-Ramírez, S.; Covarrubias, A.A.; Reyes, J.L. Origin and evolutionary dynamics of the miR2119 and ADH1 regulatory module in Legumes. Genome Biol. Evol. 2020, 12, 2355–2369. [Google Scholar] [CrossRef]
- Cakir, O.; Candar-Cakir, B.; Zhang, B. Small RNA and degradome sequencing reveals important micro RNA function in Astragalus chrysochlorus response to selenium stimuli. Plant Biotech. J. 2016, 14, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, N.; Li, H.; Liu, J.; Fu, C.; Xiao, Z.; Wei, C.; Lu, X.; Feng, J.; Zhou, Y. Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 2016, 6, 34601. [Google Scholar] [CrossRef] [Green Version]
- De la Rosa, C.; Covarrubias, A.A.; Reyes, J.L. A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant Cell Environ. 2019, 42, 133–144. [Google Scholar] [CrossRef]
- Subramanian, S.; Fu, Y.; Sunkar, R.; Barbazuk, W.B.; Zhu, J.K.; Yu, O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom. 2008, 9, 160. [Google Scholar] [CrossRef] [Green Version]
- Szittya, G.; Moxon, S.; Santos, D.M.; Jing, R.; Fevereiro, M.P.; Moulton, V.; Dalmay, T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genom. 2008, 9, 593. [Google Scholar] [CrossRef] [Green Version]
- Jagadeeswaran, G.; Zheng, Y.; Li, Y.F.; Shukla, L.I.; Matts, J.; Hoyt, P.; Macmil, S.L.; Wiley, G.B.; Roe, B.A.; Zhang, W.; et al. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 2009, 184, 85–98. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2415. [Google Scholar] [CrossRef] [Green Version]
- Barrera-Figueroa, B.E.; Gao, L.; Diop, N.N.; Wu, Z.; Ehlers, J.D.; Roberts, P.A.; Close, T.J.; Zhu, J.K.; Liu, R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 2011, 11, 127. [Google Scholar] [CrossRef] [Green Version]
- Chi, X.; Yang, Q.; Chen, X.; Wang, J.; Pan, L.; Chen, M.; Yang, Z.; He, Y.; Liang, X.; Yu, S. Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS ONE 2011, 6, e27530. [Google Scholar] [CrossRef]
- Das, A.; Nigam, D.; Junaid, A.; Tribhuvan, K.U.; Kumar, K.; Durgesh, K.; Singh, N.K.; Gaikwad, K. Expressivity of the key genes associated with seed and pod development is highly regulated via lncRNAs and miRNAs in pigeonpea. Sci. Rep. 2019, 9, 18191. [Google Scholar] [CrossRef] [Green Version]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Dos Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P.; Sun, H.; Yu, J.; Yang, Q. Genome-wide identification of microRNAs in response to salt/alkali stress in Medicago truncatula through high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef] [Green Version]
- Valdés-López, O.; Yang, S.S.; Aparicio-Fabre, R.; Graham, P.H.; Reyes, J.L.; Vance, C.P.; Hernández, G. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010, 187, 805–818. [Google Scholar] [CrossRef]
- Golicz, A.A.; Singh, M.B.; Bhalla, P.L. The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome. Plant Physiol. 2018, 176, 2133–2147. [Google Scholar] [CrossRef] [Green Version]
- Joshi, T.; Patil, K.; Fitzpatrick, M.R.; Franklin, L.D.; Yao, Q.; Cook, J.R.; Wang, Z.; Libault, M.; Brechenmacher, L.; Valliyodan, B.; et al. Soybean Knowledge Base (SoyKB): A web resource for soybean translational genomics. BMC Genom. 2012, 13, S15. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef] [Green Version]
- Xuan, H.; Zhang, L.; Liu, X.; Han, G.; Li, J.; Li, X.; Liu, A.; Liao, M.; Zhang, S. PLNlncRbase: A resource for experimentally identified lncRNAs in plants. Gene 2015, 573, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Gallart, A.P.; Pulido, A.H.; de Lagrán, I.A.M.; Sanseverino, W.; Cigliano, R.A. GREENC: A wiki-based database of plant lncRNAs. Nucleic Acid Res. 2016, 44, D1161–D1166. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar] [CrossRef]
- Tian, B.; Wang, S.; Todd, T.C.; Johnson, C.D.; Tang, G.; Trick, H.N. Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genom. 2017, 18, 572. [Google Scholar] [CrossRef]
- Kohli, D.; Joshi, G.; Deokar, A.A.; Bhardwaj, A.R.; Agarwal, M.; Katiyar-Agarwal, S.; Srinivasan, R.; Jain, P.K. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS ONE 2014, 9, e108851. [Google Scholar] [CrossRef]
- Garg, V.; Khan, A.W.; Kudapa, H.; Kale, S.M.; Chitikineni, A.; Qiwei, S.; Sharma, M.; Li, C.; Zhang, B.; Xin, L.; et al. Integrated transcriptome, small RNA and degradome sequencing approaches provide insights into Ascochyta blight resistance in chickpea. Plant Biotech. J. 2019, 17, 914–931. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. 2021. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Zipfel, C.; Robatzek, S. Pathogen-associated molecular pattern triggered immunity: Veni, vidi? Plant. Physiol. 2010, 154, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Deng, Y.W.; Ning, Y.S.; Hu, Z.H.; Wang, G.L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant. Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Fang, Y.; Chen, L.; Wang, J.; Chen, X. Role of non-coding RNAs in plant immunity. Plant. Commun. 2021, 20, 100180. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Zhang, S.; Liu, D.; Duan, Y.; Dong, W. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 2012, 7, e39650. [Google Scholar] [CrossRef] [Green Version]
- Formey, D.; Sallet, E.; Lelandais-Brière, C.; Ben, C.; Bustos-Sanmamed, P.; Niebel, A.; Frugier, F.; Combier, J.P.; Debellé, F.; Hartmann, C.; et al. The small RNA diversity from Medicago truncatula roots under biotic interactions evidences the environmental plasticity of the miRNAome. Genome Biol. 2014, 15, 457. [Google Scholar] [CrossRef] [PubMed]
- Lelandais-Brière, C.; Naya, L.; Sallet, E.; Calenge, F.; Frugier, F.; Hartmann, C.; Gouzy, J.; Crespi, M. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant. Cell 2009, 21, 2780–2796. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.S.; Zeng, H.Q.; Liu, Z.P.; Yang, Z.M. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012, 35, 86–99. [Google Scholar] [CrossRef]
- Chen, L.; Wang, T.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012, 235, 375–386. [Google Scholar] [CrossRef]
- Chen, L.; Wang, T.; Zhao, M.; Zhang, W. Ethylene-responsive miRNAs in roots of Medicago truncatula identified by high-throughput sequencing at the whole genome level. Plant. Sci. 2012, 184, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Long, R.C.; Li, M.N.; Kang, J.M.; Zhang, T.J.; Sun, Y.; Yang, Q.C. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol. Plant. 2015, 154, 13–27. [Google Scholar] [CrossRef]
- Holt, D.B.; Gupta, V.; Meyer, D.; Abel, N.B.; Andersen, S.U.; Stougaard, J.; Markmann, K. micro RNA 172 (miR172) signals epidermal infection and is expressed in cells primed for bacterial invasion in Lotus japonicus roots and nodules. New Phytol. 2015, 208, 241–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Li, M.; Zhang, H.; Duan, L.; Sun, X.; Jiang, Q.; Zhang, H.; Hu, Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genom. 2019, 20, 730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Wang, Y.; Mou, F.; Tian, Y.; Chen, L.; Zhang, S.; Jiang, Q.; Li, X. Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front. Plant. Sci. 2016, 6, 1273. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Hossain, M.S.; Arikit, S.; Valdés-López, O.; Zhai, J.; Wang, J.; Libault, M.; Ji, T.; Qiu, L.; Meyers, B.C.; et al. Identification of micro RNA s and their mRNA targets during soybean nodule development: Functional analysis of the role of miR393j-3p in soybean nodulation. New Phytol. 2015, 207, 748–759. [Google Scholar] [CrossRef]
- Turner, M.; Yu, O.; Subramanian, S. Genome organization and characteristics of soybean microRNAs. BMC Genom. 2012, 13, 169. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, C.; Hao, Q.; Sha, A.; Zhou, R.; Zhou, X.; Yuan, L. Elucidation of miRNAs-mediated responses to low nitrogen stress by deep sequencing of two soybean genotypes. PLoS ONE 2013, 8, e67423. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.X.; Liu, Y.F.; Hu, X.Y.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant. Biol. 2011, 11, 5. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Zhang, A.; Zhao, Q.; Li, Z.; Lamboro, A.; He, H.; Li, Y.; Jiao, S.; Guan, S.; Liu, S.; et al. Genome-wide identification and analysis of long non-coding RNAs involved in fatty acid biosynthesis in young soybean pods. Sci. Rep. 2021, 11, 7603. [Google Scholar] [CrossRef]
- Ding, X.; Li, J.; Zhang, H.; He, T.; Han, S.; Li, Y.; Yang, S.; Gai, J. Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genom. 2016, 17, 24. [Google Scholar] [CrossRef] [Green Version]
- Arikit, S.; Xia, R.; Kakrana, A.; Huang, K.; Zhai, J.; Yan, Z.; Valdés-López, O.; Prince, S.; Musket, T.A.; Nguyen, H.T.; et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant. Cell 2014, 26, 4584–4601. [Google Scholar] [CrossRef] [Green Version]
- Khemka, N.; Singh, V.K.; Garg, R.; Jain, M. Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci. Rep. 2016, 6, 33297. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Chevala, V.N.; Garg, R. Genome-wide discovery and differential regulation of conserved and novel microRNAs in chickpea via deep sequencing. J. Exp. Bot. 2014, 65, 5945–5958. [Google Scholar] [CrossRef] [Green Version]
- Khemka, N.; Singh Rajkumar, M.; Garg, R.; Jain, M. Genome-wide profiling of miRNAs during seed development reveals their functional relevance in seed size/weight determination in chickpea. Plant. Direct 2021, 5, e00299. [Google Scholar] [CrossRef]
- Pradhan, S.; Verma, S.; Chakraborty, A.; Bhatia, S. Identification and molecular characterization of miRNAs and their target genes associated with seed development through small RNA sequencing in chickpea. Func. Integr. Genom. 2021, 21, 283–298. [Google Scholar] [CrossRef]
- Khandal, H.; Parween, S.; Roy, R.; Meena, M.K.; Chattopadhyay, D. MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci. Rep. 2017, 7, 4632. [Google Scholar] [CrossRef] [Green Version]
- Peláez, P.; Trejo, M.S.; Iñiguez, L.P.; Estrada-Navarrete, G.; Covarrubias, A.A.; Reyes, J.L.; Sánchez, F. Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom. 2012, 13, 83. [Google Scholar] [CrossRef] [Green Version]
- Patwa, N.; Nithin, C.; Bahadur, R.P.; Basak, J. Identification and characterization of differentially expressed Phaseolus vulgaris miRNAs and their targets during mungbean yellow mosaic India virus infection reveals new insight into Phaseolus-MYMIV interaction. Genomics 2019, 111, 1333–1342. [Google Scholar] [CrossRef]
- Parreira, J.R.; Cappuccio, M.; Balestrazzi, A.; Fevereiro, P.; de Sousa Araújo, S. MicroRNAs expression dynamics reveal post-transcriptional mechanisms regulating seed development in Phaseolus vulgaris L. Hortic. Res. 2021, 8, 18. [Google Scholar] [CrossRef]
- Mendoza-Soto, A.B.; Naya, L.; Leija, A.; Hernández, G. Responses of symbiotic nitrogen-fixing common bean to aluminum toxicity and delineation of nodule responsive microRNAs. Front. Plant. Sci. 2015, 6, 587. [Google Scholar] [CrossRef]
- Formey, D.; Iñiguez, L.P.; Peláez, P.; Li, Y.F.; Sunkar, R.; Sánchez, F.; Reyes, J.L.; Hernández, G. Genome-wide identification of the Phaseolus vulgaris sRNAome using small RNA and degradome sequencing. BMC Genom. 2015, 16, 423. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; Sanseverino, W.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Guo, F.; Zhang, Z.; Ding, H.; Meng, J.; Li, X.; Peng, Z.; Wan, S. Discovery, identification, and functional characterization of long noncoding RNAs in Arachis hypogaea L. BMC Plant. Biol. 2020, 20, 308. [Google Scholar] [CrossRef]
- Zhao, X.; Gan, L.; Yan, C.; Li, C.; Sun, Q.; Wang, J.; Yuan, C.; Zhang, H.; Shan, S.; Liu, J.N. Genome-wide identification and characterization of Long non-coding RNAs in Peanut. Genes 2019, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhang, C.X.; Pan, L.J.; Wang, T.; Chen, N.; Chen, M.N.; Yang, Z.; Guo, X.G.; Yu, S.L.; Chi, X.Y. Small RNA profiling reveal regulation of microRNAs in field peanut pod rot pathogen infection. Biologia 2020, 75, 1779–1788. [Google Scholar] [CrossRef]
- Gao, C.; Wang, P.; Zhao, S.; Zhao, C.; Xia, H.; Hou, L.; Ju, Z.; Zhang, Y.; Li, C.; Wang, X. Small RNA profiling and degradome analysis reveal regulation of microRNA in peanut embryogenesis and early pod development. BMC Genom. 2017, 18, 220. [Google Scholar] [CrossRef] [Green Version]
- Ram, M.K.; Mukherjee, K.; Pandey, D.M. Identification of miRNA, their targets and miPEPs in peanut (Arachis hypogaea L.). Comput. Biol. Chem. 2019, 83, 107100. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Zhao, K.; Li, F.; Li, K.; Ning, L.; He, J.; Xin, Z.; Yin, D. Small RNA and degradome deep sequencing reveals the roles of microRNAs in seed expansion in peanut (Arachis hypogaea L.). Front. Plant Sci. 2018, 9, 349. [Google Scholar] [CrossRef]
- Figueredo, M.S.; Formey, D.; Rodríguez, J.; Ibáñez, F.; Hernández, G.; Fabra, A. Identification of miRNAs linked to peanut nodule functional processes. J. Biosci. 2020, 45, 62. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y.; et al. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 392. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ma, X.; Ning, L.; Li, Z.; Zhao, K.; Li, K.; He, J.; Yin, D. Genome-wide identification of circular RNAs in peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 653. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Traore, S.M.; Xin, Z.; Ning, L.; Li, K.; Zhao, K.; Li, Z.; He, G.; Yin, D. Genome-wide identification and analysis of long noncoding RNAs (lncRNAs) during seed development in peanut (Arachis hypogaea L.). BMC Plant Biol. 2020, 20, 192. [Google Scholar] [CrossRef]
- Martins, T.F.; Souza, P.F.; Alves, M.S.; Silva, F.D.A.; Arantes, M.R.; Vasconcelos, I.M.; Oliveira, J.T. Identification, characterization, and expression analysis of cowpea (Vigna unguiculata [L.] Walp.) miRNAs in response to cowpea severe mosaic virus (CPSMV) challenge. Plant Cell Rep. 2020, 39, 1061–1078. [Google Scholar] [CrossRef]
- Shui, X.R.; Chen, Z.W.; Li, J.X. MicroRNA prediction and its function in regulating drought-related genes in cowpea. Plant Sci. 2013, 210, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kundu, A.; Pal, A. Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress. Plant Cell Tissue Organ Cult. 2011, 105, 233–242. [Google Scholar] [CrossRef]
- Nithin, C.; Thomas, A.; Basak, J.; Bahadur, R.P. Genome-wide identification of miRNAs and lncRNAs in Cajanus cajan. BMC Genom. 2017, 18, 878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzahrani, S.M.; Alaraidh, I.A.; Khan, M.A.; Migdadi, H.M.; Alghamdi, S.S.; Alsahli, A.A. Identification and characterization of salt-responsive microRNAs in Vicia faba by high-throughput sequencing. Genes 2019, 10, 303. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Kundu, A.; Pal, A. Identification and expression profiling of Vigna mungo microRNAs from leaf small RNA transcriptome by deep sequencing. J. Integr. Plant Biol. 2014, 56, 15–23. [Google Scholar] [CrossRef] [PubMed]
- DeBoer, K.; Melser, S.; Sperschneider, J.; Kamphuis, L.G.; Garg, G.; Gao, L.L.; Frick, K.; Singh, K.B. Identification and profiling of narrow-leafed lupin (Lupinus angustifolius) microRNAs during seed development. BMC Genom. 2019, 20, 135. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Zeng, H.Q.; Dong, C.X.; Yin, X.M.; Shen, Q.R.; Yang, Z.M. microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L.). Plant Sci. 2010, 178, 23–29. [Google Scholar] [CrossRef]
- Glazińska, P.; Kulasek, M.; Glinkowski, W.; Wojciechowski, W.; Kosiński, J. Integrated analysis of small RNA, transcriptome and degradome sequencing provides new insights into floral development and abscission in yellow lupine (Lupinus luteus l.). Int. J. Mol. Sci. 2019, 20, 5122. [Google Scholar] [CrossRef] [Green Version]
- Bhat, K.V.; Mondal, T.K.; Gaikwad, A.B.; Kole, P.R.; Chandel, G.; Mohapatra, T. Genome-wide identification of drought-responsive miRNAs in grass pea (Lathyrus sativus L.). Plant Gene 2020, 21, 100210. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Wang, S. MicroRNAs associated with drought response in the pulse crop common bean (Phaseolus vulgaris L.). Gene 2017, 628, 78–86. [Google Scholar] [CrossRef]
- Zheng, Y.; Hivrale, V.; Zhang, X.; Valliyodan, B.; Lelandais-Brière, C.; Farmer, A.D.; May, G.D.; Crespi, M.; Nguyen, H.T.; Sunkar, R. Small RNA profiles in soybean primary root tips under water deficit. BMC Syst. Biol. 2016, 10, 126. [Google Scholar] [CrossRef] [Green Version]
- Jovanović, Ž.; Stanisavljević, N.; Mikić, A.; Radović, S.; Maksimović, V. Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiol. Biochem. 2014, 83, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Bian, X.H.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Wang, T.; Sun, T.; Yu, X.; Tian, R.; Zhang, W.H. Identification of tissue-specific and cold-responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing. BMC Plant Biol. 2020, 20, 99. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Hossain, M.S.; Valdés-López, O.; Hoang, N.T.; Zhai, J.; Wang, J.; Libault, M.; Brechenmacher, L.; Findley, S.; Joshi, T.; et al. Identification and functional characterization of soybean root hair micro RNA s expressed in response to Bradyrhizobium japonicum infection. Plant Biotech. J. 2016, 14, 332–341. [Google Scholar] [CrossRef]
- Li, H.; Deng, Y.; Wu, T.; Subramanian, S.; Yu, O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010, 153, 1759–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuma, N.; Soyano, T.; Suzaki, T.; Kawaguchi, M. MIR2111-5 locus and shoot-accumulated mature miR2111 systemically enhance nodulation depending on HAR1 in Lotus japonicus. Nat. Commun. 2020, 11, 5192. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact root architecture 2 promotes root competence for nodulation through the miR2111 systemic effector. Curr. Biol. 2020, 30, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Laporte, P.; Jovanović, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008, 54, 876–887. [Google Scholar] [CrossRef]
- Combier, J.P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernié, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Hossain, M.S.; Wang, J.; Valdés-López, O.; Liang, Y.; Libault, M.; Qiu, L.; Stacey, G. miR172 regulates soybean nodulation. Mol. Plant-Microbe Int. 2013, 26, 1371–1377. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, L.; Zou, Y.; Chen, L.; Cai, Z.; Zhang, S.; Zhao, F.; Tian, Y.; Jiang, Q.; Ferguson, B.J.; et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 2014, 26, 4782–4801. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, Z.; Amyot, L.; Tian, L.; Xu, Z.; Gruber, M.Y.; Hannoufa, A. Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol. Genet. Genom. 2015, 290, 471–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.C.; Katinakis, P.; Hendriks, P.; Smolders, A.; de Vries, F.; Spee, J.; van Kammen, A.; Bisseling, T.; Franssen, H. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993, 3, 573–585. [Google Scholar] [CrossRef]
- De Luis, A.; Markmann, K.; Cognat, V.; Holt, D.B.; Charpentier, M.; Parniske, M.; Stougaard, J.; Voinnet, O. Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 2012, 160, 2137–2154. [Google Scholar] [CrossRef] [Green Version]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodríguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Lauressergues, D.; Delaux, P.M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.P. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef]
- Martín-Rodríguez, J.Á.; Leija, A.; Formey, D.; Hernández, G. The MicroRNA319d/TCP10 node regulates the common bean–rhizobia nitrogen-fixing symbiosis. Front. Plant Sci. 2018, 9, 1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sós-Hegedűs, A.; Domonkos, Á.; Tóth, T.; Gyula, P.; Kaló, P.; Szittya, G. Suppression of NB-LRR genes by miRNAs promotes nitrogen-fixing nodule development in Medicago truncatula. Plant Cell Environ. 2020, 43, 1117–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, A.; Paul, S.; Dey, A.; Pal, A. High throughput sequencing reveals modulation of microRNAs in Vigna mungo upon Mungbean Yellow Mosaic India Virus inoculation highlighting stress regulation. Plant Sci. 2017, 257, 96–105. [Google Scholar] [CrossRef]
- Liu, J.Q.; Allan, D.L.; Vance, C.P. Systemic signaling and local sensing of phosphate in common bean: Cross-talk between photosynthate and microRNA399. Mol. Plant 2010, 3, 428–437. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Wahid, A.; Kobayashi, N.S.M.A.; Fujita, D.B.S.M.A.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 153–188. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Nayyar, H. Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 2020, 139, 1–27. [Google Scholar] [CrossRef]
- Dasmandal, T.; Rao, A.R.; Sahu, S. Identification and characterization of circular RNAs regulating genes responsible for drought stress tolerance in chickpea and soybean. Indian J. Genet. 2020, 80, 1–8. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Jha, R.; Parida, S.K. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019, 38, 255–277. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A. Genomics enabled breeding approaches for improving cadmium stress tolerance in plants. Euphytica 2016, 208, 1–31. [Google Scholar] [CrossRef]
- Feng, S.J.; Zhang, X.D.; Liu, X.S.; Tan, S.K.; Chu, S.S.; Meng, J.G.; Zhao, K.X.; Zheng, J.F.; Yang, Z.M. Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus. RSC Adv. 2016, 6, 82157. [Google Scholar] [CrossRef]
- Zhou, Z.S.; Huang, S.Q.; Yang, Z.M. Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem. Biophys. Res. Commun. 2008, 374, 538–542. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [Green Version]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [Green Version]
- Smith, F.W.; Mudge, S.R.; Rae, A.L.; Glassop, D. Phosphate transport in plants. Plant Soil 2003, 248, 71–83. [Google Scholar] [CrossRef]
- Fan, X.; Naz, M.; Fan, X.; Xuan, W.; Miller, A.J.; Xu, G. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 2017, 68, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Uchida, R. Essential nutrients for plant growth: Nutrient functions and deficiency symptoms. Plant Nutr. Manag. Hawaiis Soils 2000, 4, 31–55. [Google Scholar]
- Simons, M.; Saha, R.; Guillard, L.; Clément, G.; Armengaud, P.; Cañas, R.; Maranas, C.D.; Lea, P.J.; Hirel, B. Nitrogen-use efficiency in maize (Zea mays L.): From ‘omics’ studies to metabolic modelling. J. Exp. Bot. 2014, 65, 5657–5671. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [Green Version]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Zhang, Y.; Dong, J.; Sun, Y.; Lim, B.L.; Liu, D.; Lu, Z.J. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genom. 2016, 17, 655. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.Q.; Zhu, Y.Y.; Huang, S.Q.; Yang, Z.M. Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J. Plant Physiol. 2010, 167, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. Ectopic expression of LjmiR156 delays flowering, enhances shoot branching, and improves forage quality in alfalfa. Plant Biotech. Rep. 2015, 9, 379–393. [Google Scholar] [CrossRef]
- Gao, R.; Austin, R.S.; Amyot, L.; Hannoufa, A. Comparative transcriptome investigation of global gene expression changes caused by miR156 overexpression in Medicago sativa. BMC Genom. 2016, 17, 658. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huen, A.K.; Rodriguez-Medina, C.; Ho, A.Y.Y.; Atkins, C.A.; Smith, P.M.C. Long-distance movement of phosphate starvation-responsive microRNAs in Arabidopsis. Plant Biol. 2017, 19, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Yang, Z.; Fei, Y.; Feng, J.; Zhu, H.; Huang, F.; Zhang, H.; Huang, J. Construction and analysis of degradome-dependent microRNA regulatory networks in soybean. BMC Genom. 2019, 20, 534. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Shin, C. Regulatory non-coding RNAs in plants: Potential gene resources for the improvement of agricultural traits. Plant Biotechnol. Rep. 2016, 10, 35–47. [Google Scholar] [CrossRef]
- Yan, J.; Cai, X.; Luo, J.; Sato, S.; Jiang, Q.; Yang, J.; Cao, X.; Hu, X.; Tabata, S.; Gresshoff, P.M.; et al. The REDUCED LEAFLET genes encode key components of the trans-acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus. Plant Physiol. 2010, 152, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Han, L.; Fu, C.; Wen, J.; Cheng, X.; Nakashima, J.; Ma, J.; Tang, Y.; Tan, Y.; Tadege, M.; et al. The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 2013, 25, 4845–4862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.Y.; Zhang, Z.G.; Huang, S.Y.; Han, X.; Wang, X.Y.; Pan, W.J.; Qin, H.T.; Qi, H.D.; Yin, Z.G.; Qu, K.X.; et al. Analysis of miRNAs targeted storage regulatory genes during soybean seed development based on transcriptome sequencing. Genes 2019, 10, 408. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Ji, R.; Li, Z.; Yu, Y.; Nakano, M.; Long, Y.; Feng, L.; Qin, C.; Lu, D.; Zhan, J.; et al. Soybean DICER-LIKE2 Regulates Seed Coat Color via Production of Primary 22-Nucleotide Small Interfering RNAs from Long Inverted Repeats. Plant Cell 2020, 32, 3662–3673. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Costas, A.G.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.H.; Oldroyd, G.E.; Poole, P.S.; et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, P.; Cao, X.; Wang, X.; Zhang, A.; Li, X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009, 378, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Yan, Z.; Libault, M.; Jeong, D.H.; Park, S.; Green, P.J.; Sherrier, D.J.; Farmer, A.; May, G.; Meyers, B.C.; et al. Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinform. 2010, 11, S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulcheski, F.R.; de Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, L.G.; Cordenonsi da Fonseca, G.; Morais, G.L.D.; de Oliveira, L.F.V.; Carvalho, J.B.D.; Kulcheski, F.R.; Margis, R. Metatranscriptomic analysis of small RNAs present in soybean deep sequencing libraries. Genet. Mol. Biol. 2012, 35, 292–303. [Google Scholar] [CrossRef]
- Zabala, G.; Campos, E.; Varala, K.K.; Bloomfield, S.; Jones, S.I.; Win, H.; Tuteja, J.H.; Calla, B.; Clough, S.J.; Hudson, M.; et al. Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biol. 2012, 12, 177. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.S.; Hoang, N.T.; Yan, Z.; Tóth, K.; Meyers, B.C.; Stacey, G. Characterization of the spatial and temporal expression of two soybean miRNAs identifies SCL6 as a novel regulator of soybean nodulation. Front. Plant Sci. 2019, 10, 475. [Google Scholar] [CrossRef]
- Branscheid, A.; Devers, E.A.; May, P.; Krajinski, F. Distribution pattern of small RNA and degradome reads provides information on miRNA gene structure and regulation. Plant Signal. Bhav. 2011, 6, 1609–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devers, E.A.; Branscheid, A.; May, P.; Krajinski, F. Stars and symbiosis: microRNA-and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011, 156, 1990–2010. [Google Scholar] [CrossRef] [Green Version]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, W.; Qin, E.; Liu, S.; Liu, H.; Grennan, A.K.; Liu, H.; Qin, R. Comprehensive Identification and Expression Profiling of Circular RNAs During Nodule Development in Phaseolus vulgaris. Front. Plant Sci. 2020, 11, 1638. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Pandey, V.; Singh, B.; Bhatia, S. Dynamics of miRNA mediated regulation of legume symbiosis. Plant Cell Environ. 2021, 44, 1279–1291. [Google Scholar] [CrossRef]
- Hoang, N.T.; Tóth, K.; Stacey, G. The role of microRNAs in the legume–Rhizobium nitrogen-fixing symbiosis. J. Exp. Bot. 2020, 71, 1668–1680. [Google Scholar] [CrossRef]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef] [PubMed]

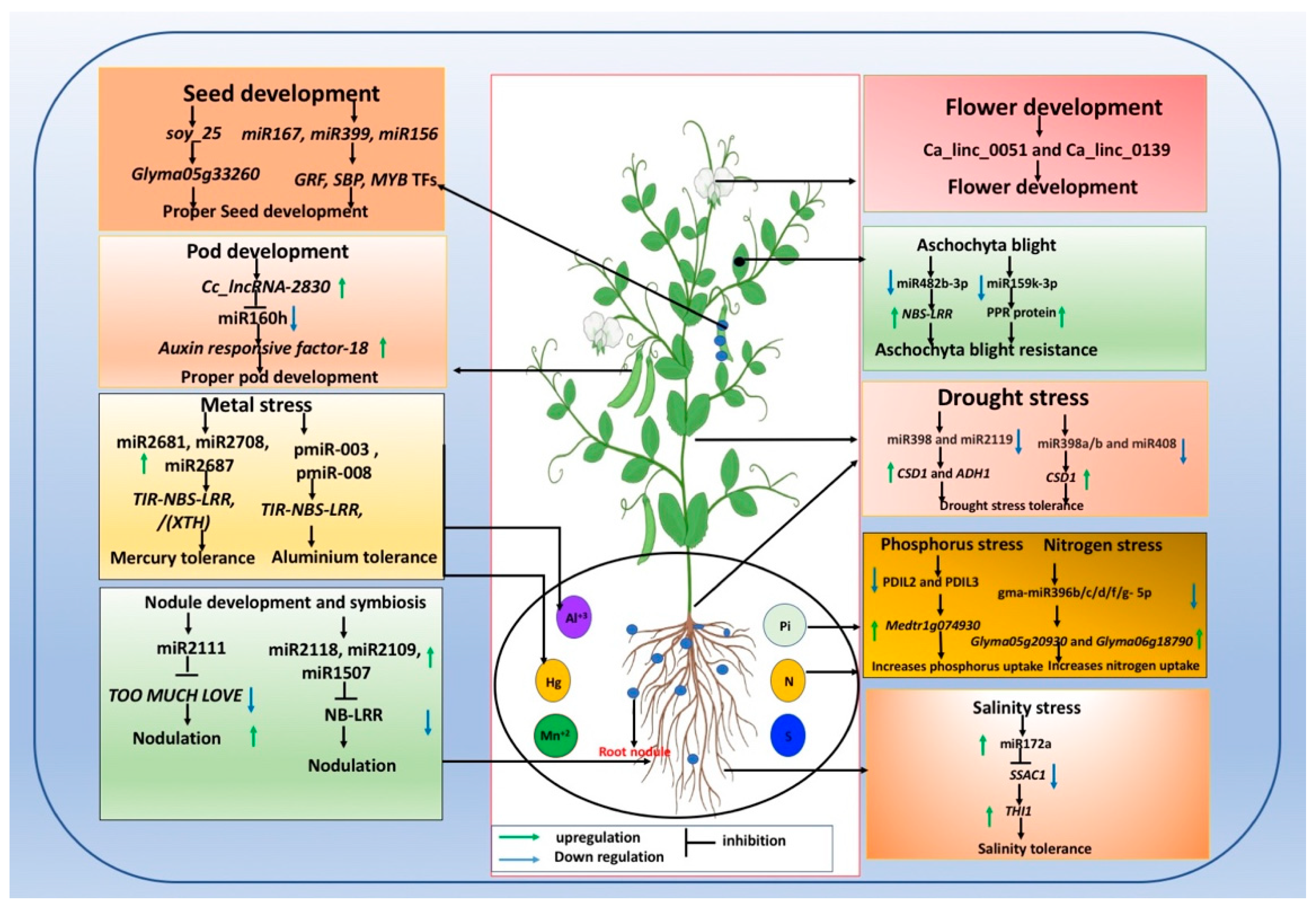

| Number of ncRNA | Crop | Genotype | Trait | Tissue | References |
|---|---|---|---|---|---|
| 416 miRNAs | M. truncatula | Jemalong A17 | Symbiosis and pathogenic interactions | Roots | [75] |
| 100 novel candidate miRNAs | M. truncatula | Root and nodule development | – | [76] | |
| 201 individual miRNAs | M. truncatula | Jemalong | Heavy metal | Seedlings | [77] |
| 326 known miRNAs and 21 new miRNAs | M. truncatula | Jemalong A17 | Aluminium toxicity | Root apices | [78] |
| 301 known miRNAs and identified 3 new miRNAs | M. truncatula | – | Ethylene response | Roots | [79] |
| 26 novel miRNAs | M. truncatula | Jemalong | – | Leaves | [50] |
| 385 conserved miRNAs and 68 novel miRNAs | M. truncatula Medicago sativa | Jemalong A17, Zhongmu-1 | Salinity stress | Roots | [80] |
| 876 miRNAs | M. truncatula | R108 | Salinity | Seedlings | [59] |
| 100 novel candidate miRNAs | M. truncatula | Jemalong A17 | Root and nodule development | Roots | [76] |
| 8 miRNAs | M. truncatula | Jemalong | – | Roots, shoots | [51] |
| 219 novel L. japonicus micro RNAs | Lotus japonicus | Gifu wild-type | Epidermal and cortical signalling events | – | [81] |
| 3030 long intergenic noncoding RNAs (lincRNAs), 275 natural antisense transcripts (lncNATs) | Soybean | Williams 82 | Salinity | Roots | [82] |
| 55 families of miRNAs | Soybean | Williams82 | Nodulation | Roots | [49] |
| 5372 circRNAs | Soybean | – | Developmental process | Stems, roots, mature leaves | [16] |
| 537 known and 70 putative novel miRNAs | Soybean | KS4607, KS4313N | Soybean cyst nematode | Roots | [67] |
| 71 miRNAs | Soybean | Williams 82 | Salinity | Roots | [83] |
| 364 + 21 | Soybean | Hairbin xiaoheidou, Liaodou 10 | Soybean cyst nematode | Roots | [74] |
| 284 miRNAs | Soybean | Williams 82 | Nodulation | Roots | [84] |
| 120 miRNA genes | Soybean | Williams82 | Root, nodule, organ development | Roots, stems, young leaves | [85] |
| 362 known miRNAs | Soybean | No.116, No.84-70 | Nitrogen stress | Roots, shoots | [86] |
| 38+8 miRNAs | Soybean | Heinong44 | Seed development | Seeds | [87] |
| 6018 lincRNAs | Soybean | – | Various agronomic trait | Flower buds, unopened flowers, florescence, pods, seeds | [61] |
| 46 lncRNAs | Soybean | MT72 and JN18 | Fatty acid synthesis | Pods | [88] |
| 158 novel miRNAs and 160 high-confidence soybean miRNAs | Soybean | NJCMS1A, NJCMS1B | Male sterility | Flower buds | [89] |
| 500 loci generating phasiRNAs from PHAS loci | Soybean | Williams 82 | Reproductive development | Anther and ovary tissues | [90] |
| 2248 lincRNAs | Chickpea | Flower development | Vegetative tissues, shoot apical meristem, young leaves | [91] | |
| 59 novel miRNAs | Chickpea | ICC4958 | Fusarium wilt, salinity | Roots | [68] |
| 157 miRNA loci | Chickpea | ICC4958 | Stress response | Leaves, inflorescence | [12] |
| 440 conserved miRNAs + 178 novel miRNAs | Chickpea | ICC4958 | Diverse cellular processes and metabolism | Leaves, stems, flower buds, young pods | [92] |
| 651 miRNAs | Chickpea | C 214, Pb 7, ILC 3279, ICCV 05530, BC3F6 | Aschochyta blight | Seedlings | [69] |
| 113 +243 miRNAs | Chickpea | JGK3 and Himchana1 | Seed size and development | Seeds | [93] |
| 74 known and 26 novel miRNAs | Chickpea | – | Seed development | Seeds | [94] |
| 3457 high-confidence lncRNAs | Chickpea | ICC4958, ICC1882, ICCV2, JG62 | Drought and salinity | – | [66] |
| 284 unique miRNAs | Chickpea | BGD72 | Drought and salinity | Roots | [95] |
| 114 miRNAs | Common bean | Leaves, flower, roots | [96] | ||
| 422 miRNAs | Common bean | MYMIV | Leaves | [97] | |
| 68 miRNAs | Common bean | Nutrient deficiency and manganese toxicity stress | Leaves, roots, nodules | [60] | |
| 72 known and 39 new miRNAs | Common bean | SER16 | Seed development | Seeds | [98] |
| 28 miRNAs | Common bean | Negro Jamapa 81 | Aluminium toxicity | Nodules | [99] |
| 185 mature miRNAs | Common bean | Negro Jamapa, Pinto Villa | N2-fixing symbiotic nodules | Flowers, leaves, roots, seedlings | [100] |
| 197 lncRNAs | Common bean | BAT93 | Fruit development | Flowers, pods, seeds, leaves, roots, stems | [101] |
| 16 conserved miRNAs | Common bean | Negro Jamapa, Pinto Villa | Different stress | – | [43] |
| 1442+ 189 lncRNAs | Groundnut | Fenghua-1 | Development, growth and stress tolerance | Roots, leaves, seeds | [102] |
| 50,873 lncRNAs | Groundnut | Growth and development | 15 different tissues | [103] | |
| 334 peanut miRNAs | Groundnut | Huayu 20 | Pod rot | [104] | |
| 70 known and 24 novel miRNAs | Groundnut | Luhua-14 | Pod development | Gynophores | [105] |
| 126 known miRNAs + 25 novel peanut | Groundnut | Development | Leaves, stems, roots, seeds | [56] | |
| 18 miRNAs | Groundnut | Disease resistant proteins, auxin responsive proteins | – | [106] | |
| 1,082 miRNAs | Groundnut | 8106, 8107 | Seed expansion | Seeds | [107] |
| 32 miRNAs | Groundnut | Nodule development | Nodules | [108] | |
| 29 known and 132 potential novel miRNAs | Groundnut | Baisha1016 | Ca deficiency | – | [109] |
| 347 circRNAs | Groundnut | RIL 8106, RIL 8107 | Seed development and size | – | [110] |
| 9388 known and 4037 novel lncRNAs | Groundnut | Huayou 7, Huayou 4 | Seed development | Seeds | [111] |
| 617 mature microRNAs | Cowpea | Cowpea severe mosaic virus | Leaves | [112] | |
| 17 new miRNAs | Cowpea | Dan lla, Tvu7778 | Drought | Leaves, roots | [113] |
| 157 miRNA genes | Cowpea | CB46, IT93K503-1 | Drought | Leaves | [55] |
| 18 miRNAs | Cowpea | Salinity stress | Roots | [114] | |
| 616 mature miRNAs + 3919 lncRNAs | Pigeonpea | – | – | – | [115] |
| 3919 lncRNAs | Pigeonpea | – | – | [115] | |
| 3019 lncRNAs and 227 miRNAs | Pigeonpea | Asha | Seed and pod development | Seeds, pods | [57] |
| 298 upregulated and 395 downregulated 284 upregulated and 243 downregulated | Faba bean | Hassawi-3 ILB4347 | Salinity | Leaves | [116] |
| 66 miRNAs | Urd bean | Leaves, stems, roots | [117] | ||
| 56miRNAs | Narrow-leafed lupin | Tanjil | Seed development | Stems, leaves, seeds | [118] |
| 167 miRNAs | White lupin | Phosphate deficiency | Roots, stems, leaves | [119] | |
| 394 known and 28 novel miRNAs and 316 phased siRNAs | Yellow lupine | Taper | Floral development and abscission | Flowers | [120] |
| 143 and 128 | Lathyrus | IC-143067 | Drought | – | [121] |
| 47 and 44 miRNAs | Alfalfa | Phosphorus deficiency | Roots, shoots | [13] | |
| 371 circRNAs | Soybean | Bogao, Nannong 94156 | Phosphorus deficiency | Roots | [14] |
| Name of ncRNA | Crop | Trait/Stress | Target Gene(s)/Protein Coding Gene(s) | Function | References |
|---|---|---|---|---|---|
| miR408 | Chickpea | Drought | DREB | Overexpression represses plantacyanin encoding genes and controls DREB regulation under water stress | [122] |
| 16 drought-responsive miRNAs | Common bean | Drought | TFs and protein kinases | Control drought stress by targeting various TFs and protein kinases | [123] |
| 6 downregulated and 6 upregulated miRNAs | Soybean | Drought | Auxin signalling, plantacyanin, Cu/Zn superoxide dismutases | Control drought stress by targeting auxin signalling, plantacyanin and Cu/Zn superoxide dismutases encoding genes | [124] |
| 44 drought-responsive miRNAs | Cowpea | Drought | Zinc finger family protein, serine/threonine protein kinase | Involved in development and stress response | [55] |
| vun-miR5021, vun-miR156b-3p, vun-miR5021, vun-miR156b, vun-miR156f | Cowpea | Drought | Kelch repeat-containing F-box protein, CPRD86, P5CS, multicystatin gene, and glutathione reductase | Induce genes PLD (phospholipase D), APX (ascorbate peroxidase) and P5CS (delta 1-pyrroline-5-carboxylate synthase) under stress | [113] |
| miR162, miR164, miR319, miR403, miR828, miR160a, miR160b, miR171e, vun_cand015, vun_cand033, vun_cand048, miR171b, miR171d, miR2111b, miR390b, and miR393, vun_cand001, vun_cand010, vun_cand041, vun_cand057 | Cowpea | Drought | ARF10, ARF8, zinc finger protein, basic-helix-loop-helix (bHLH), TF leucine-rich repeat transmembrane protein kinase, pentatricopeptide repeat-containing protein | Involved in development and stress response | [55] |
| miR398a/b, miR408 | Pea | Drought | Copper superoxide dismutase, CSD1 | Reduce oxidative stress | [125] |
| lsa-miR169b, lsa-miR1508a, lsa-miR319a, lsa-miR156a, lsa-miR398b, lsa-miR396d, lsa-miR166b, lsa-miR390a, lsa-miR167b, lsa-miR186, lsa-miR786, lsa-miR897, lsa- miR969 and lsa-miR1361, miR397, miR398, miR164, miR399 | Lathyrus | Drought | F-box, U-Box or protein coding genes involved in proline, betain, and osmolyte biosynthesis pathway | Induce osmo-protective compounds under stress | [121] |
| Chickpea | Drought and salinity | LACCASE4, COPPER SUPEROXIDE DISMUTASE (Cu-SOD), NAC1 and PHO2/UBC24 | Increase lateral root formation and improves uptake of K+ under salinity stress | [95] | |
| MIR2119 and MIR398a | Common bean | Drought | ALCOHOL DEHYDROGENASE 1 (ADH1) and COPPER-ZINC SUPEROXIDE DISMUTASE 1 (CSD1) | By reducing oxidative stress | [45,48] |
| pvu-miR2118 | Common bean | Drought | – | Controls drought stress | [43] |
| miR169, miR398a/b and miR408 | M. truncatula | Drought stress | Copper proteins COX5b, copper superoxide dismutase, and plantacyanin | [58] | |
| miR172a | Soybean | Salinity | Glyma.10G116600, Glyma.02G087400, Glyma.13G329700, Glyma.12G073300, Glyma.15G044400, Glyma.11G053800, AP2/EREBP-type TF gene SSAC1, thiamine biosynthesis gene THI1 | Induction cleaves mRNA transcripts of salt-suppressed AP2 domain-containing genes increasing expression of thiamine biosynthesis gene THI1 and resulting salinity tolerance | [126] |
| 18 conserved miRNAs | Cowpea | Salinity | 15 target genes | Control plant development and root growth under stress conditions by targeting various TF genes viz., SBP, ARF, SPL, TCP, NFY, and AP2 | [114] |
| miR156_1, miR156_10, car-miR008, car-miR011, car-miR015 | Chickpea | Salinity | Squamosa promoter-binding protein | Target protein-encoding gene to control salinity stress | [68] |
| lncRNA TCONS_ 00097188, TCONS_00046739, TCONS_00100258, TCONS_ 00118328, TCONS_00047650, lncRNA TCONS_ 00020253, TCONS_00116877 | Medicago truncatula | Salinity | Medtr6g006990, cytochrome P450, Medtr3g069280, Medtr1g081900, Medtr7g094600 | Upregulate various gene expression contributing to salinity stress adaptation | [15] |
| TCONS_ 00292946, TCONS_00176941, TCONS_00011551 | Groundnut | Salinity | – | Control salinity stress tolerance | [102] |
| pvu-miR159.2 | Common bean | Salinity | – | – | [43] |
| miR160, miR156/157, miR159, miR169, miR172, miR408 | Cowpea | Salinity stress | Auxin response factor (ARF), squamosa promoter-binding protein (SBP), TCP family transcription factor, CCAAT-binding transcription factor (CBF), PHAP2B protein, APETALA2 protein (AP2), Basic blue copper protein/Plantacyanin | Target TFs and control salinity stress | [114] |
| lncRNA MtCIR1 | Medicago truncatula | Cold stress | MtCBF genes | Controls cold tolerance | [127] |
| soy_25 | Soybean | Seed development | Glyma05g33260 | Controls seed development | [87] |
| gma-miR168 | Soybean | – | Glyma16g34300 | ||
| miR167, miR399, miR156, miR319, miR164, miR166, miR1507 and miR396 | Narrow leaf lupin | Seed development | GROWTH-REGULATING FACTOR (GRF) TF, SBP-box transcription factors, MYB transcription factors, Zinc finger domain proteins, molybdate transporter 1, calcium-transporting ATPase 8, TMV resistance protein N, lysine-specific demethylase JMJ16, nudix hydrolase protein | Target TF (Class III HD-Zip, NAC) related to seed development process | [118] |
| ahy_novel_miRn1 to ahy_novel_miRn132, miR3509, miR3511, and miR3512, miR159 and miR167, miR3514, miR3518 | Groundnut | Ca deficiency driven embryo abortion | TCP3, AP2, EMB2750, GRFs, HsfB4, DIVARICATA, CYP707A1, CYP707A3 | Regulate embryo abnormality under Ca deficiency by modulating the target genes | [109] |
| miR_18, miR_6, miR_11, miR_29, miR_6, miR_38, miR_6, pvu-miR399a, miR_18, miR_33, miR_16, pvu-miR156i | Common bean | Seed development | DEHYDRIN FAMILY PROTEIN (RAB18), DEAD BOX RNA HELICASE (PRH75) CESA3, LEUCINE-RICH PROTEIN KINASE FAMILY PROTEIN, PRH75, MEE9, EM1, PHO2, RAB18, PROTEIN KINASE SUPERFAMILY PROTEIN, DUF827, and SPL2 | Regulate these genes during various stages of seed development, viz., seed filling, maturation, and dormancy | [98] |
| XR_001593099.1, MSTRG.18462.1, MSTRG.34915.1, MSTRG.41848.1, MSTRG.22884.1, MSTRG.12404.1, MSTRG.26719.1, MSTRG.35761.1, MSTRG.20033.1, MSTRG.13500.1, MSTRG.9304.1 | Groundnut | Seed development | XM_016114848.1, XM_ 016087708.1, XM_016309191.1, XM_ 016324297.1, XM_016327810.1, XM_016116309.1, XM_ 016335443.1, XM_ 016310265.1, XM_ 016091385.1 | Regulate groundnut seed development by modulating the target genes encoding MADS-box transcription factor 23-like, protein transport protein sec31-like, squamosa promoter-binding-like protein 14 | [111] |
| Ca_linc_0051 and Ca_linc_0139 | Chickpea | Flower development | [91] | ||
| miR156/157, miR164, miR167, miR1088, miR172, miR396 | Groundnut | Pod development | SPL, NAC, PPRP, AP2, GRF | Control pod development | [105] |
| Cc_lncRNA-2830 | Pigeonpea | Pod development | miR160h- Auxin responsive factor-18 | Upregulates Cc_lncRNA-2830, sequesters miR160h promoting expression of auxin responsive factor-18 and helps in pod formation | [57] |
| gma-miR156b and gma-miR156f, gma-miR162a, gma-miR162b, gma- miR162c, gma-miR399d, gma-miR399e, gma- miR399f gma-miR399g | Soybean | Male sterility | MADS-box transcription factor, NADP-dependent isocitrate dehydrogenase, 6-phosphogluconate dehydrogenase, NADH-ubiquinone oxidoreductase | Target these genes and cause programmed cell death, ROS toxicity and energy deficiency | [89] |
| lncRNA MSTRG.45502.1, lncRNAs MSTRG.40968.1 | Soybean | Lipid metabolic processes | XM_003538388.3,XM_006588497.2 00,061 | [88] | |
| miR393j-3p | Soybean | Nodule development | Early Nodulin 93 (ENOD93) | Targets Early Nodulin 93 (ENOD93) gene and regulates nodule formation | [84] |
| gma-miR2606b, gma-miR4416 | Soybean | Nodule development | Mannosyl- oligosaccharide 1, 2-alpha-mannosidase, Rhizobium-induced peroxidase 1 (RIP1)-like peroxidase gene | Target these genes to positively and negatively regulate the nodulation process | [128] |
| miR482, miR1512, miR1515 | Soybean | Nodule development | Gm12g28730, Gm17g04060, Gm04g05920, Glyma09g27690 | Regulates nodulation process | [129] |
| miR2111 | Lotus japonicus | Nodulation | TOO MUCH LOVE, a nodulation suppressor | Low expression after rhizobial infection relying on shoot-acting HYPERNODULATION ABERRANT ROOT FORMATION1 (HAR1) receptor | [130] |
| miR2111 | M. truncatula | Nodulation and symbiosis | Too Much Love 1, Too Much Love 2 | Positively controls root symbiotic nodulation, which is systemic from shoots and depends on the CRA2 receptor | [131] |
| MIR166 | M. truncatula | Root and nodule development | Class-III HD-ZIP genes | Overexpression reduced the number of symbiotic nodules and lateral roots | [132] |
| microRNA169 | M. truncatula | Nodule development | MtHAP2-1 | Regulates MtHAP2-1 gene controlling symbiotic nodule formation | [133] |
| ahy-mi399, ahy-miR159, ahy-miR3508 | Groundnut | Nodule infection | Pectinesterase gene | Regulate nodulation development process | [108] |
| miRNA 172 | Soybean | Nodulation | AP2 transcription factor | Controls miR172 expression and regulates AP2 TF activity | [134] |
| miRNA 172c | Soybean | Nodulation | Nodule Number Control1 | Controls nodule formation by repressing its target gene | [135] |
| miRNA156 | Lotus japonicus | Nodulation | ENOD genes, SymPK, POLLUX, CYCLOPS, Cerberus, and Nsp1, SPLs | Represses downstream target SPLs and other nodulation genes | [136] |
| MtENOD40 | M. truncatula | Nodule development | – | Regulates re-localization of proteins | [38] |
| GmENOD40 | Soybean | Nodule development | – | Regulates re-localization of proteins | [137] |
| miR156e, miR156g, miR167b | M. truncatula | Symbiosis signals | Induced by Myc-LCO and repressed by Nod signals | [75] | |
| miR172a | Lotus japonicus | Epidermal infection during symbiosis | APETALA2-type (AP2) transcription factors | Targets AP2 TF and regulates bacterial symbiosis | [81] |
| miR171 isoform, miR397 | Lotus japonicus | N2 fixation | Laccase copper protein family, Nodulation Signalling Pathway2 | Respond to symbiotic infection and nodule function | [138] |
| miR396 | M. truncatula | Root growth and mycorrhizal associations | Growth-regulating factor genes (MtGRF) and two bHLH79-like target genes | Regulates root growth and mycorrhizal associations | [139] |
| miR171h | M. truncatula | Mycorrhizal colonization | NSP2 | Targets NSP2 and modulates mycorrhizal colonization | [140] |
| miR1507, miR2118, miR2119, miR2199 | M. truncatula | Pathogen infection | TIR-NBS-LRR proteins targeted by miR2118 auxin response factor (ARF) | miRNA-mediated plant defence response | [51] |
| miR319d | Common bean | Rhizobium N2 fixation | TCP10 (Phvul.005G067950) | [141] | |
| miR1507, miR2109, miR2118 | M. truncatula | Nodulation and symbiosis | NB-LRR genes | Suppress activity of NB-LRR genes and allow nodulation process | [142] |
| ENOD40 | Soybean | Nodule development | – | – | [137] |
| ENOD40 | and M. truncatula | Nodule development | - | – | [38] |
| 617 mature microRNAs | Cowpea | Cowpea severe mosaic virus | Kat-p80, DEAD-Box, GST, and SPB9 | Involved in defence response to CSMV | [112] |
| vun-miR156a, vun-miR156b, vun-miR156b-3p, vun-miR156b-5p, vun-miR156f, vun-miR156 g, vun-miR157d, vun-miR2610a, vun-miR2673b, vun-miR5021 | Ted2 protein gene, Glutathione reductase, R3H domain protein gene, P5CS, Phosphoribosylpyrophosphate amidotransferase, 5-aminoimidazole ribonucleotide carboxylase, R3H domain protein gene, Ted2 protein, 5-aminoimidazole ribonucleotide carboxylase, Vigna unguiculata extensine-like protein 3, Aspartic proteinase, CPRD86 | ||||
| miR156, miR159, miR160, miR166, miR398, miR1511, miR1514, miR2118, and novel vmu-miRn7, vmu-miRn8, vmu-miRn13, vmu-miRn14 | Urdbean | MYMIV | NB-LRR, NAC, MYB, Zinc finger, CCAAT-box transcription factor, fructose 2-6 bisphosphate, HDZIP protein | Participate in defence/immune response to MYMIV | [143] |
| miR530 | Chickpea | Fusarium wilt infection | Zinc knuckle- and microtubule-associated proteins | Regulates plant defence against pathogen attack | [68] |
| miR166 | Chickpea | – | HD-ZIPIII transcription factor | – | [68] |
| car-miRNA008 | Chickpea | Natural defence | Chalcone synthase (CHS) gene | Regulates plant defence against pathogen attack | [68] |
| car-miR2118, car-miR5213 | Chickpea | Defence response | TIR-NBS-LRR | Regulate plant defence against pathogen attack | [68] |
| miR156, miR159, miR160, miR162, miR164, miR168, miR172, miR393, miR408 | Chickpea | Stress response and development processes | SPB factor, MYB transcription factor, ARFs, DCL1, HD-Zip, Arg onaute 1, AP2, F-box protein, plantacyanin | Target superoxide dismutases, plantacyanin, laccases and F-box proteins genes during stress | [12] |
| ahy-miR396e-5, ahy-miR3509-5p, ahy-miR166f, ahy-miR159b | Groundnut | Pod rot | c39419_g1_i1, c40055_g1_i3, c31393_g1_i1, c41016_g4_i1 | Related to defence response | [104] |
| miR482b-3p, miR159k-3p, nov_miR66, miR171, miR162, miR167c, miR171b | Chickpea | Ascochyta blight resistance | NBS-LRR, PR protein, serine-threonine kinase, PPR protein, Dicer-like gene (Ca_01367), Dof zinc finger (Ca_19433), ERF (Ca_00359) gene | Produce pathogenesis-related protein, ROS activity, cell wall synthesis, hormone synthesis, R gene activation | [69] |
| miR171, miR159, miR399, miR398, miR408, miR9750, miR2119, miR1512 | Soybean | Rootknot nematode | ATPase, Glycosyl hydrolases, multicopper oxidase, SOD, peroxidase, Glucose-6-phosphate dehydrogenase encoding genes | Regulate PR genes, oxidative stress and defence response | [67] |
| miR156/157, miR164, miR167 and miR1088, miR172, miR396 | Groundnut | – | SPL, NAC, PPRP, AP2 GRF | Control seed development | [105] |
| miR157, miR156, miR170, miR172, miR319, miR398, pvu-miR159.2, pvu-miR2118, gma-miR1508, gma-miR1526, gma-miR1532, miR160, miR397, miR399, miR408, pvu-miR1509, pvu-miR1514a | Common bean | Manganese toxicity | – | Upregulated miR157, miR156, miR170, miR172 and downregulated pvu-miR2118, gma-miR1508, gma-miR1526, and gma-miR1532, etc. | [60] |
| miR2681, miR2708, miR2687 | M. truncatula | Mercury tolerance | TIR-NBS-LRR, TC114805, xyloglucan endotransglucosylase/ hydrolase (XTH) | XTH helps in cell wall development under heavy metal stress | [77] |
| Gm03circRNA1785 | Soybean | gma-miR167c and GmARF6 and GmARF8 | [16] | ||
| PDIL1, PDIL2, PDIL3 | M. truncatula | Phosphate starvation | MtPHO2, Medtr1g074930 | Regulate phosphate uptake | [17] |
| miR399 | Common bean | Phosphorus deficiency | PvHAD1 | [144] | |
| gma-miR156b/6f-5p, gma-miR396b∼g-5p, gma-miR5372-5p, gma-miR159d-3p, gma-miR396b∼g-5p | Soybean | Nitrogen deficiency | Glyma07g31580, Glyma05g20930, Glyma06g18790, Glyma09g02600, Glyma05g23280, Glyma07g05550, Glyma16g02090, Glyma17g16750, Glyma19g44930, Glyma15g08010, Glyma19g01200 | Play role in protein degradation | [86] |
| miR399, miR398, miR156, miR159, miR164, miR168, miR172, miR393, miR408 | Alfalfa | Phosphate starvation | Phosphate transporter, Copper chaperone for SOD, Squamosa promoter-binding-like (SPL), MYB TF, auxin response factor (ARF), GRAS, MATE | Regulate phosphate uptake | [13] |
| circ_000232 | Soybean | Phosphorus deficiency | Glyma.13G117700 | Regulates P use efficiency | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chand Jha, U.; Nayyar, H.; Mantri, N.; Siddique, K.H.M. Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development. Cells 2021, 10, 1674. https://doi.org/10.3390/cells10071674
Chand Jha U, Nayyar H, Mantri N, Siddique KHM. Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development. Cells. 2021; 10(7):1674. https://doi.org/10.3390/cells10071674
Chicago/Turabian StyleChand Jha, Uday, Harsh Nayyar, Nitin Mantri, and Kadambot H. M. Siddique. 2021. "Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development" Cells 10, no. 7: 1674. https://doi.org/10.3390/cells10071674






