Piezo1 Mechanosensitive Ion Channel Mediates Stretch-Induced Nppb Expression in Adult Rat Cardiac Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Cardiac Fibroblasts and Cardiomyocytes
2.2. Experimental Stretch Protocols
2.3. Experimental Stimuli
2.4. Gene Expression Analysis
2.5. BNP ELISA
2.6. Gene Silencing
2.7. Statistics
3. Results
3.1. Mechanical Stretch Induces BNP Expression in Cardiac Fibroblasts
3.2. Recombinant BNP Inhibits Profibrotic Gene Expression in Cardiac Fibroblasts
3.3. Both Cardiomyocytes and Cardiac Fibroblasts Express Nppb
3.4. Stretch-Induced Nppb and Tgfb1 Expression Are Mediated by Piezo1
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoffman, B.D.; Grashoff, C.; Schwartz, M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature 2011, 475, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Orr, A.W.; Helmke, B.P.; Blackman, B.R.; Schwartz, M.A. Mechanisms of mechanotransduction. Dev. Cell 2006, 10, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.S. Mechanotransduction–a field pulling together? J. Cell Sci 2008, 121, 3285–3292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, W.H. Biomechanical regulation of in vitro cardiogenesis for tissue-engineered heart repair. Stem Cell Res. 2013, 4, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, W.; Jones, D.; McLaurin, L.P. Wall stress and patterns of hypertrophy in the human left ventricle. J. Clin. Investig. 1975, 56, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Toischer, K.; Rokita, A.G.; Unsold, B.; Zhu, W.; Kararigas, G.; Sossalla, S.; Reuter, S.P.; Becker, A.; Teucher, N.; Seidler, T.; et al. Differential cardiac remodeling in preload versus afterload. Circulation 2010, 122, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Frangogiannis, N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Invest. 2017, 127, 1600–1612. [Google Scholar] [CrossRef] [Green Version]
- Van Nieuwenhoven, F.A.; Munts, C.; Op ‘t Veld, R.C.; Gonzalez, A.; Diez, J.; Heymans, S.; Schroen, B.; van Bilsen, M. Cartilage intermediate layer protein 1 (CILP1): A novel mediator of cardiac extracellular matrix remodelling. Sci Rep. 2017, 7, 16042. [Google Scholar] [CrossRef] [Green Version]
- Tsuruda, T.; Boerrigter, G.; Huntley, B.K.; Noser, J.A.; Cataliotti, A.; Costello-Boerrigter, L.C.; Chen, H.H.; Burnett, J.C., Jr. Brain natriuretic Peptide is produced in cardiac fibroblasts and induces matrix metalloproteinases. Circ. Res. 2002, 91, 1127–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imanaka-Yoshida, K. Tenascin-C in cardiovascular tissue remodeling: From development to inflammation and repair. Circ. J. 2012, 76, 2513–2520. [Google Scholar] [CrossRef] [Green Version]
- Daniels, A.; van Bilsen, M.; Goldschmeding, R.; van der Vusse, G.J.; van Nieuwenhoven, F.A. Connective tissue growth factor and cardiac fibrosis. Acta Physiol (Oxf) 2009, 195, 321–338. [Google Scholar] [CrossRef]
- Powell, D.W.; Mifflin, R.C.; Valentich, J.D.; Crowe, S.E.; Saada, J.I.; West, A.B. Myofibroblasts. I. Paracrine cells important in health and disease. Am. J. Physiol 1999, 277, C1–C9. [Google Scholar] [CrossRef] [PubMed]
- van den Borne, S.W.; Diez, J.; Blankesteijn, W.M.; Verjans, J.; Hofstra, L.; Narula, J. Myocardial remodeling after infarction: The role of myofibroblasts. Nat. Rev. Cardiol 2010, 7, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Tarbit, E.; Singh, I.; Peart, J.N.; Rose’Meyer, R.B. Biomarkers for the identification of cardiac fibroblast and myofibroblast cells. Heart Fail. Rev. 2019, 24, 1–15. [Google Scholar] [CrossRef]
- Swaney, J.S.; Roth, D.M.; Olson, E.R.; Naugle, J.E.; Meszaros, J.G.; Insel, P.A. Inhibition of cardiac myofibroblast formation and collagen synthesis by activation and overexpression of adenylyl cyclase. Proc. Natl Acad Sci U S A 2005, 102, 437–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, B. The myofibroblast: Paradigm for a mechanically active cell. J. Biomech 2010, 43, 146–155. [Google Scholar] [CrossRef]
- Yong, K.W.; Li, Y.; Huang, G.; Lu, T.J.; Safwani, W.K.; Pingguan-Murphy, B.; Xu, F. Mechanoregulation of cardiac myofibroblast differentiation: Implications for cardiac fibrosis and therapy. Am. J. Physiol Heart Circ. Physiol 2015, 309, H532–H542. [Google Scholar] [CrossRef] [Green Version]
- Stewart, L.; Turner, N.A. Channelling the Force to Reprogram the Matrix: Mechanosensitive Ion Channels in Cardiac Fibroblasts. Cells 2021, 10, 990. [Google Scholar] [CrossRef]
- Kinnunen, P.; Vuolteenaho, O.; Ruskoaho, H. Mechanisms of Atrial and Brain Natriuretic Peptide Release From Rat Ventricular Myocardium: Effect of Stretching. Endocrinology 1993, 132, 1961–1970. [Google Scholar] [CrossRef] [PubMed]
- Toth, M.; Vuorinen, K.H.; Vuolteenaho, O.; Hassinen, I.E.; Uusimaa, P.A.; Leppaluoto, J.; Ruskoaho, H. Hypoxia stimulates release of ANP and BNP from perfused rat ventricular myocardium. Am. J. Physiol 1994, 266, H1572–H1580. [Google Scholar] [CrossRef] [PubMed]
- Zois, N.E.; Bartels, E.D.; Hunter, I.; Kousholt, B.S.; Olsen, L.H.; Goetze, J.P. Natriuretic peptides in cardiometabolic regulation and disease. Nat. Rev. Cardiol 2014, 11, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Ping, P.; Wang, F.; Luo, L. Synthesis, secretion, function, metabolism and application of natriuretic peptides in heart failure. J. Biol Eng 2018, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Jarai, R.; Kaun, C.; Weiss, T.W.; Speidl, W.S.; Rychli, K.; Maurer, G.; Huber, K.; Wojta, J. Human cardiac fibroblasts express B-type natriuretic peptide: Fluvastatin ameliorates its up-regulation by interleukin-1alpha, tumour necrosis factor-alpha and transforming growth factor-beta. J. Cell Mol. Med. 2009, 13, 4415–4421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Ry, S.; Cabiati, M.; Lionetti, V.; Emdin, M.; Recchia, F.A.; Giannessi, D. Expression of C-type natriuretic peptide and of its receptor NPR-B in normal and failing heart. Peptides 2008, 29, 2208–2215. [Google Scholar] [CrossRef] [PubMed]
- Maisel, A.S.; Duran, J.M.; Wettersten, N. Natriuretic Peptides in Heart Failure: Atrial and B-type Natriuretic Peptides. Heart Fail. Clin. 2018, 14, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Forte, M.; Madonna, M.; Schiavon, S.; Valenti, V.; Versaci, F.; Zoccai, G.B.; Frati, G.; Sciarretta, S. Cardiovascular Pleiotropic Effects of Natriuretic Peptides. Int J. Mol. Sci 2019, 20, 3874. [Google Scholar] [CrossRef] [Green Version]
- He, X.L.; Dukkipati, A.; Garcia, K.C. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J. Mol. Biol 2006, 361, 698–714. [Google Scholar] [CrossRef]
- Matsuo, A.; Nagai-Okatani, C.; Nishigori, M.; Kangawa, K.; Minamino, N. Natriuretic peptides in human heart: Novel insight into their molecular forms, functions, and diagnostic use. Peptides 2019, 111, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Potter, L.R.; Yoder, A.R.; Flora, D.R.; Antos, L.K.; Dickey, D.M. Natriuretic peptides: Their structures, receptors, physiologic functions and therapeutic applications. Handb Exp. Pharm. 2009, 341–366. [Google Scholar] [CrossRef] [Green Version]
- Moyes, A.J.; Chu, S.M.; Aubdool, A.A.; Dukinfield, M.S.; Margulies, K.B.; Bedi, K.C.; Hodivala-Dilke, K.; Baliga, R.S.; Hobbs, A.J. C-type natriuretic peptide co-ordinates cardiac structure and function. Eur Heart J. 2020, 41, 1006–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaauw, E.; van Nieuwenhoven, F.A.; Willemsen, P.; Delhaas, T.; Prinzen, F.W.; Snoeckx, L.H.; van Bilsen, M.; van der Vusse, G.J. Stretch-induced hypertrophy of isolated adult rabbit cardiomyocytes. Am. J. Physiol Heart Circ. Physiol 2010, 299, H780–H787. [Google Scholar] [CrossRef] [Green Version]
- Ovchinnikova, E.; Hoes, M.; Ustyantsev, K.; Bomer, N.; de Jong, T.V.; van der Mei, H.; Berezikov, E.; van der Meer, P. Modeling Human Cardiac Hypertrophy in Stem Cell-Derived Cardiomyocytes. Stem Cell Rep. 2018, 10, 794–807. [Google Scholar] [CrossRef] [Green Version]
- Kapoun, A.M.; Liang, F.; O’Young, G.; Damm, D.L.; Quon, D.; White, R.T.; Munson, K.; Lam, A.; Schreiner, G.F.; Protter, A.A. B-type natriuretic peptide exerts broad functional opposition to transforming growth factor-beta in primary human cardiac fibroblasts: Fibrosis, myofibroblast conversion, proliferation, and inflammation. Circ. Res. 2004, 94, 453–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, N.; Ogawa, Y.; Chusho, H.; Nakamura, K.; Nakao, K.; Suda, M.; Kasahara, M.; Hashimoto, R.; Katsuura, G.; Mukoyama, M.; et al. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc. Natl Acad Sci U S A 2000, 97, 4239–4244. [Google Scholar] [CrossRef] [Green Version]
- Ichiki, T.; Dzhoyashvili, N.; Burnett, J.C., Jr. Natriuretic peptide based therapeutics for heart failure: Cenderitide: A novel first-in-class designer natriuretic peptide. Int J. Cardiol 2019, 281, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Jhund, P.S.; McMurray, J.J. The neprilysin pathway in heart failure: A review and guide on the use of sacubitril/valsartan. Heart 2016, 102, 1342–1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, N.; Sugano, M.; Satoh, S.; Oyama, J.; Maeda, T. Peroxisome Proliferator-Activated Receptor-γ Ligands Attenuate Brain Natriuretic Peptide Production and Affect Remodeling in Cardiac Fibroblasts in Reoxygenation After Hypoxia. Cell Biochem. Biophys. 2006, 44, 065–072. [Google Scholar] [CrossRef]
- Morita, E.; Yasue, H.; Yoshimura, M.; Ogawa, H.; Jougasaki, M.; Matsumura, T.; Mukoyama, M.; Nakao, K. Increased plasma levels of brain natriuretic peptide in patients with acute myocardial infarction. Circulation 1993, 88, 82–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikimi, T.; Yoshihara, F.; Morimoto, A.; Ishikawa, K.; Ishimitsu, T.; Saito, Y.; Kangawa, K.; Matsuo, H.; Omae, T.; Matsuoka, H. Relationship Between Left Ventricular Geometry and Natriuretic Peptide Levels in Essential Hypertension. Hypertension 1996, 28, 22–30. [Google Scholar] [CrossRef]
- Koglin, J.; Pehlivanli, S.; Schwaiblmair, M.; Vogeser, M.; Cremer, P.; vonScheidt, W. Role of brain natriuretic peptide in risk stratification of patients with congestive heart failure. J. Am. Coll. Cardiol. 2001, 38, 1934–1941. [Google Scholar] [CrossRef] [Green Version]
- Nishikimi, T.; Maeda, N.; Matsuoka, H. The role of natriuretic peptides in cardioprotection. Cardiovasc Res. 2006, 69, 318–328. [Google Scholar] [CrossRef]
- Watson, C.J.; Phelan, D.; Xu, M.; Collier, P.; Neary, R.; Smolenski, A.; Ledwidge, M.; McDonald, K.; Baugh, J. Mechanical stretch up-regulates the B-type natriuretic peptide system in human cardiac fibroblasts: A possible defense against transforming growth factor-beta mediated fibrosis. Fibrogenesis Tissue Repair 2012, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blythe, N.M.; Muraki, K.; Ludlow, M.J.; Stylianidis, V.; Gilbert, H.T.J.; Evans, E.L.; Cuthbertson, K.; Foster, R.; Swift, J.; Li, J.; et al. Mechanically activated Piezo1 channels of cardiac fibroblasts stimulate p38 mitogen-activated protein kinase activity and interleukin-6 secretion. J. Biol Chem 2019, 294, 17395–17408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.A.; Porter, K.E.; Smith, W.H.T.; White, H.L.; Ball, S.G.; Balmforth, A.J. Chronic β2-adrenergic receptor stimulation increases proliferation of human cardiac fibroblasts via an autocrine mechanism. Cardiovasc. Res. 2003, 57, 784–792. [Google Scholar] [CrossRef] [Green Version]
- Van Nieuwenhoven, F.A.; Hemmings, K.E.; Porter, K.E.; Turner, N.A. Combined effects of interleukin-1α and transforming growth factor-β1 on modulation of human cardiac fibroblast function. Matrix Biol. 2013, 32, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Blaauw, E.; Lorenzen-Schmidt, I.; Babiker, F.A.; Munts, C.; Prinzen, F.W.; Snoeckx, L.H.; van Bilsen, M.; van der Vusse, G.J.; van Nieuwenhoven, F.A. Stretch-induced upregulation of connective tissue growth factor in rabbit cardiomyocytes. J. Cardiovasc Transl Res. 2013, 6, 861–869. [Google Scholar] [CrossRef]
- McMurray, J.J.; Packer, M.; Desai, A.S.; Gong, J.; Lefkowitz, M.P.; Rizkala, A.R.; Rouleau, J.; Shi, V.C.; Solomon, S.D.; Swedberg, K.; et al. Dual angiotensin receptor and neprilysin inhibition as an alternative to angiotensin-converting enzyme inhibition in patients with chronic systolic heart failure: Rationale for and design of the Prospective comparison of ARNI with ACEI to Determine Impact on Global Mortality and morbidity in Heart Failure trial (PARADIGM-HF). Eur J. Heart Fail. 2013, 15, 1062–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderone, A.; Bel-Hadj, S.; Drapeau, J.; El-Helou, V.; Gosselin, H.; Clement, R.; Villeneuve, L. Scar myofibroblasts of the infarcted rat heart express natriuretic peptides. J. Cell Physiol 2006, 207, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Volker, K.; Schwarz, K.; Carbajo-Lozoya, J.; Flogel, U.; Jacoby, C.; Stypmann, J.; van Eickels, M.; Gambaryan, S.; Hartmann, M.; et al. The natriuretic peptide/guanylyl cyclase--a system functions as a stress-responsive regulator of angiogenesis in mice. J. Clin. Invest. 2009, 119, 2019–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herum, K.M.; Lunde, I.G.; McCulloch, A.D.; Christensen, G. The Soft- and Hard-Heartedness of Cardiac Fibroblasts: Mechanotransduction Signaling Pathways in Fibrosis of the Heart. J. Clin. Med. 2017, 6, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.F.; Engler, A.J.; Woo, Y.J.; Pirolli, T.J.; Bish, L.T.; Jayasankar, V.; Morine, K.J.; Gardner, T.J.; Discher, D.E.; Sweeney, H.L. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol Heart Circ. Physiol 2006, 290, H2196–H2203. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Carag-Krieger, C.; Johnson, C.P.; Raab, M.; Tang, H.Y.; Speicher, D.W.; Sanger, J.W.; Sanger, J.M.; Discher, D.E. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: Scar-like rigidity inhibits beating. J. Cell Sci 2008, 121, 3794–3802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wanggou, S.; Bodalia, A.; Zhu, M.; Dong, W.; Fan, J.J.; Yin, W.C.; Min, H.K.; Hu, M.; Draghici, D.; et al. A Feedforward Mechanism Mediated by Mechanosensitive Ion Channel PIEZO1 and Tissue Mechanics Promotes Glioma Aggression. Neuron 2018, 100, 799–815 e797. [Google Scholar] [CrossRef] [Green Version]
- Emig, R.; Knodt, W.; Krussig, M.J.; Zgierski-Johnston, C.M.; Gorka, O.; Gross, O.; Kohl, P.; Ravens, U.; Peyronnet, R. Piezo1 Channels Contribute to the Regulation of Human Atrial Fibroblast Mechanical Properties and Matrix Stiffness Sensing. Cells 2021, 10, 663. [Google Scholar] [CrossRef] [PubMed]
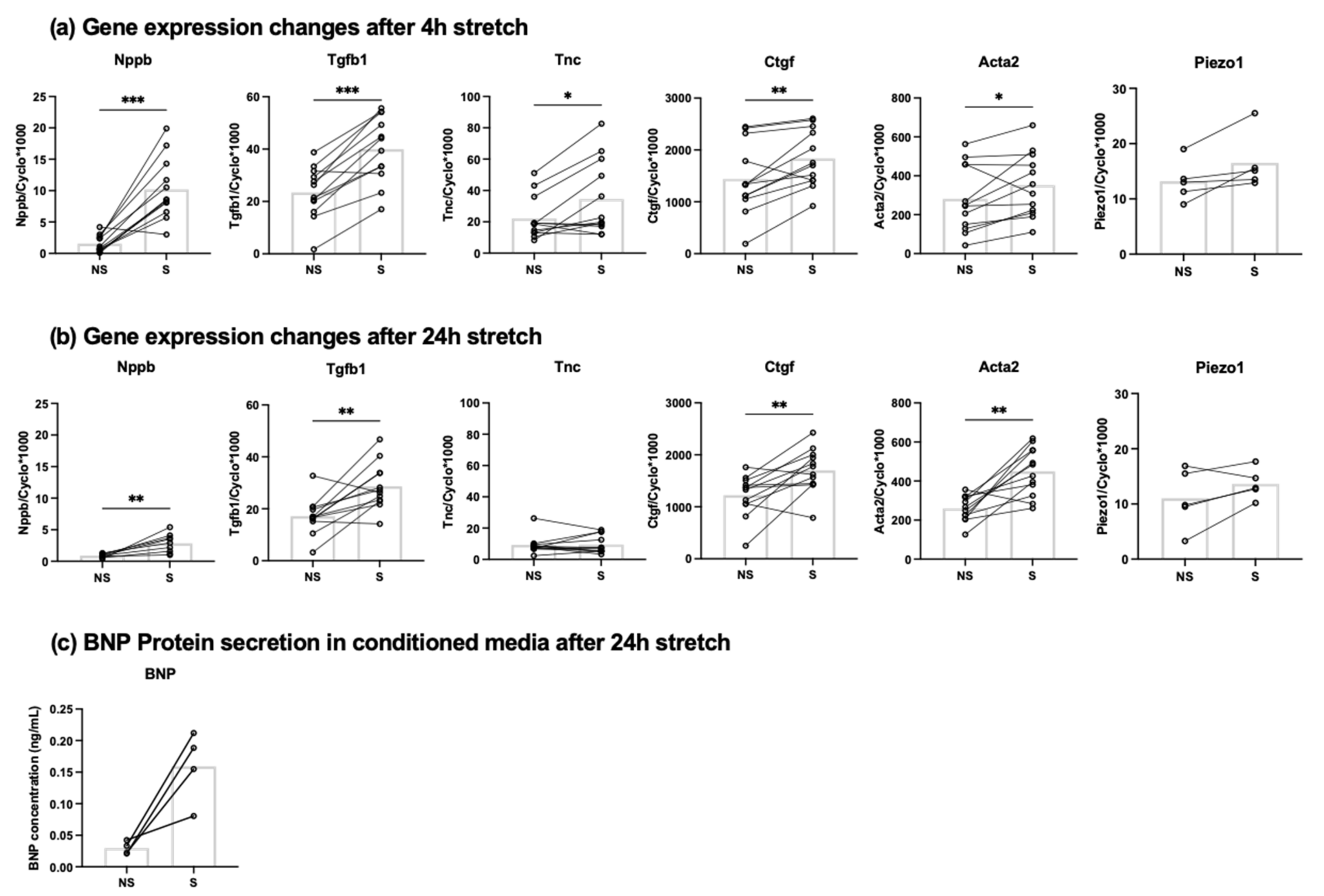
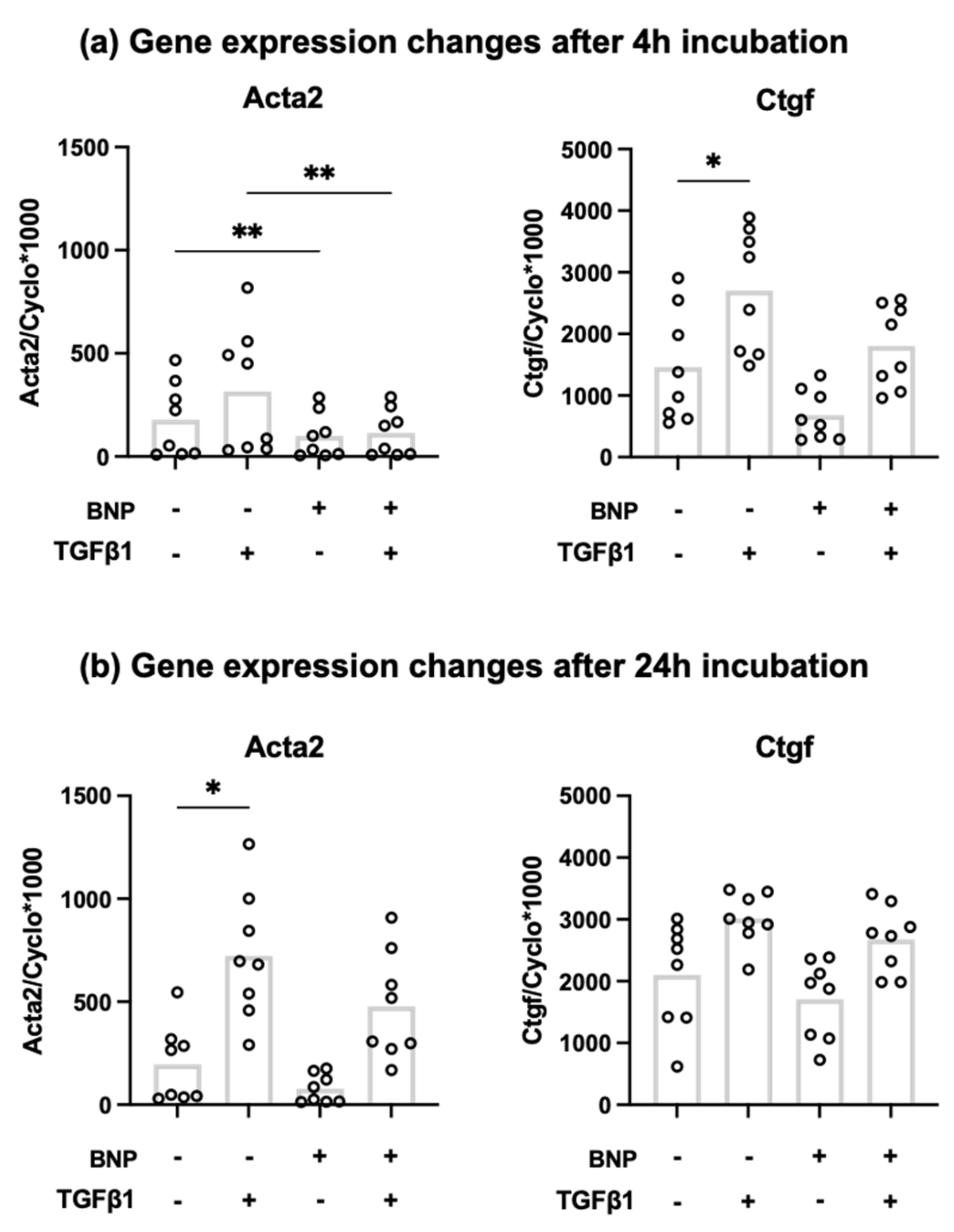
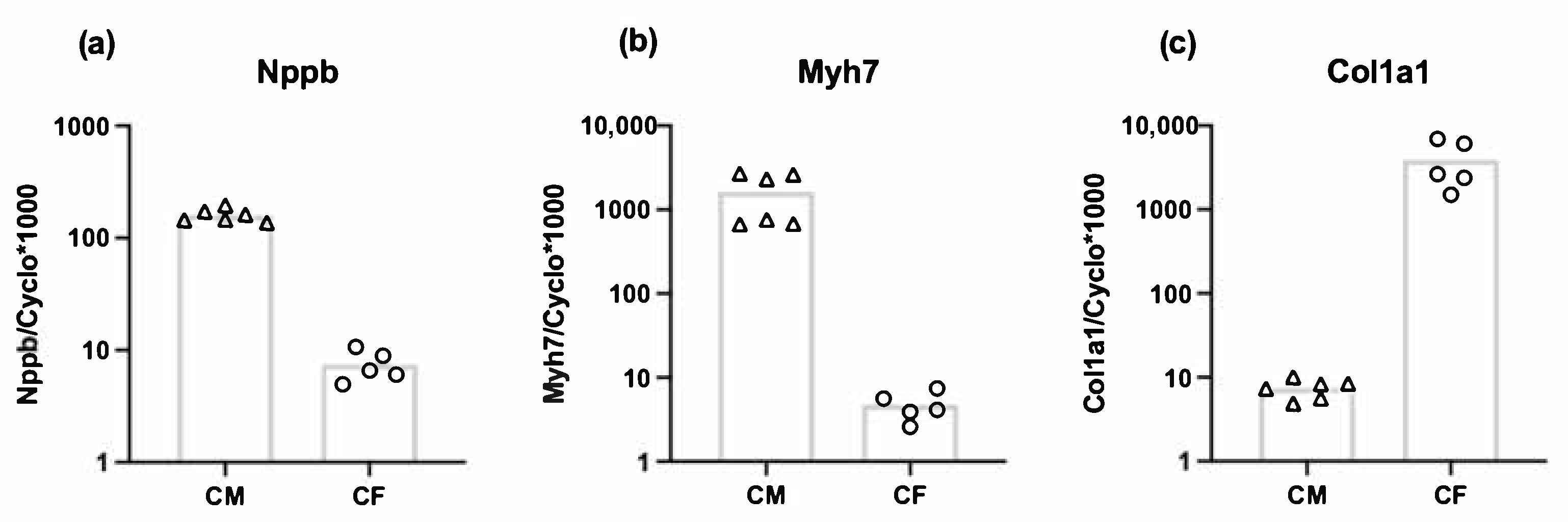

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Alpha-smooth muscle actin (Acta2) | AAGGCCAACCGGGAGAAAAT | AGTCCAGCACAATACCAGTTGT |
| Connective tissue growth factor (Ctgf) | CACAGAGTGGAGCGCCTGTTC | GATGCACTTTTTGCCCTTCTTAATG |
| Transforming growth factor, beta 1 (Tgfb1) | GCACCATCCATGACATGAAC | GCTGAAGCAGTAGTTGGTATC |
| Tenascin C (Tnc) | TCTGTCCTGGACTGCTGATG | TGGCCTCTCTGAGACCTGTT |
| Piezo1 | TTGCGTACGTTCACGAAGGA | TTCGCTCACGTAAAGCTGGT |
| Atrial Natriuretic peptide (Nppa) | ATCACCAAGGGCTTCTTCCT | TGTTGGACACCGCACTGTAT |
| Brain Natriuretic Peptide (Nppb) | AGACAGCTCTCAAAGGACCA | CTATCTTCTGCCCAAAGCAG |
| C-Type natriuretic peptide (Nppc) | ACAAAGGCGGCAACAAGAAG | GCAGTTCCCAATCCGCCG |
| Cyclophilin-A (Cyclo) | CAAATGCTGGACCAAACACAA | TTCACCTTCCCAAAGACCACAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ploeg, M.C.; Munts, C.; Prinzen, F.W.; Turner, N.A.; van Bilsen, M.; van Nieuwenhoven, F.A. Piezo1 Mechanosensitive Ion Channel Mediates Stretch-Induced Nppb Expression in Adult Rat Cardiac Fibroblasts. Cells 2021, 10, 1745. https://doi.org/10.3390/cells10071745
Ploeg MC, Munts C, Prinzen FW, Turner NA, van Bilsen M, van Nieuwenhoven FA. Piezo1 Mechanosensitive Ion Channel Mediates Stretch-Induced Nppb Expression in Adult Rat Cardiac Fibroblasts. Cells. 2021; 10(7):1745. https://doi.org/10.3390/cells10071745
Chicago/Turabian StylePloeg, Meike C., Chantal Munts, Frits W. Prinzen, Neil A. Turner, Marc van Bilsen, and Frans A. van Nieuwenhoven. 2021. "Piezo1 Mechanosensitive Ion Channel Mediates Stretch-Induced Nppb Expression in Adult Rat Cardiac Fibroblasts" Cells 10, no. 7: 1745. https://doi.org/10.3390/cells10071745
APA StylePloeg, M. C., Munts, C., Prinzen, F. W., Turner, N. A., van Bilsen, M., & van Nieuwenhoven, F. A. (2021). Piezo1 Mechanosensitive Ion Channel Mediates Stretch-Induced Nppb Expression in Adult Rat Cardiac Fibroblasts. Cells, 10(7), 1745. https://doi.org/10.3390/cells10071745







