Transcriptomic Hallmarks of Ischemia-Reperfusion Injury
Abstract
:1. Introduction
2. Methods
2.1. Systematic Review
2.2. Data Extraction and Handling
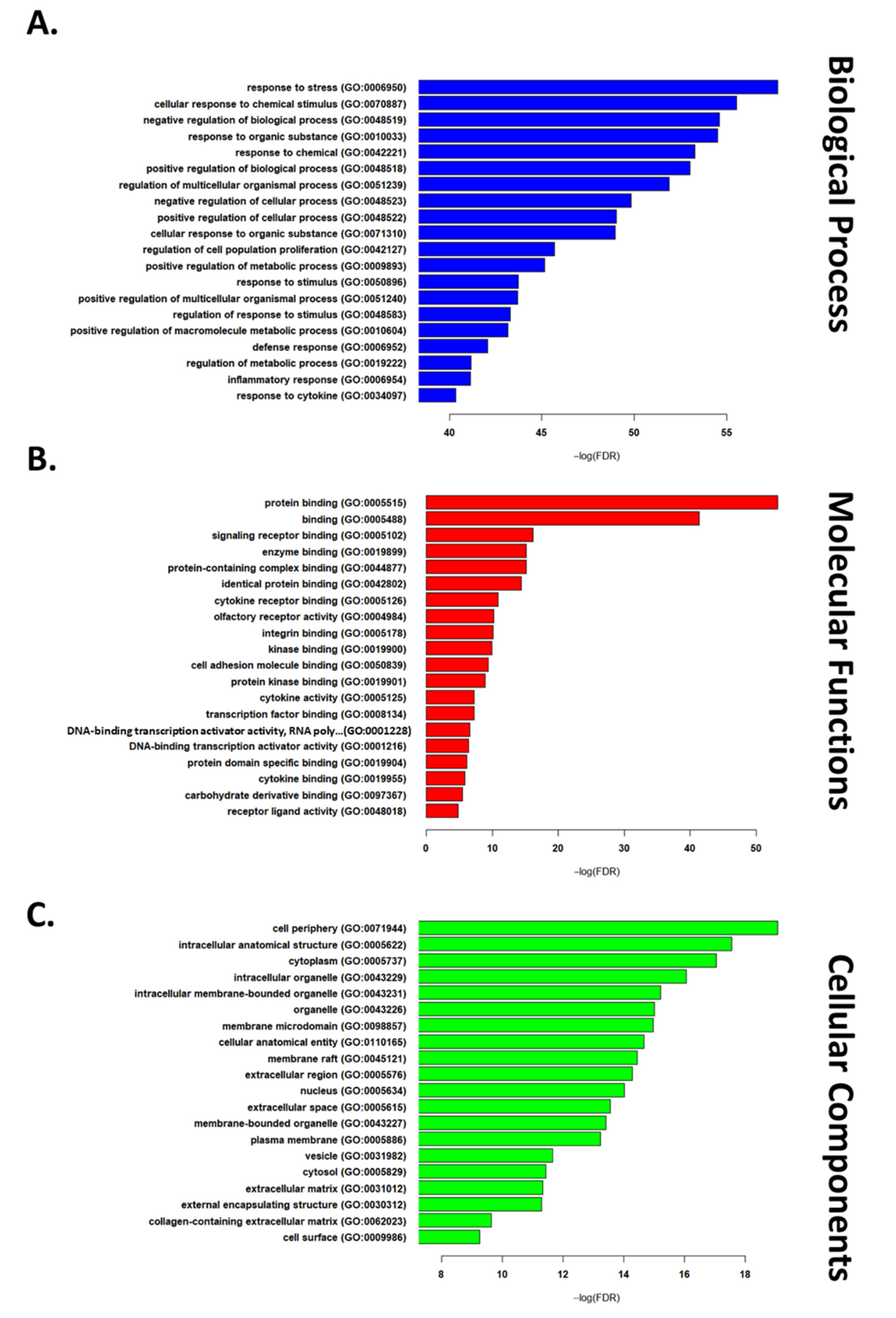
2.3. GGO Enrichment Analysis
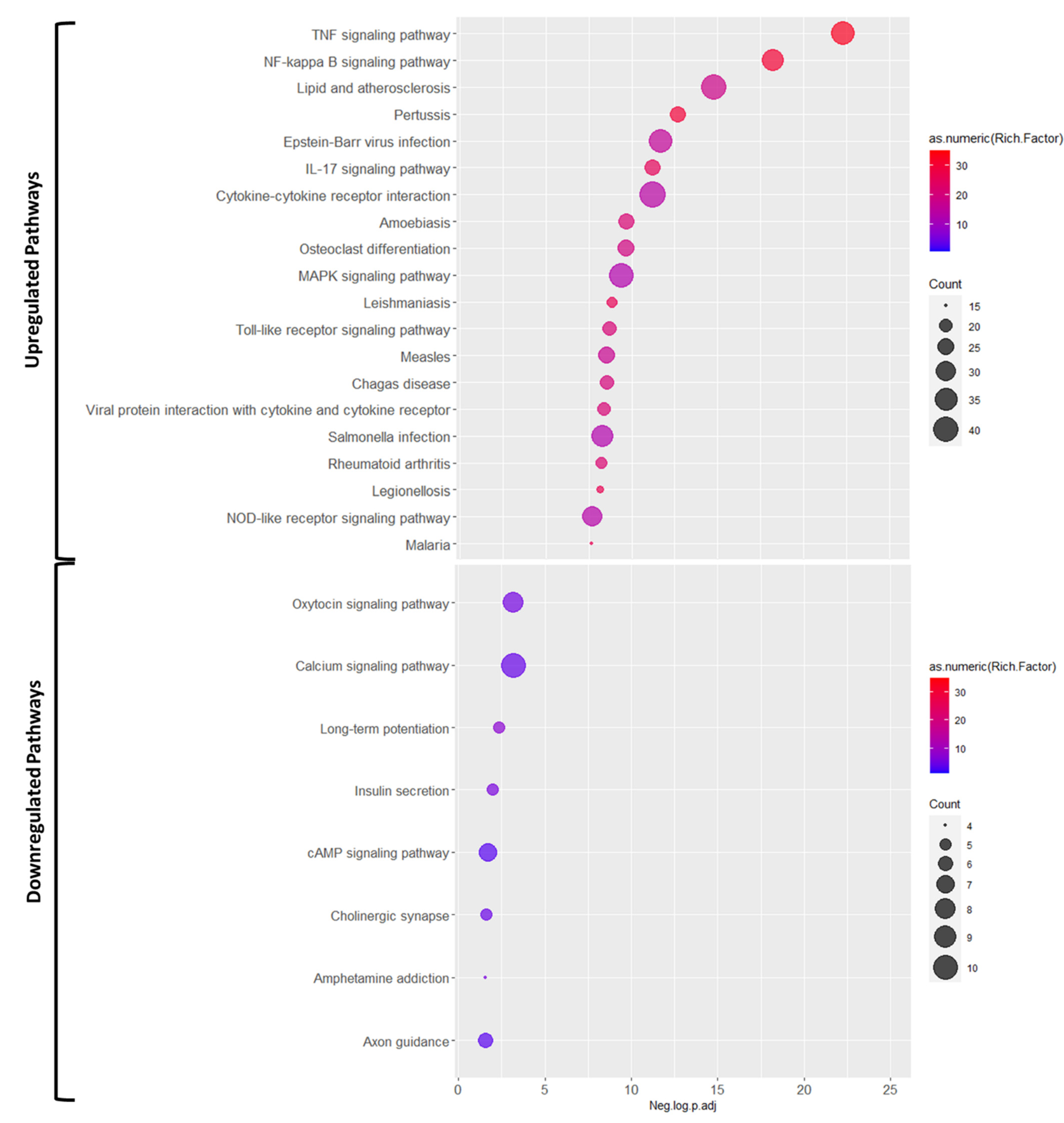
2.4. KEGG Analysis
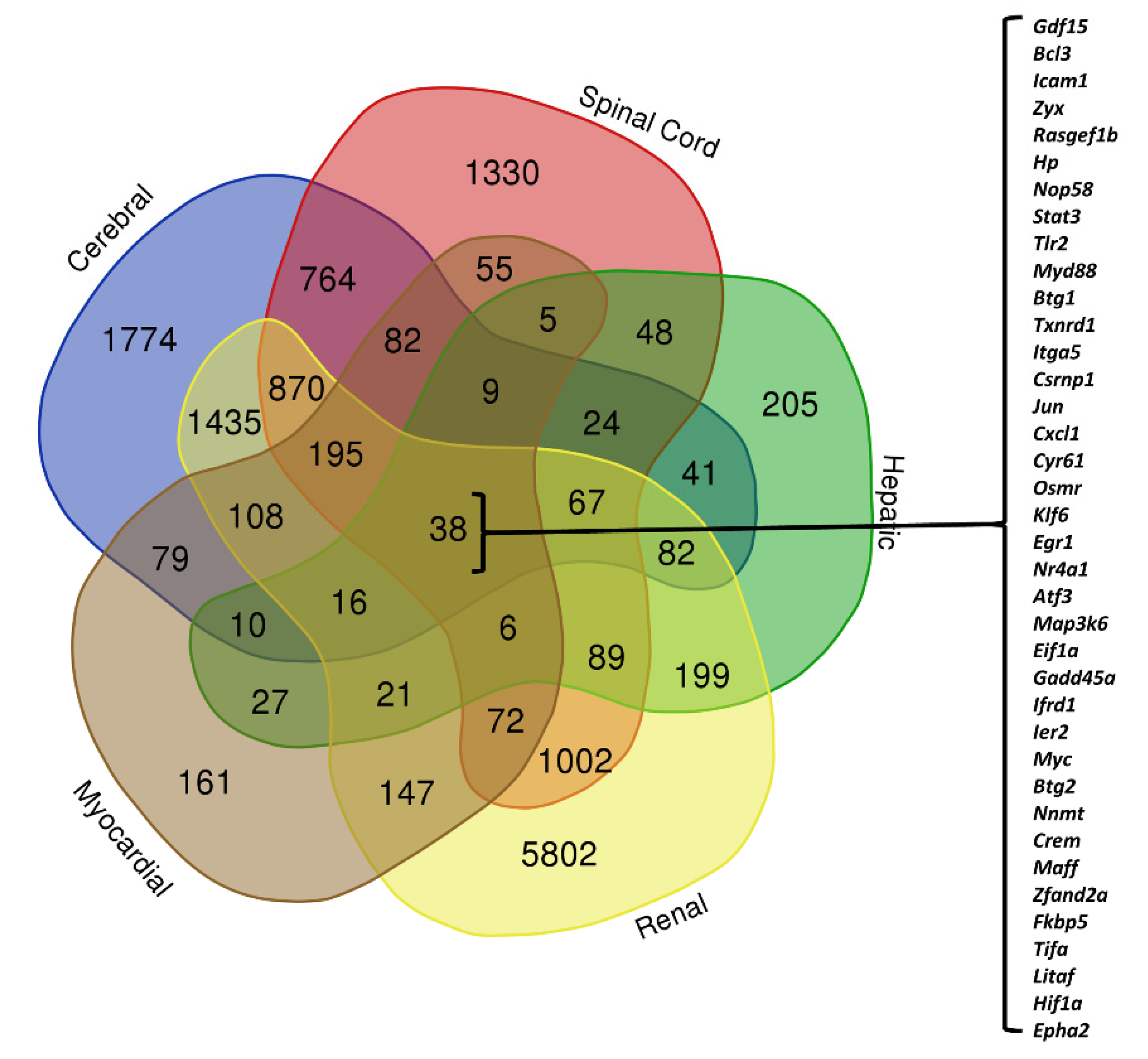
2.5. Venn Diagram
3. Results
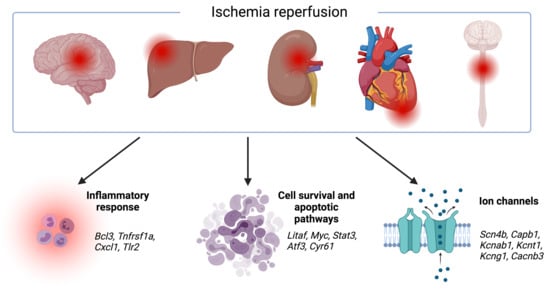
3.1. Transcriptomic Hallmark of Ischemic Reperfusion Injury
3.2. Pathways Involved in Ischemia Reperfusion Injury
3.3. Organ-Specific Differential Expression in Response to IRI
4. Discussion
4.1. Therapeutic Interventions Act on Genes and Pathways Involved in IRI
4.2. Ischemic Postconditioning
4.3. Caloric and Hypoxic Preconditioning
4.4. Stem Cell Therapy Approaches
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Explanation |
| ATF | activating transcription factors |
| BP | biological process |
| CaMK | calcium/calmodulin-dependent protein kinase |
| cAMP | cyclic adenosine monophosphate |
| CCN | CYR61 (cysteine-rich angiogenic protein 61), CTGF (connective tissue growth factor), NOV (nephroblastoma overexpressed) |
| CC | cellular components |
| CR | caloric restriction |
| DEGs | differentially expressed genes |
| DENN | differentially expressed in normal and neoplastic cells |
| ERK | extracellular signal-regulated kinases |
| FDR | false discovery rate |
| GO | Gene Ontology |
| HP | hypoxic preconditioning |
| HSPC | pluripotent hematopoietic stem/progenitor cells |
| IL-10 | Interleukin-10 |
| IL-12 | Interleukin-12 |
| IL-17 | Interleukin-17 |
| IL-6 | Interleukin-6 |
| IPO | ischemic postconditioning |
| IRI | ischemia reperfusion injury |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | mitogen-activated protein kinase |
| MCAO | middle cerebral artery occlusion |
| MF | molecular function |
| MSC | mesenchymal stem cells |
| MT-1 | metallothionein |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| PANTHER | protein analysis through evolutionary relationships |
| ROS | reactive oxidative species |
| TNF | tumor necrosis factor alpha |
| UC-MSC | umbilical cord-derived mesenchymal stem cells |
References
- Kalogeris, T.; Baines, C.; Krenz, M.; Korthuis, R.J. Cell Biology of Ischemia/Reperfusion Injury. Int. Rev. Cell Mol. Biol. 2012, 298, 229–317. [Google Scholar] [CrossRef] [Green Version]
- Hausenloy, D.J.; Yellon, D.M. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J. Clin. Investig. 2013, 123, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Nour, M.; Scalzo, F.; Liebeskind, D.S. Ischemia-Reperfusion Injury in Stroke. Interv. Neurol. 2012, 1, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Kosieradzki, M.; Rowiński, W. Ischemia/Reperfusion Injury in Kidney Transplantation: Mechanisms and Prevention. In Proceedings of the Transplantation Proceedings; Elsevier BV: Toronto, ON, Canada, 2008; Volume 40, pp. 3279–3288. [Google Scholar]
- Kupiec-Weglinski, J.; Busuttil, R. Ischemia and Reperfusion Injury in Liver Transplantation. In Proceedings of the Transplantation Proceedings; Elsevier BV: Toronto, ON, Canada, 2005; Volume 37, pp. 1653–1656. [Google Scholar]
- Honda, H.M.; Korge, P.; Weiss, J.N. Mitochondria and Ischemia/Reperfusion Injury. Ann. New York Acad. Sci. 2005, 1047, 248–258. [Google Scholar] [CrossRef]
- Minamino, T.; Komuro, I.; Kitakaze, M.; Minamino, T.; Komuro, I.; Kitakaze, M. Endoplasmic Reticulum Stress as a Therapeutic Target in Cardiovascular Disease. Circ. Res. 2010, 107, 1071–1082. [Google Scholar] [CrossRef]
- Lillig, C.H.; Berndt, C.; Holmgren, A. Glutaredoxin systems. Biochim. Biophys. Acta (BBA) Gen. Subj. 2008, 1780, 1304–1317. [Google Scholar] [CrossRef]
- Flück, M.; Carson, J.A.; Gordon, S.E.; Ziemiecki, A.; Booth, F.W. Focal adhesion proteins FAK and paxillin increase in hypertrophied skeletal muscle. Am. J. Physiol. Content 1999, 277, C152–C162. [Google Scholar] [CrossRef]
- Gundewar, S.; Calvert, J.; Elrod, J.; Lefer, D.J. Cytoprotective effects of N,N,N-trimethylsphingosine during ischemia- reperfusion injury are lost in the setting of obesity and diabetes. Am. J. Physiol. Circ. Physiol. 2007, 293, H2462–H2471. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Steenbergen, C. Ion Transport and Energetics During Cell Death and Protection. Physiology 2008, 23, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Satkunendrarajah, K.; Teng, Y.; Chow, D.S.-L.; Buttigieg, J.; Fehlings, M.G. Delayed Post-Injury Administration of Riluzole Is Neuroprotective in a Preclinical Rodent Model of Cervical Spinal Cord Injury. J. Neurotrauma 2013, 30, 441–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Zhuang, S. Histone acetylation and DNA methylation in ischemia/reperfusion injury. Clin. Sci. 2019, 133, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Han, B.; Jin, H.; Chen, A.; Zhu, L. Changes in long non-coding RNA transcriptomic profiles after ischemia-reperfusion injury in rat spinal cord. PeerJ 2020, 8, e8293. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Y.; Ke, X.; Guo, Y.; Yao, C.; Tang, N.; Pang, P.; Xie, G.; Fang, L.; Zhang, Z.; et al. Transcriptome Sequencing Unravels Potential Biomarkers at Different Stages of Cerebral Ischemic Stroke. Front. Genet. 2019, 10, 814. [Google Scholar] [CrossRef] [PubMed]
- Dergunova, L.V.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Mozerov, S.A.; Gubsky, L.V.; Limborska, S.A. Genome-wide transcriptome analysis using RNA-Seq reveals a large number of differentially expressed genes in a transient MCAO rat model. BMC Genom. 2018, 19, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Ming, Y.; Cheng, K.; Niu, Y.; Ye, Q. Gene Expression Profiling in Ischemic Postconditioning to Alleviate Mouse Liver Ischemia/Reperfusion Injury. Int. J. Med Sci. 2019, 16, 343–354. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Chen, X.; Li, H.; Wu, Y.; Wang, S.; Shi, W.; Chen, J.; Ni, Y. Neuron-autonomous transcriptome changes upon ischemia/reperfusion injury. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Johnsen, M.; Kubacki, T.; Yeroslaviz, A.; Späth, M.R.; Mörsdorf, J.; Göbel, H.; Bohl, K.; Ignarski, M.; Meharg, C.; Habermann, B.; et al. The Integrated RNA Landscape of Renal Preconditioning against Ischemia-Reperfusion Injury. J. Am. Soc. Nephrol. 2020, 31, 716–730. [Google Scholar] [CrossRef]
- Li, X.; Zheng, S.; Tan, W.; Chen, H.; Li, X.; Wu, J.; Luo, T.; Ren, X.; Pyle, G.; Wang, L.; et al. Slit2 Protects Hearts Against Ischemia-Reperfusion Injury by Inhibiting Inflammatory Responses and Maintaining Myofilament Contractile Properties. Front. Physiol. 2020, 11, 228. [Google Scholar] [CrossRef]
- Mo, D.; Tian, W.; Zhang, H.-N.; Feng, Y.-D.; Sun, Y.; Quan, W.; Hao, X.-W.; Wang, X.-Y.; Liu, X.-X.; Li, C.; et al. Cardioprotective effects of galectin-3 inhibition against ischemia/reperfusion injury. Eur. J. Pharmacol. 2019, 863, 172701. [Google Scholar] [CrossRef]
- Kestner, R.-I.; Mayser, F.; Vutukuri, R.; Hansen, L.; Günther, S.; Brunkhorst, R.; Devraj, K.; Pfeilschifter, W. Gene Expression Dynamics at the Neurovascular Unit During Early Regeneration After Cerebral Ischemia/Reperfusion Injury in Mice. Front. Neurosci. 2020, 14, 280. [Google Scholar] [CrossRef] [Green Version]
- Kölling, M.; Genschel, C.; Kaucsar, T.; Hübner, A.; Rong, S.; Schmitt, R.; Sörensen-Zender, I.; Haddad, G.; Kistler, A.; Seeger, H.; et al. Hypoxia-induced long non-coding RNA Malat1 is dispensable for renal ischemia/reperfusion-injury. Sci. Rep. 2018, 8, 3438. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-M.; Pan, Y.-Y.; Liu, M.-H.; Cheng, B.-H.; Bai, B.; Chen, J. RNA-seq expression profiling of rat MCAO model following reperfusion Orexin-A. Oncotarget 2017, 8, 113066–113081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.; Kwon, C.H.; Ha, H.K.; Han, M.; Song, S.H. RNA-Seq identifies condition-specific biological signatures of ischemia-reperfusion injury in the human kidney. BMC Nephrol. 2020, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Giraud, S.; Steichen, C.; Allain, G.; Couturier, P.; Labourdette, D.; Lamarre, S.; Ameteau, V.; Tillet, S.; Hannaert, P.; Thuillier, R.; et al. Dynamic transcriptomic analysis of Ischemic Injury in a Porcine Pre-Clinical Model mimicking Donors Deceased after Circulatory Death. Sci. Rep. 2018, 8, 5986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.; Essandoh, K.; Wang, X.; Li, Y.; Huang, W.; Chen, J.; Peng, J.; Jiang, D.-S.; Mu, X.; Wang, C.; et al. Tsg101 positively regulates P62-Keap1-Nrf2 pathway to protect hearts against oxidative damage. Redox Biol. 2020, 32, 101453. [Google Scholar] [CrossRef]
- Smith, H.K.; Omura, S.; Vital, S.A.; Becker, F.; Senchenkova, E.Y.; Kaur, G.; Tsunoda, I.; Peirce, S.M.; Gavins, F.N.E. Metallothionein I as a direct link between therapeutic hematopoietic stem/progenitor cells and cerebral protection in stroke. FASEB J. 2018, 32, 2381–2394. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wang, Y.; Zhou, L.; Zou, Y.; Huang, G.; Gao, G.; Ting, S.; Lei, X.; Ding, X. Spheroid-cultured human umbilical cord-derived mesenchymal stem cells attenuate hepatic ischemia-reperfusion injury in rats. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Liu, J.; Kumar, S.; Dolzhenko, E.; Alvarado, G.F.; Guo, J.; Lu, C.; Chen, Y.; Li, M.; Dessing, M.C.; Parvez, R.K.; et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2017, 2, e94716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 July 2021).
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic Analysis of Reactive Astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzeszkiewicz, T.M.; Lindner, V.; Chen, N.; Lam, S.C.-T.; Lau, L.F. The Angiogenic Factor Cysteine-Rich 61 (CYR61, CCN1) Supports Vascular Smooth Muscle Cell Adhesion and Stimulates Chemotaxis through Integrin α6β1 and Cell Surface Heparan Sulfate Proteoglycans. Endocrinology 2002, 143, 1441–1450. [Google Scholar] [CrossRef]
- Chen, Y.; Du, X. Functional properties and intracellular signaling of CCN1/Cyr61. J. Cell. Biochem. 2007, 100, 1337–1345. [Google Scholar] [CrossRef]
- De Nardo, D.; Labzin, L.; Kono, H.; Seki, R.; Schmidt, S.V.; Beyer, M.; Xu, D.; Zimmer, S.; Lahrmann, C.; Schildberg, F.A.; et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 2014, 15, 152–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-F.; Lin, H. Acute kidney injury and the potential for ATF3-regulated epigenetic therapy. Toxicol. Mech. Methods 2011, 21, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Ruttkay-Nedecky, B.; Nejdl, L.; Gumulec, J.; Zitka, O.; Masarik, M.; Eckschlager, T.; Stiborova, M.; Adam, V.; Kizek, R. The Role of Metallothionein in Oxidative Stress. Int. J. Mol. Sci. 2013, 14, 6044–6066. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Liu, R.; Luan, Y.-Y.; Yao, Y.-M. Tumor Necrosis Factor-α Induced Protein 8: Pathophysiology, Clinical Significance, and Regulatory Mechanism. Int. J. Biol. Sci. 2018, 14, 398–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, M.L.; De La Vega, A.; Mills, C.; Andrews-Hanna, J.; Spreng, R.N.; Cole, M.W.; Christoff, K. Heterogeneity within the frontoparietal control network and its relationship to the default and dorsal attention networks. Proc. Natl. Acad. Sci. USA 2018, 115, E1598–E1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| References | Reperfusion Model | Species | Reperfusion Duration |
|---|---|---|---|
| Zhou et al. 2020 [15] * | Spinal cord IRI | Sprague-Dawley rat | 48 h |
| Cai et al. 2019 [16] * | Cerebral IRI | C57BL/6J mice | 1, 3, 7, 14, 28 d |
| Dergunova et al. 2018 [17] * | Cerebral IRI | Wistar rats | 4.5, 24 h |
| Zhang et al. 2019 [18] * | Hepatic IRI | C57BL/6J mice | 4 h |
| Shi et al. 2017 [19] * | Cerebral IRI | Sprague-Dawley rat | 45 min, 6 h, 12 h and 18 h |
| Johnsen et al. 2020 [20] * | Renal IRI | C57BL/6J mice | 4, 24 h |
| Li et al. 2020 [21] * | Myocardial IRI | C57BL/6J mice | 40 min |
| Mo et al. 2019 [22] | Myocardial IRI | C57BL/6J mice | 1 h |
| Kestner et al. 2020 [23] | Cerebral IRI | C57BL/6J mice | 24 h |
| Kolling et al. 2018 [24] | Renal IRI | C57BL/6J mice | 24 h |
| Wang et al. 2017 [25] | Cerebral IRI | Wistar rats | 24 h |
| Park et al. 2020 [26] | Renal IRI | Human | 10 min |
| Giraud et al. 2018 [27] | Renal IRI | Porcine | 6, 24 h |
| Deng et al. 2020 [28] | Myocardial IRI | C57BL/6J mice | 24 h |
| Smith et al. 2018 [29] | Cerebral IRI | C57BL/6J mice | 24 h, 48 h, 1 w, 2 w |
| Sun et al. 2018 [30] | Hepatic IRI | Sprague-Dawley rats | 6, 24, 48 h |
| Liu et al. 2017 [31] | Renal IRI | C57BL/6J mice | 2 h, 4 h, 1 d, 2 d, 3 d, 1 w, 2 w, 4 w, 6 months, 12 months |
| Upregulated | Comments |
|---|---|
| Itga5 | Programmed cell death |
| Atf3 | Cell proliferation, programmed cell death |
| Cyr61 | Cell proliferation, programmed cell death |
| Litaf | Programmed cell death |
| Stat3 | Programmed cell death, inflammatory response |
| Icam1 | Inflammatory response, programmed cell death |
| Sik1 | Programmed cell death |
| Tlr2 | Inflammatory response |
| Zc3h12a | Inflammatory response |
| Epha2 | Inflammatory response |
| Maff | Response to stress |
| Nes | Cell proliferation, programmed cell death |
| Hmox1 | Response to stress, inflammatory response |
| Myc | Programmed cell death, response to stress |
| Cd44 | Inflammatory response |
| Hbegf | Cell proliferation |
| Ifrd1 | Developmental process |
| Pdpn | Cell proliferation, programmed cell death |
| Cd14 | Inflammatory response |
| Jun | Cell proliferation, programmed cell death |
| Csf1 | Inflammatory response |
| Gadd45a | Programmed cell death, response to stress |
| Ccl2 | Inflammatory response |
| Klf6 | Developmental process |
| Hspb1 | Response to stress, programmed cell death |
| Olr1 | Inflammatory response |
| Mafk | Response to stress |
| Zyx | Inflammatory response |
| Osmr | Inflammatory response |
| Tifa | Response to stress |
| Bcl3 | Inflammatory response, programmed cell death |
| Irf1 | Cell proliferation, programmed cell death |
| Tnfrsf1a | Inflammatory response |
| Cxcl1 | Inflammatory response |
| Txnrd1 | Cell proliferation, response to stress |
| Spon2 | Cell surface adhesion and signaling, response to stress |
| Myd88 | Inflammatory response |
| Bach1 | Response to stress |
| Downregulated | Comments |
|---|---|
| Cabp1 | Voltage-gated calcium ion channel regulator |
| Pclo | Presynaptic cytoskeletal matrix |
| Npy1r | Synaptic signaling |
| Tenm2 | Formation of growth cone in neural cells |
| Cbx7 | Cellular lifespan regulator |
| Camk4 | Transcriptional regulation |
| Kcng1 | Voltage-gated potassium (Kv) channels subunit |
| L1cam | Cell surface adhesion and signaling |
| Dlg2 | Response to oxidative stress and DNA damage |
| Scn4b | Sodium channel beta subunit |
| Kcnab1 | Cytoplasmic potassium channel subunit |
| Rasgrp1 | T cell/B cell regulator |
| Kcnt1 | Potassium-sodium activated channel subunit |
| Adora2a | Cell surface adhesion and signaling |
| Phactr1 | Endothelial cell survival |
| Lzts3 | Maturation of dendritic spines |
| Stard8 | Cell surface adhesion and signaling |
| Agrn | Cytoplasmic Ca ion regulator |
| Cacnb3 | Voltage-dependent calcium channel regulatory subunit |
| Cobl | Reorganization of the actin cytoskeleton |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Movahed, M.; Brockie, S.; Hong, J.; Fehlings, M.G. Transcriptomic Hallmarks of Ischemia-Reperfusion Injury. Cells 2021, 10, 1838. https://doi.org/10.3390/cells10071838
Movahed M, Brockie S, Hong J, Fehlings MG. Transcriptomic Hallmarks of Ischemia-Reperfusion Injury. Cells. 2021; 10(7):1838. https://doi.org/10.3390/cells10071838
Chicago/Turabian StyleMovahed, Mandana, Sydney Brockie, James Hong, and Michael G. Fehlings. 2021. "Transcriptomic Hallmarks of Ischemia-Reperfusion Injury" Cells 10, no. 7: 1838. https://doi.org/10.3390/cells10071838
APA StyleMovahed, M., Brockie, S., Hong, J., & Fehlings, M. G. (2021). Transcriptomic Hallmarks of Ischemia-Reperfusion Injury. Cells, 10(7), 1838. https://doi.org/10.3390/cells10071838