Deoxynivalenol and Zearalenone: Different Mycotoxins with Different Toxic Effects in the Sertoli Cells of Equus asinus
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals
2.3. Dose Selection
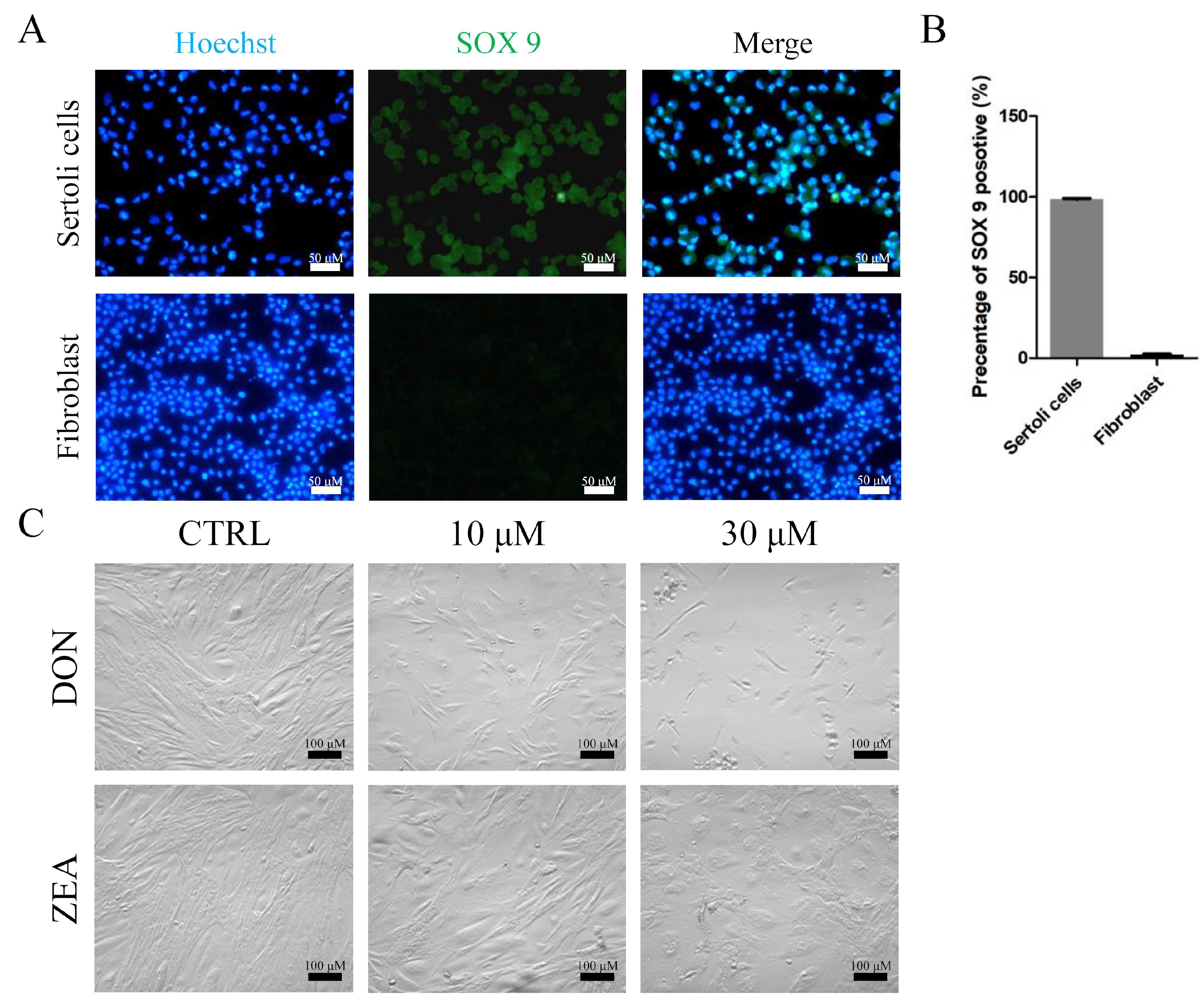
2.4. Isolation and Culture of Equus asinus SCs
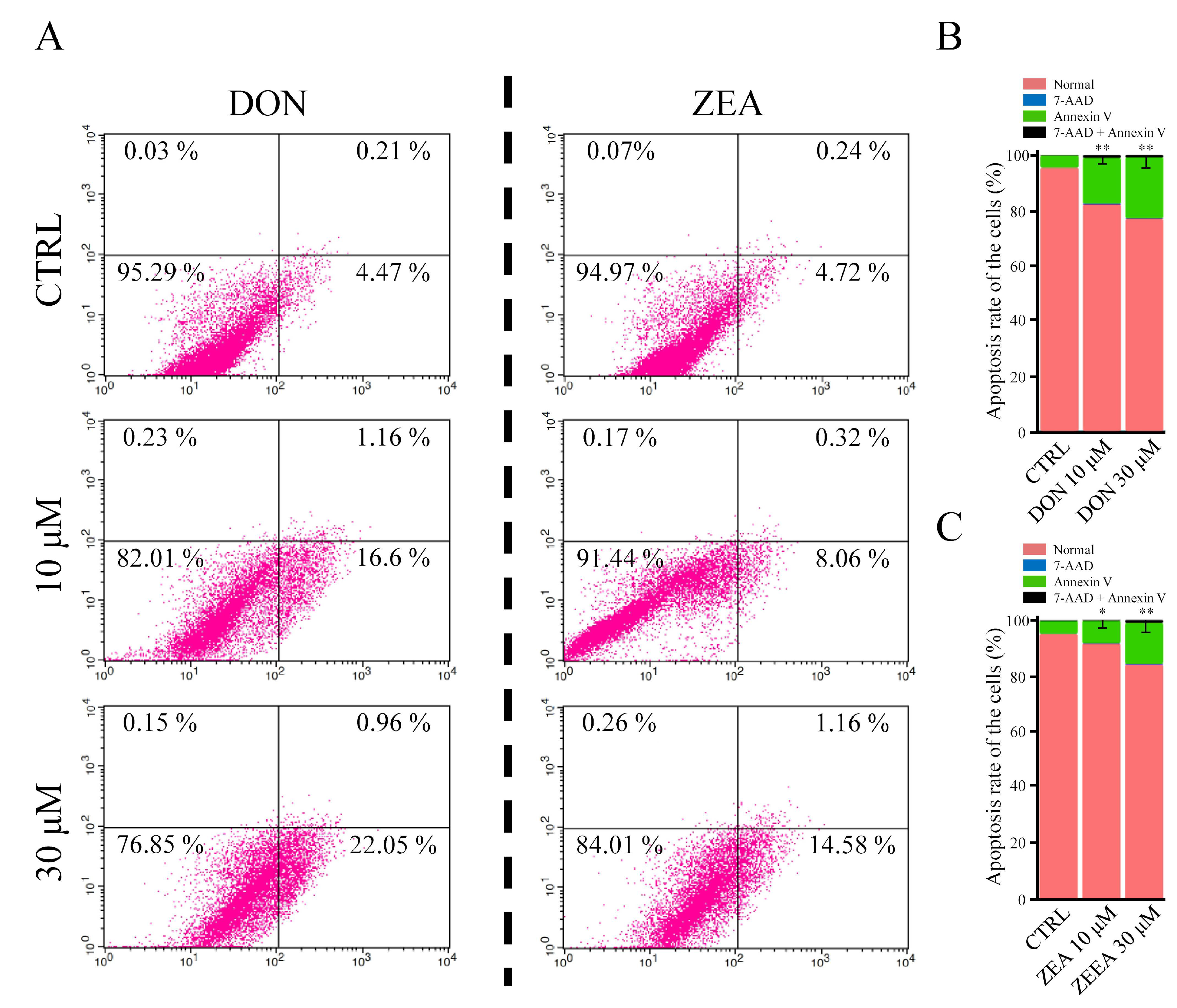
2.5. Flow Cytometry Analysis of Apoptosis
2.6. Immunofluorescence and Cell Counting
2.7. Western Blot Analysis
2.8. TUNEL Staining
2.9. RNA Extraction, Reverse Transcription, and RNA-Seq
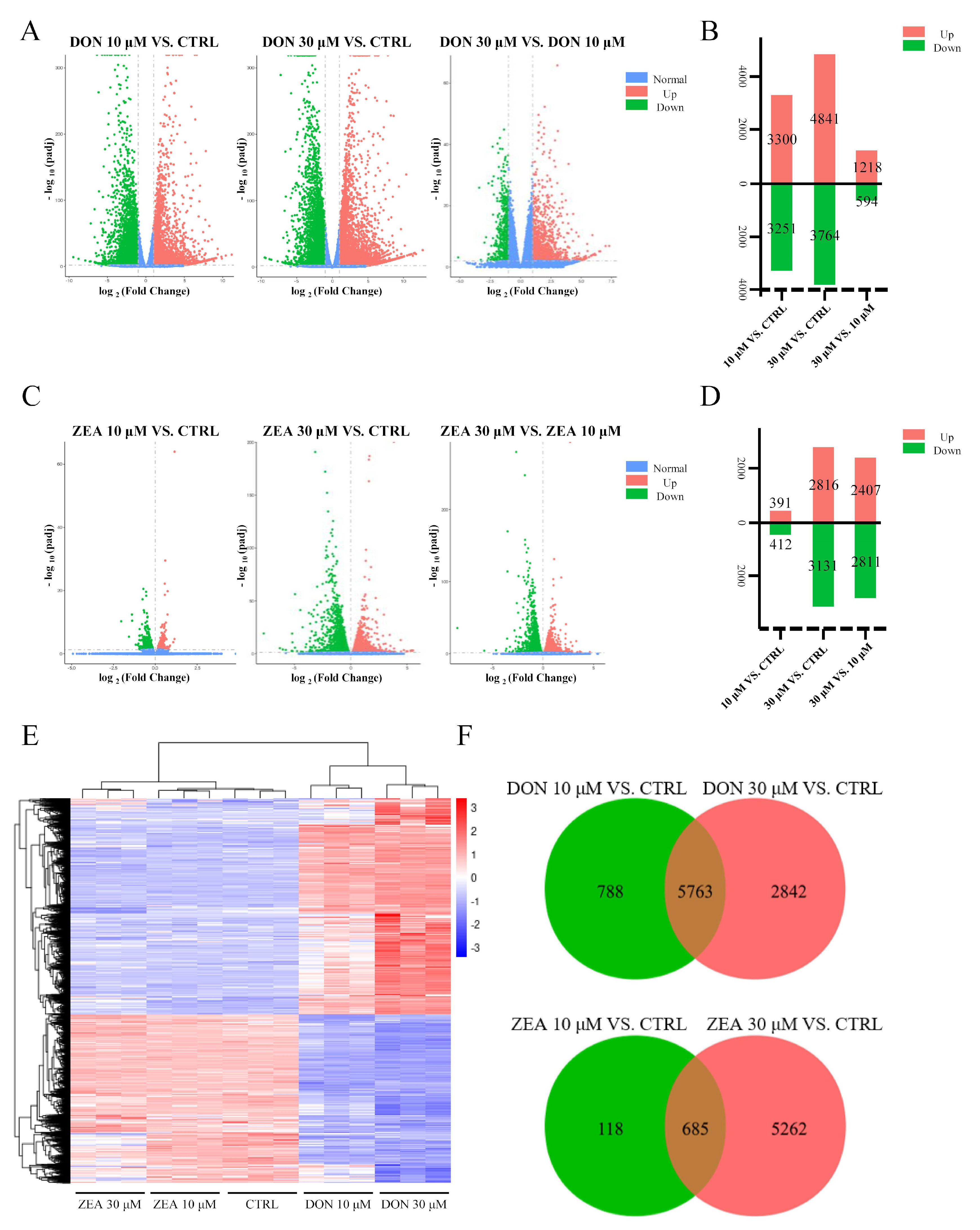
2.10. Identification of Differentially Expressed Genes
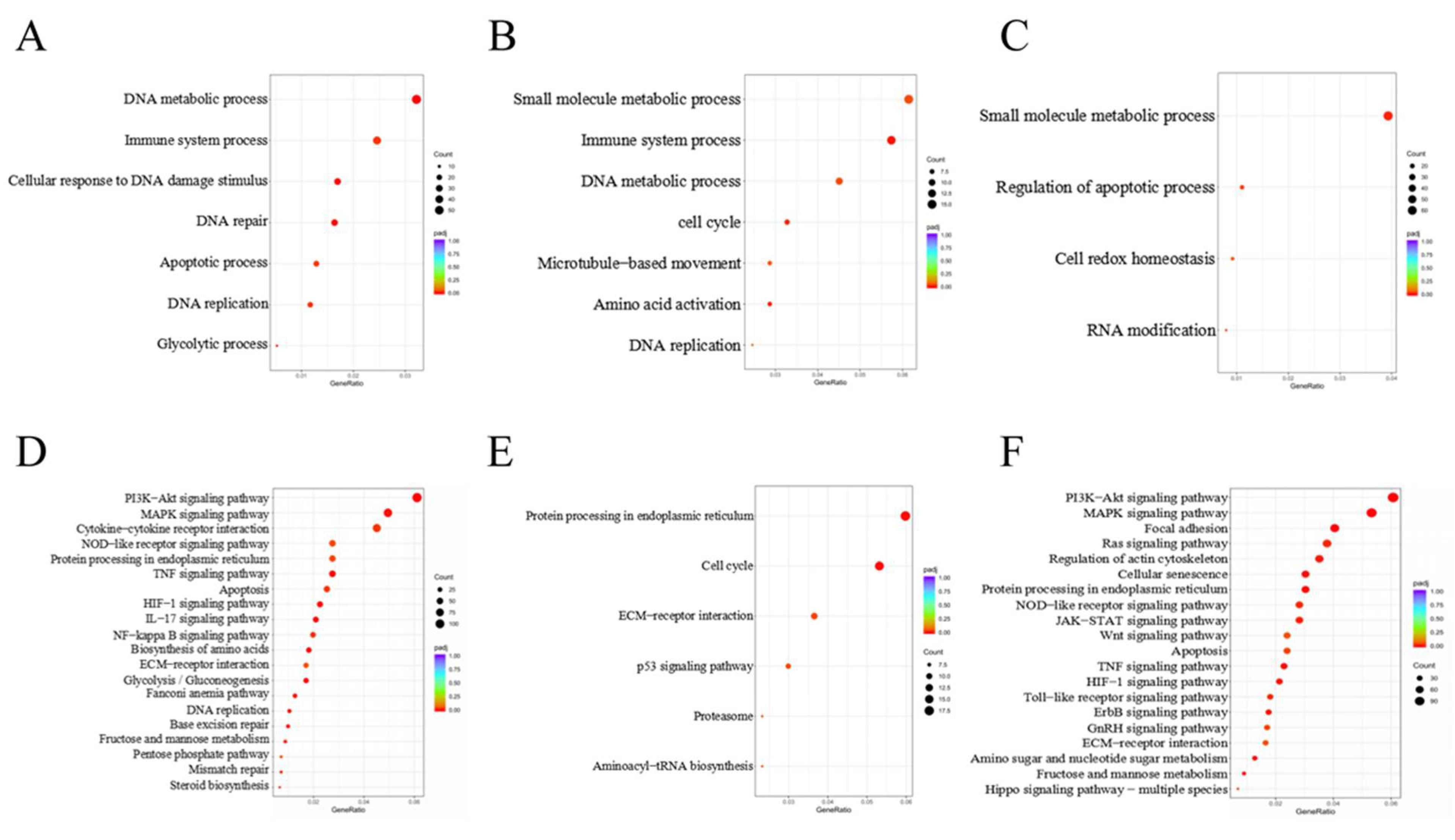
2.11. GO and KEGG Enrichment Analysis
2.12. Quantitative Real-Time PCR
2.13. Scanning Electron Microscopy (SEM)
2.14. Statistical Methods
3. Results
3.1. Apoptosis Rates and DEGs of Equus asinus SCs Exposed to DON and ZEA
3.2. DEGs Involved in GO Classification and KEGG Pathways
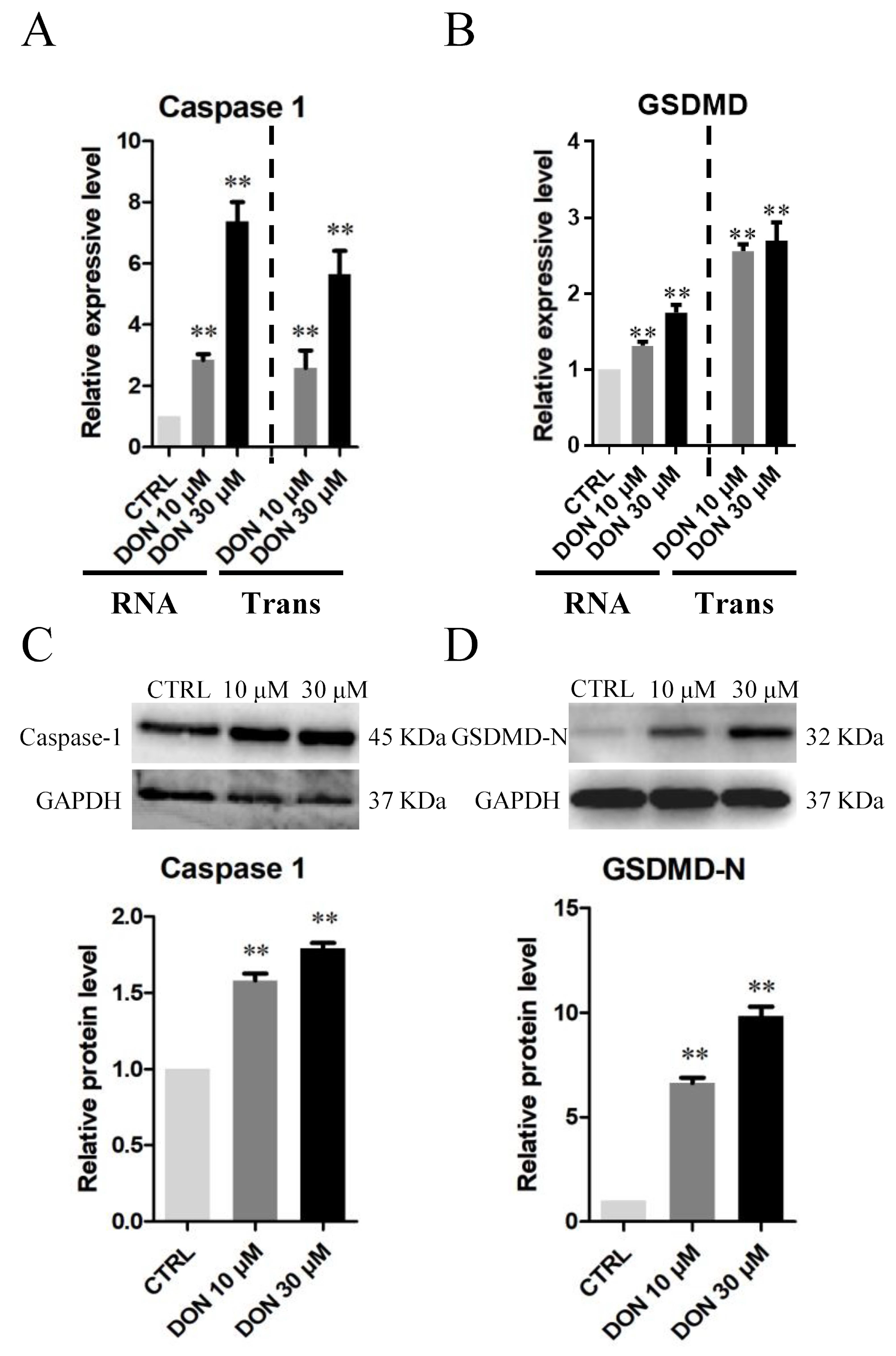
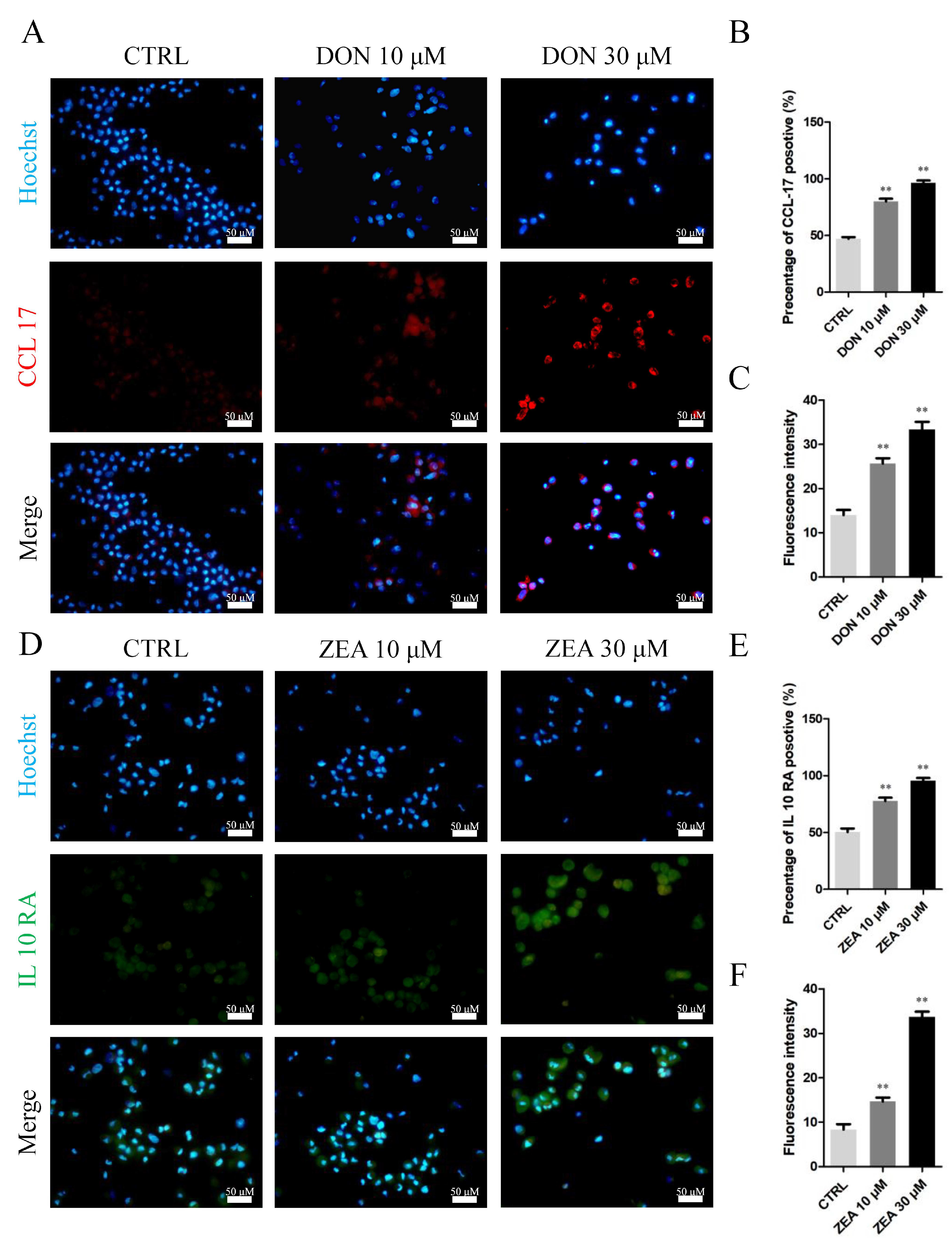
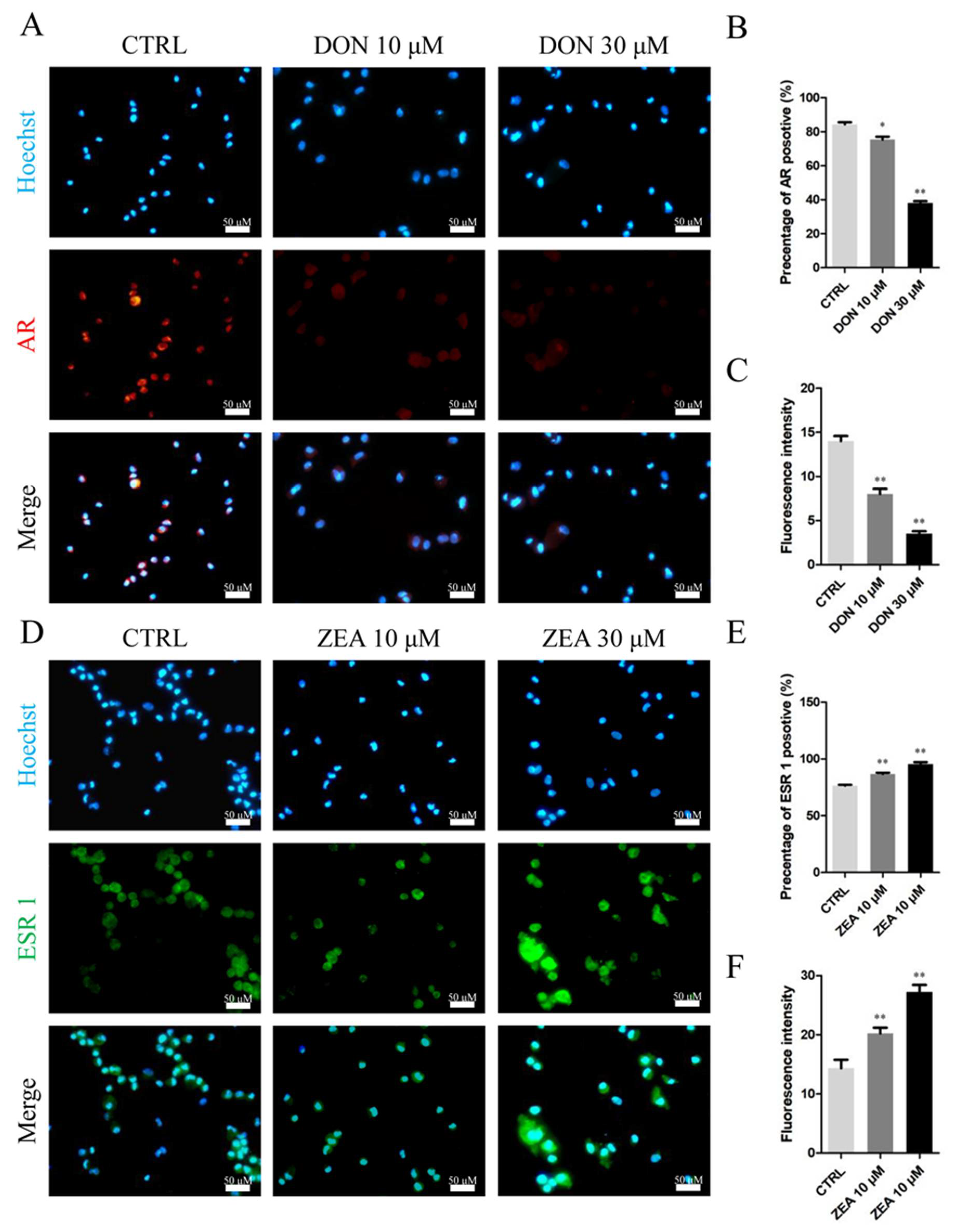
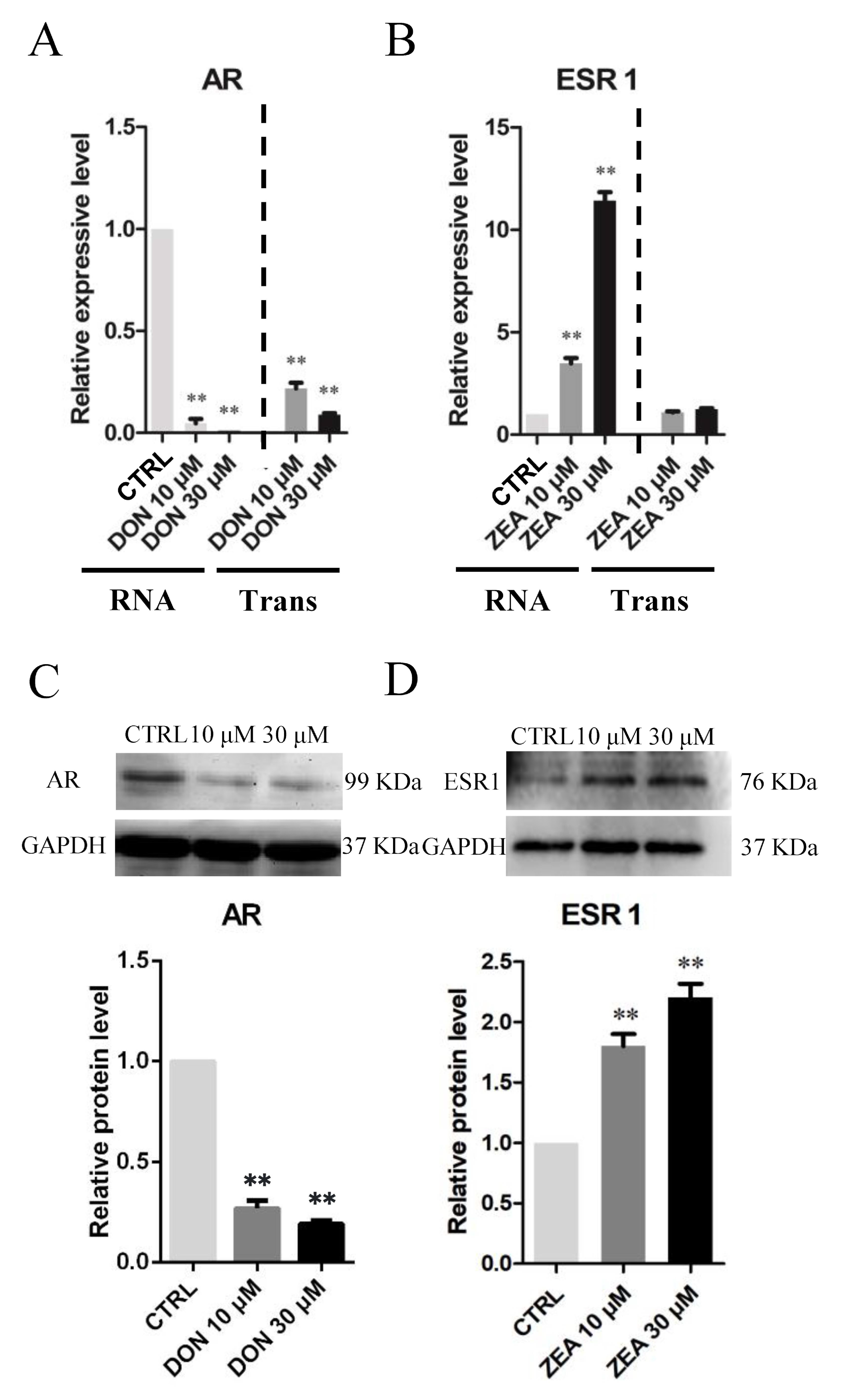
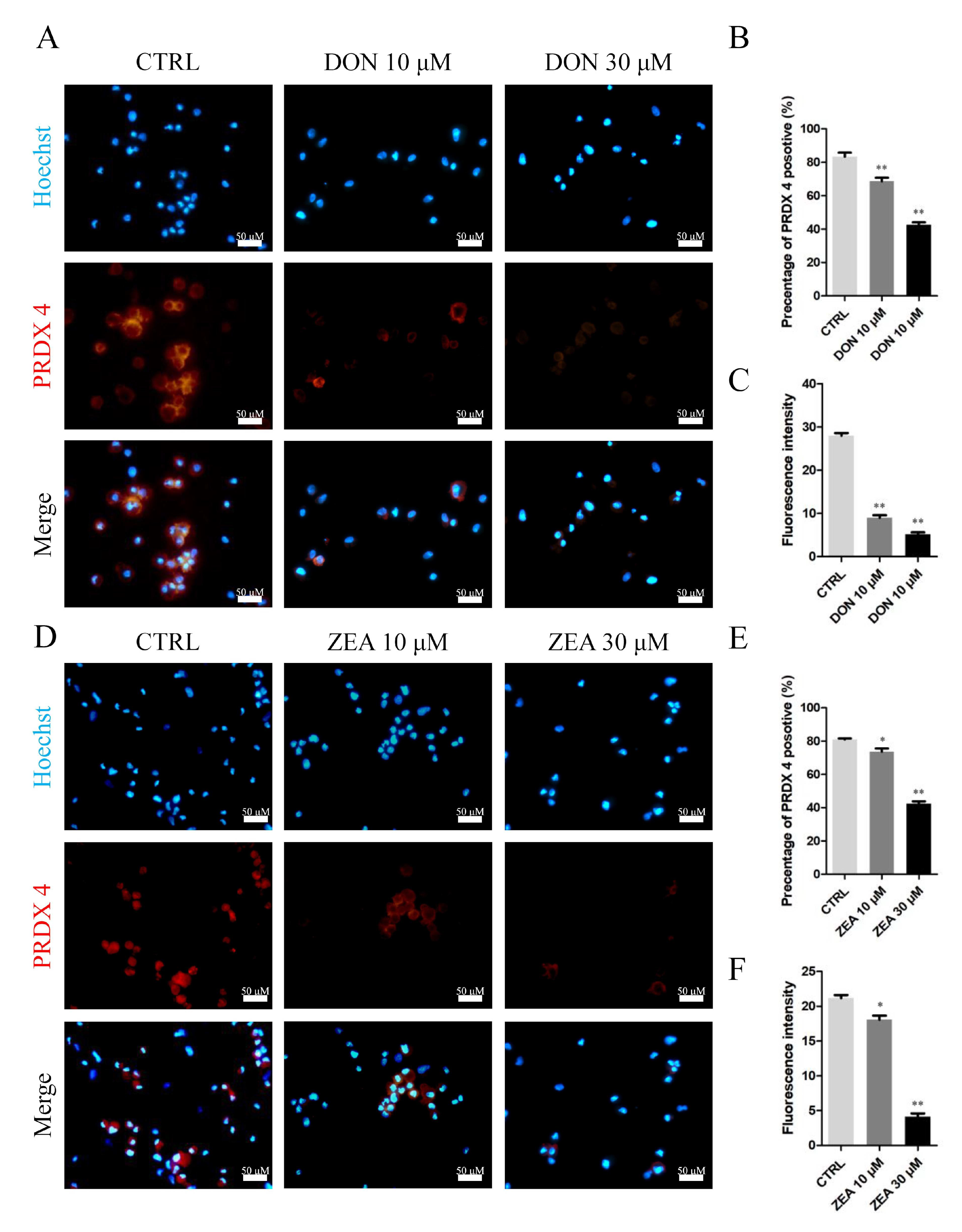
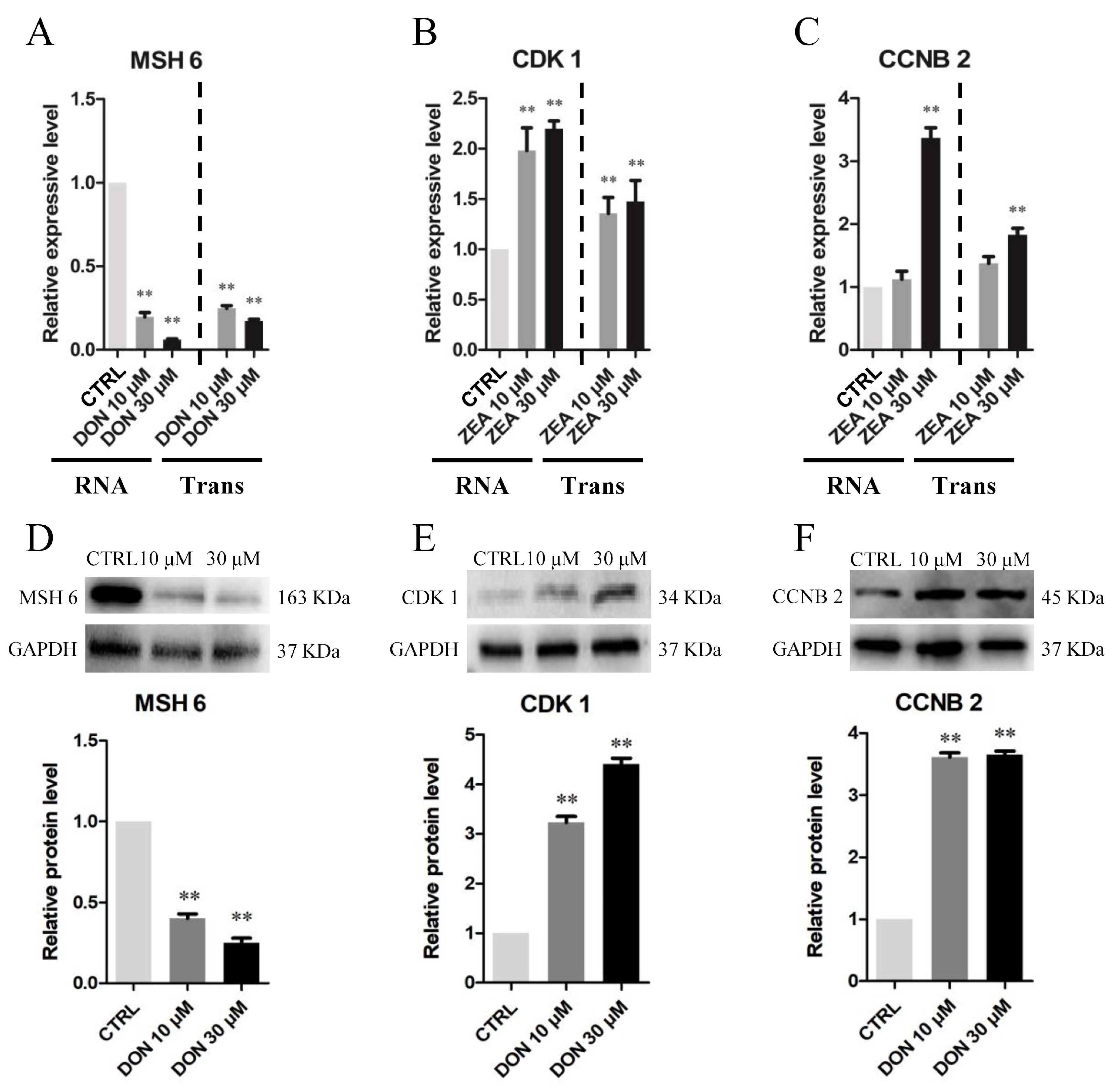
3.3. Cellular and Molecular Effects of DON and ZEA Exposure on SCs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, Q.; Lohrey, L.; Cramer, B.; Yuan, Z.; Humpf, H.U. Impact of physicochemical parameters on the decomposition of deoxynivalenol during extrusion cooking of wheat grits. J. Agric. Food Chem. 2011, 59, 12480–12485. [Google Scholar] [CrossRef]
- Pinton, P.; Oswald, I.P. Effect of deoxynivalenol and other Type B trichothecenes on the intestine: A review. Toxins 2014, 6, 1615–1643. [Google Scholar] [CrossRef]
- Doll, S.; Danicke, S. The Fusarium toxins deoxynivalenol (DON) and zearalenone (ZON) in animal feeding. Prev. Vet. Med. 2011, 102, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Mousavi Khaneghah, A.; Fakhri, Y.; Sant’Ana, A.S. Impact of unit operations during processing of cereal-based products on the levels of deoxynivalenol, total aflatoxin, ochratoxin A, and zearalenone: A systematic review and meta-analysis. Food Chem. 2018, 268, 611–624. [Google Scholar] [CrossRef]
- Cortinovis, C.; Pizzo, F.; Spicer, L.J.; Caloni, F. Fusarium mycotoxins: Effects on reproductive function in domestic animals--A review. Theriogenology 2013, 80, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Halenar, M.; Medvedova, M.; Maruniakova, N.; Kolesarova, A. Assessment of a potential preventive ability of amygdalin in mycotoxin-induced ovarian toxicity. J. Environ. Sci. Health B 2015, 50, 411–416. [Google Scholar] [CrossRef]
- Kolesarova, A.; Medvedova, M.; Halenar, M.; Sirotkin, A.V.; Bulla, J. The influence of deoxynivalenol and zearalenone on steroid hormone production by porcine ovarian granulosa cells in vitro. J. Environ. Sci. Health B 2017, 52, 823–832. [Google Scholar] [CrossRef]
- Zhang, G.L.; Feng, Y.L.; Song, J.L.; Zhou, X.S. Zearalenone: A Mycotoxin with Different Toxic Effect in Domestic and Laboratory Animals’ Granulosa Cells. Front. Genet. 2018, 9, 667. [Google Scholar] [CrossRef]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Poor, M.; Kunsagi-Mate, S.; Sali, N.; Koszegi, T.; Szente, L.; Peles-Lemli, B. Interactions of zearalenone with native and chemically modified cyclodextrins and their potential utilization. J. Photochem. Photobiol. B 2015, 151, 63–68. [Google Scholar] [CrossRef]
- Vanyi, A.; Bata, A.; Glavits, R.; Kovacs, F. Perinatal oestrogen syndrome in swine. Acta Vet. Hung. 1994, 42, 433–446. [Google Scholar] [PubMed]
- Dacasto, M.; Rolando, P.; Nachtmann, C.; Ceppa, L.; Nebbia, C. Zearalenone mycotoxicosis in piglets suckling sows fed contaminated grain. Vet. Hum. Toxicol. 1995, 37, 359–361. [Google Scholar]
- Osweiler, G.D.; Stahr, H.M.; Beran, G.W. Relationship of mycotoxins to swine reproductive failure. J. Vet. Diagn. Investig. 1990, 2, 73–75. [Google Scholar] [CrossRef]
- Zwierzchowski, W.; Przybylowicz, M.; Obremski, K.; Zielonka, L.; Skorska-Wyszynska, E.; Gajecka, M.; Polak, M.; Jakimiuk, E.; Jana, B.; Rybarczyk, L.; et al. Level of zearalenone in blood serum and lesions in ovarian follicles of sexually immature gilts in the course of zearalenone micotoxicosis. Pol. J. Vet. Sci. 2005, 8, 209–218. [Google Scholar]
- Lai, F.N.; Ma, J.Y.; Liu, J.C.; Wang, J.J.; Cheng, S.F.; Sun, X.F.; Li, L.; Li, B.; Nyachoti, C.M.; Shen, W. The influence of N-acetyl-l-cysteine on damage of porcine oocyte exposed to zearalenone in vitro. Toxicol. Appl. Pharmacol. 2015, 289, 341–348. [Google Scholar] [CrossRef]
- Canisso, I.F.; Carvalho, G.R.; Morel, M.C.; Guimaraes, J.D.; McDonnell, S.M. Sexual behavior and ejaculate characteristics in Pega donkeys (Equus asinus) mounting estrous horse mares (Equus caballus). Theriogenology 2010, 73, 56–63. [Google Scholar] [CrossRef]
- Canisso, I.F.; Carvalho, G.R.; Morel, M.D.; Ker, P.G.; Rodrigues, A.L.; Silva, E.C.; Coutinho Da Silva, M.A. Seminal parameters and field fertility of cryopreserved donkey jack semen after insemination of horse mares. Equine Vet. J. 2011, 43, 179–183. [Google Scholar] [CrossRef]
- Cong, W.; Chen, L.; Shan, X.F.; Qian, A.D.; Meng, Q.F. First genetic characterization of Toxoplasma gondii infection in donkey meat slaughtered for human consumption in Shandong province, eastern China. Infect. Genet. Evol. 2018, 61, 1–3. [Google Scholar] [CrossRef]
- Zhu, M.; Weng, Q. China: Will the donkey become the next pangolin? Equine Vet. J. 2018, 50, 276. [Google Scholar] [CrossRef]
- Zhang, G.L.; Zhang, R.Q.; Sun, X.F.; Cheng, S.F.; Wang, Y.F.; Ji, C.L.; Feng, Y.Z.; Yu, J.; Ge, W.; Zhao, Y.; et al. RNA-seq based gene expression analysis of ovarian granulosa cells exposed to zearalenone in vitro: Significance to steroidogenesis. Oncotarget 2017, 8, 64001–64014. [Google Scholar] [CrossRef]
- Minervini, F.; Giannoccaro, A.; Fornelli, F.; Dell’Aquila, M.E.; Minoia, P.; Visconti, A. Influence of mycotoxin zearalenone and its derivatives (alpha and beta zearalenol) on apoptosis and proliferation of cultured granulosa cells from equine ovaries. Reprod. Biol. Endocrinol. 2006, 4, 62. [Google Scholar] [CrossRef]
- Christine Arnesen, M.; Marie-Lisa Eich, M.; Maria, D.C.; Rodriguez Pena, M.; Jaclyn, R.; Cappel, M.; Lauren Schwartz, M.; Soroush Rais-Bahrami, M.; Sheila, F.; Faraj, M.; et al. NKX3.1 and prostein expression in testicular tissue and sex cord-stromal tumors. Am. J. Surg. Pathol. 2020, 44, 61–67. [Google Scholar] [CrossRef]
- Khaleel, I.; Al-Obaidy, M.; Muhammad, T.; Idrees, M.; David, J.; Grignon, M.; Thomas, M.; Ulbright, M. Adenocarcinoma of the rete testis clinicopathologic and immunohistochemical characterization of 6 cases and review of the literature. Am. J. Surg. Pathol. 2019, 43, 670–681. [Google Scholar]
- Gelmann, E.P.; Bowen, C.; Bubendorf, L. Expression of NKX3.1 in normal and malignant tissues. Prostate 2003, 55, 111–117. [Google Scholar] [CrossRef]
- Food and Drug Administration. Food and Drug Administration (FDA) Mycotoxin Regulatory Guidance; National Grain and Feed Association: Washington, DC, USA, 2011; pp. 1–9. [Google Scholar]
- EFSA Contam Panel. Scientific opinion on risk for animal health related to the presence of zearalenone and its modified forms in feed. EFSA J. 2017, 15, 4851. [Google Scholar]
- Chang, Y.F.; Lee-Chang, J.S.; Panneerdoss, S.; MacLean, J.A., 2nd; Rao, M.K. Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. Biotechniques 2011, 51, 341–342. [Google Scholar] [CrossRef]
- Dance, A.; Kastelic, J.; Thundathil, J. A combination of insulin-like growth factor I (IGF-I) and FSH promotes proliferation of prepubertal bovine Sertoli cells isolated and cultured in vitro. Reprod. Fertil. Dev. 2017, 29, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Jegou, B. The Sertoli cell in vivo and in vitro. Cell Biol. Toxicol. 1992, 8, 49–54. [Google Scholar] [CrossRef]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef]
- Tarulli, G.A.; Stanton, P.G.; Meachem, S.J. Is the Adult Sertoli Cell Terminally Differentiated? Biol. Reprod. 2012, 87, 1–11. [Google Scholar] [CrossRef]
- Orth, J.M.; Gunsalus, G.L.; Lamperti, A.A. Evidence from Sertoli cell-depleted rats indicates that spermatid number in adults depends on numbers of Sertoli cells produced during perinatal development. Endocrinology 1988, 122, 787–794. [Google Scholar] [CrossRef]
- Chang, L.; Wang, J.; She, R.; Ma, L.; Wu, Q. In vitro toxicity evaluation of melamine on mouse TM4 Sertoli cells. Environ. Toxicol. Pharmacol. 2017, 50, 111–118. [Google Scholar] [CrossRef]
- Mariana, R.; Marcelo, R.G.; Noel, G.M.; Herminia, P.E.; Beatriz, C.S.; Beatriz, M.S.; Fernanda, R.M. Germ cells regulate 3-hydroxybutyrate production in rat Sertoli cells. Gen. Comp. Endocrinol. 2017, 248, 5–15. [Google Scholar]
- Zhang, G.L.; Song, J.L.; Ji, C.L.; Feng, Y.L.; Yu, J.; Nyachoti, C.M.; Yang, G.S. Zearalenone Exposure Enhanced the Expression of Tumorigenesis Genes in Donkey Granulosa Cells via the PTEN/PI3K/AKT Signaling Pathway. Front. Genet. 2018, 9, 293. [Google Scholar] [CrossRef]
- Zhang, P.; Chao, H.; Sun, X.; Li, L.; Shi, Q.; Shen, W. Murine folliculogenesis in vitro is stage-specifically regulated by insulin via the Akt signaling pathway. Histochem. Cell Biol. 2010, 134, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.H.; Zhang, X.F.; Chen, B.; Pan, B.; Zhang, L.J.; Li, L.; Sun, X.F.; Shi, Q.H.; Shen, W. Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem. Cell Biol. 2012, 137, 249–259. [Google Scholar] [CrossRef]
- Pan, B.; Chao, H.; Chen, B.; Zhang, L.; Li, L.; Sun, X.; Shen, W. DNA methylation of germ-cell-specific basic helix-loop-helix (HLH) transcription factors, Sohlh2 and Figlalpha during gametogenesis. Mol. Hum. Reprod. 2011, 17, 550–561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, G.L.; Song, J.L.; Zhou, Y.; Zhang, R.Q.; Cheng, S.F.; Sun, X.F.; Qin, G.Q.; Shen, W.; Li, L. Differentiation of sow and mouse ovarian granulosa cells exposed to zearalenone in vitro using RNA-seq gene expression. Toxicol. Appl. Pharmacol. 2018, 350, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Hemendinger, R.A.; Gores, P.; Blacksten, L.; Harley, V.; Halberstadt, C. Identification of a specific Sertoli cell marker, Sox9, for use in transplantation. Cell Transplant. 2002, 11, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Mering, C.v.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; He, W.T.; Hu, L.C.; Li, J.X.; Fang, Y.; Wang, X.; Xu, X.Z.; Wang, Z.; Huang, K.; Han, J.H. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 2016, 26, 1007–1020. [Google Scholar] [CrossRef]
- Kouadio, J.H.; Mobio, T.A.; Baudrimont, I.; Moukha, S.; Dano, S.D.; Creppy, E.E. Comparative study of cytotoxicity and oxidative stress induced by deoxynivalenol, zearalenone or fumonisin B1 in human intestinal cell line Caco-2. Toxicology 2005, 213, 56–65. [Google Scholar] [CrossRef]
- Larsen, J.C.; Hunt, J.; Perrin, I.; Ruckenbauer, P. Workshop on trichothecenes with a focus on DON: Summary report. Toxicol. Lett. 2004, 153, 1–22. [Google Scholar] [CrossRef]
- Sprando, R.L.; Pestka, J.; Collins, T.F.; Rorie, J.; O’Donnell, M.; Hinton, D.; Chirtel, S. The effect of vomitoxin (Deoxnivalenol) on testicular morphology, testicular spermatid counts and epididymal sperm counts in IL-6KO [B6129-IL6 [TmlKopf] (IL-6 gene deficient)] and WT [B6129F2 (wild type to B6129-IL6 with an intact IL-6 gene)] mice. Food Chem. Toxicol. 1999, 37, 1073–1079. [Google Scholar] [CrossRef]
- Kiang, D.T.; Kennedy, B.J.; Pathre, S.V.; Mirocha, C.J. Binding characteristics of zearalenone analogs to estrogen receptors. Cancer Res. 1978, 38 Pt 1, 3611–3615. [Google Scholar]
- Nikov, G.N.; Hopkins, N.E.; Boue, S.; Alworth, W.L. Interactions of dietary estrogens with human estrogen receptors and the effect on estrogen receptor-estrogen response element complex formation. Environ. Health Perspect. 2000, 108, 867–872. [Google Scholar] [CrossRef]
- Mehmood, Z.; Smith, A.G.; Tucker, M.J.; Chuzel, F.; Carmichael, N.G. The development of methods for assessing the in vivo oestrogen-like effects of xenobiotics in CD-1 mice. Food Chem. Toxicol. 2000, 38, 493–501. [Google Scholar] [CrossRef]
- Mueller, S.O.; Simon, S.; Chae, K.; Metzler, M.; Korach, K.S. Phytoestrogens and their human metabolites show distinct agonistic and antagonistic properties on estrogen receptor alpha (ERalpha) and ERbeta in human cells. Toxicol. Sci. 2004, 80, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Albonico, M.; Schutz, L.F.; Caloni, F.; Cortinovis, C.; Spicer, L.J. Toxicological effects of fumonisin B1 alone and in combination with other fusariotoxins on bovine granulosa cells. Toxicon 2016, 118, 47–53. [Google Scholar] [CrossRef]
- Abbes, S.; Ouanes, Z.; ben Salah-Abbes, J.; Houas, Z.; Oueslati, R.; Bacha, H.; Othman, O. The protective effect of hydrated sodium calcium aluminosilicate against haematological, biochemical and pathological changes induced by Zearalenone in mice. Toxicon 2006, 47, 567–574. [Google Scholar] [CrossRef]
- Abbes, S.; Salah-Abbes, J.B.; Ouanes, Z.; Houas, Z.; Othman, O.; Bacha, H.; Abdel-Wahhab, M.A.; Oueslati, R. Preventive role of phyllosilicate clay on the Immunological and Biochemical toxicity of zearalenone in Balb/c mice. Int. Immunopharmacol. 2006, 6, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Luongo, D.; Severino, L.; Bergamo, P.; De Luna, R.; Lucisano, A.; Rossi, M. Interactive effects of fumonisin B1 and alpha-zearalenol on proliferation and cytokine expression in Jurkat T cells. Toxicol. In Vitro 2006, 20, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Ling, A.; Sun, L.; Guo, W.; Sun, S.; Yang, J.; Zhao, Z. Individual and combined cytotoxic effects of T-2 toxin and its four metabolites on porcine Leydig cells. Food Chem. Toxicol. 2020, 139, 111277. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jia, B.P.; Cao, L.; Xu, J.R.; Zhao, J.; Feng, S.B.; Li, Y.; Wu, J.J.; Wang, X.C. Effect of sertoli cells apoptosis in piglets induced by single or combined administration of zearalenone and deoxynivalenol. Acta Agric. Zhejiangensis 2020, 32, 1954–1962. (In Chinese) [Google Scholar]
- Cao, L.; Zhao, J.; Xu, J.; Zhu, L.; Rahman, S.U.; Feng, S.; Li, Y.; Wu, J.; Wang, X. N-acetylcysteine ameliorate cytotoxic injury in piglets sertoli cells induced by zearalenone and deoxynivalenol. Environ. Sci. Pollut. Res. Int. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Wan, L.Y.; Woo, C.S.; Turner, P.C.; Wan, J.M.; El-Nezami, H. Individual and combined effects of Fusarium toxins on the mRNA expression of pro-inflammatory cytokines in swine jejunal epithelial cells. Toxicol. Lett. 2013, 220, 238–246. [Google Scholar] [CrossRef]
- Wan, L.Y.; Turner, P.C.; El-Nezami, H. Individual and combined cytotoxic effects of Fusarium toxins (deoxynivalenol, nivalenol, zearalenone and fumonisins B1) on swine jejunal epithelial cells. Food Chem. Toxicol. 2013, 57, 276–283. [Google Scholar] [CrossRef]
- Kesavardhana, S.; Malireddi, R.K.S.; Kanneganti, T.D. Caspases in Cell Death, Inflammation, and Pyroptosis. Ann. Rev. Immunol. 2020, 38, 567–595. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Tian, S.; Pan, Y.; Li, W.; Wang, Q.; Tang, Y.; Yu, T.; Wu, X.; Shi, Y.; Ma, P.; et al. Pyroptosis: A new frontier in cancer. Biomed. Pharmacother. 2020, 121, 109595. [Google Scholar] [CrossRef] [PubMed]
- Trump, B.F.; Goldblatt, P.J.; Stowell, R.E. Studies of necrosis in vitro of mouse hepatic parenchymal cells. Ultrastructural alterations in endoplasmic reticulum, Golgi apparatus, plasma membrane, and lipid droplets. Lab. Investig. 1965, 14, 2000–2028. [Google Scholar] [PubMed]
- Xue, Y.; Enosi Tuipulotu, D.; Tan, W.H.; Kay, C.; Man, S.M. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019, 40, 1035–1052. [Google Scholar] [CrossRef]
- Szarek, M.; Bergmann, M.; Konrad, L.; Schuppe, H.C.; Kliesch, S.; Hedger, M.P.; Loveland, K.L. Activin A target genes are differentially expressed between normal and neoplastic adult human testes: Clues to gonocyte fate choice. Andrology 2019, 7, 31–41. [Google Scholar] [CrossRef]
- Kim, B.; Lee, H.J.; Im, N.R.; Lee, D.Y.; Kim, H.K.; Kang, C.Y.; Park, I.H.; Lee, S.H.; Lee, H.M.; Lee, S.H.; et al. Decreased expression of CCL17 in the disrupted nasal polyp epithelium and its regulation by IL-4 and IL-5. PLoS ONE 2018, 13, e0197355. [Google Scholar] [CrossRef]
- Naumann, S.C.; Roos, W.P.; Jost, E.; Belohlavek, C.; Lennerz, V.; Schmidt, C.W.; Christmann, M.; Kaina, B. Temozolomide- and fotemustine-induced apoptosis in human malignant melanoma cells: Response related to MGMT, MMR, DSBs, and p53. Br. J. Cancer 2009, 100, 322–333. [Google Scholar] [CrossRef]
- Felsberg, J.; Thon, N.; Eigenbrod, S.; Hentschel, B.; Sabel, M.C.; Westphal, M.; Schackert, G.; Kreth, F.W.; Pietsch, T.; Loffler, M.; et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 2011, 129, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Kramer, A.; Matthess, Y.; Yan, R.; Spankuch, B.; Gatje, R.; Knecht, R.; Kaufmann, M.; Strebhardt, K. Stable gene silencing of cyclin B1 in tumor cells increases susceptibility to taxol and leads to growth arrest in vivo. Oncogene 2006, 25, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Yoshimi, N.; Ino, N.; Tanaka, T.; Mori, H. Overexpression of cyclin B1 in human colorectal cancers. J. Cancer Res. Clin. Oncol. 1997, 123, 124–127. [Google Scholar] [CrossRef]
- Mashal, R.D.; Lester, S.; Corless, C.; Richie, J.P.; Chandra, R.; Propert, K.J.; Dutta, A. Expression of cell cycle-regulated proteins in prostate cancer. Cancer Res. 1996, 56, 4159–4163. [Google Scholar] [PubMed]
- Kushner, J.; Bradley, G.; Young, B.; Jordan, R.C. Aberrant expression of cyclin A and cyclin B1 proteins in oral carcinoma. J. Oral Pathol Med. 1999, 28, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, H.; Koizumi, H.; Uchikoshi, T. Expression of the G2-M checkpoint regulators cyclin B1 and cdc2 in nonmalignant and malignant human breast lesions: Immunocytochemical and quantitative image analyses. Am. J. Pathol. 1997, 150, 15–23. [Google Scholar] [PubMed]
- Suzuki, T.; Urano, T.; Miki, Y.; Moriya, T.; Akahira, J.; Ishida, T.; Horie, K.; Inoue, S.; Sasano, H. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007, 98, 644–651. [Google Scholar] [CrossRef]




| Proteins | Article Number | Producer | Origin |
|---|---|---|---|
| GAPDH | AC001 | ABclone | Wuhan, China |
| GSDMD-N | A20197 | ABclone | Wuhan, China |
| Caspase-1 | A0964 | ABclone | Wuhan, China |
| CCL 17 | A2854 | ABclone | Wuhan, China |
| IL 10 RA | A1830 | ABclone | Wuhan, China |
| ESR 1 | A1668 | ABclone | Wuhan, China |
| PRDX 4 | A1486 | ABclone | Wuhan, China |
| SOX 9 | A2479 | ABclone | Wuhan, China |
| MSH 6 | A0983 | ABclone | Wuhan, China |
| CDK 1 | A0220 | ABclone | Wuhan, China |
| CCNB 2 | A3351 | ABclone | Wuhan, China |
| AR | A2053 | ABclone | Wuhan, China |
| NOX 1 | A12309 | ABclone | Wuhan, China |
| Genes | Sequences of Primers (5′ to 3′) | Products | Genbank |
|---|---|---|---|
| GAPDH | F: GAAAGCTGCCAAATACGATGAG | 136 | XM_014866500.1 |
| R: GAAGGTGGAAGAGTGGATGTC | |||
| Caspase-1 | F: GGGCACGGGTACAGTAAATAG | 114 | XM_014851328.1 |
| R: CGGGCCTTATCCATAACTGTAG | |||
| GSDMD | F: GTTATTGGCTCTGACTGGGAC | 148 | XM_014858171.1 |
| R: TGAATCCTGACACGCTCTTG | |||
| CCL 17 | F: ACTGAAGATGCTGTTCCTGG | 106 | XM_014827258.1 |
| R: GAAGTACTCCAGACAGCACTC | |||
| IL 10 RA | F: GTGGATGAAGTGACTCTGACAG | 139 | XM_014855577.1 |
| R: CTCGGAAGTTAGGGAAGATGC | |||
| IL 32 | F: CAACTCAAGACACCCTCCC | 136 | XM_014833201.1 |
| R: AAGTAGCTCGAAACAGGCG | |||
| IL 23 A | F: GATGGCTGTGATCCTGAAGG | 113 | XM_014843912.1 |
| R: TCCCCTGTGAAAATGTCTGAG | |||
| AR | F: AGAGTTGTGTAAGGCAGTGTC | 96 | XM_014832024.1 |
| R: TACATGCAATCTCCCCGAAG | |||
| AIG 1 | F: AGTGGGAACCAAGAACAAGAG | 72 | XM_014850699.1 |
| R: CAGTACGGCTAACATCCAGTC | |||
| ESR 1 | F: GAAGAGACAAACCAAAGCCAG | 83 | XM_014827966.1 |
| R: GCCTCCCCAGTGATGTAATATG | |||
| ESR 2 | F: AGAGGGAAAATGCGTAGAAGG | 143 | XM_014853708.1 |
| R: CAGGGTACATACTGGAGTTGAG | |||
| MSH 6 | F: TTCTCTGGTGCTTGTGGATG | 130 | XM_014833021.1 |
| R:ATGGTAGTGGTAGAAAACAGTG | |||
| MCM 6 | F: GTGAAGGAGTGGGAGAAAGTG | 129 | XM_014849526.1 |
| R: AAAGAGCATCAGAAGGACACC | |||
| POLD 1 | F: TCCGTCATGTGCCGATTC | 123 | XM_014830334.1 |
| R: GTAGACCTTCTCAAACTCCAGC | |||
| CDK 1 | F: CTTGCCAGAGCTTTTGGAATAC | 129 | XM_014833411.1 |
| R: ACCTATACTCCAAATGTCAACCG | |||
| CCNB 2 | F: AACAGAGTTACAACCAGAGCC | 143 | XM_014835324.1 |
| R: GCCAATATTTCCATCTGCACTG | |||
| PFKM | F: TCCAACTACCTGAACATCGTG | 116 | XM_014850046.1 |
| R: GCGTCTACAATCTCTATGATCCG | |||
| PRDX 4 | F: CTCTGAATGACCTTCCTGTGG | 73 | XM_014867883.1 |
| R: TCGGTGTACTGGAATGCTTG | |||
| NOX 1 | F: ACTACCGTCTCTTCCTTACCG | 142 | XM_014859678.1 |
| R: GCAGAAAACTCATTGTCCCAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.-L.; Zhang, G.-L. Deoxynivalenol and Zearalenone: Different Mycotoxins with Different Toxic Effects in the Sertoli Cells of Equus asinus. Cells 2021, 10, 1898. https://doi.org/10.3390/cells10081898
Song J-L, Zhang G-L. Deoxynivalenol and Zearalenone: Different Mycotoxins with Different Toxic Effects in the Sertoli Cells of Equus asinus. Cells. 2021; 10(8):1898. https://doi.org/10.3390/cells10081898
Chicago/Turabian StyleSong, Jun-Lin, and Guo-Liang Zhang. 2021. "Deoxynivalenol and Zearalenone: Different Mycotoxins with Different Toxic Effects in the Sertoli Cells of Equus asinus" Cells 10, no. 8: 1898. https://doi.org/10.3390/cells10081898
APA StyleSong, J.-L., & Zhang, G.-L. (2021). Deoxynivalenol and Zearalenone: Different Mycotoxins with Different Toxic Effects in the Sertoli Cells of Equus asinus. Cells, 10(8), 1898. https://doi.org/10.3390/cells10081898





