Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases
Abstract
:1. Introduction
2. Circular RNAs in Chronic Liver Diseases
2.1. Alcoholic Liver Disease (ALD)
2.2. Metabolic-Associated Fatty Liver Disease (MAFLD)
2.3. Viral Hepatitis (Hepatitis B and Hepatitis C)
2.4. Liver Injury and Liver Regeneration
2.5. Liver Fibrosis/Cirrhosis
2.6. Autoimmune Liver Disease
3. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Sato, K.; Kennedy, L.; Liangpunsakul, S.; Kusumanchi, P.; Yang, Z.; Meng, F.; Glaser, S.; Francis, H.; Alpini, G. Intercellular Communication between Hepatic Cells in Liver Diseases. Int. J. Mol. Sci. 2019, 20, 2180. [Google Scholar] [CrossRef] [Green Version]
- Jarido, V.; Kennedy, L.; Hargrove, L.; Demieville, J.; Thomson, J.; Stephenson, K.; Francis, H. The emerging role of mast cells in liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G89–G101. [Google Scholar] [CrossRef] [PubMed]
- Francis, H.; Meininger, C.J. A review of mast cells and liver disease: What have we learned? Dig. Liver Dis. 2010, 42, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.W.; Dorrell, C.; Grompe, M. Stem cells and liver regeneration. Gastroenterology 2009, 137, 466–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.S.; Fan, J.G.; Zhang, Z.; Gao, B.; Wang, H.Y. The global burden of liver disease: The major impact of China. Hepatology 2014, 60, 2099–2108. [Google Scholar] [CrossRef]
- Mokdad, A.A.; Lopez, A.D.; Shahraz, S.; Lozano, R.; Mokdad, A.H.; Stanaway, J.; Murray, C.J.; Naghavi, M. Liver cirrhosis mortality in 187 countries between 1980 and 2010: A systematic analysis. BMC Med. 2014, 12, 145. [Google Scholar] [CrossRef] [Green Version]
- Hardy, T.; Mann, D.A. Epigenetics in liver disease: From biology to therapeutics. Gut 2016, 65, 1895–1905. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.A. Epigenetics in liver disease. Hepatology 2014, 60, 1418–1425. [Google Scholar] [CrossRef] [Green Version]
- Blum, H.E. Gastrointestinal and liver diseases: Genetic and epigenetic markers. Gut 2011, 60, 1630–1634. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Q.; Chang, C. Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 2020, 1253, 3–55. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Iwakiri, Y. Is miR-21 a potent target for liver fibrosis? Hepatology 2018, 67, 2082–2084. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Shabgah, A.G.; Norouzi, F.; Hedayati-Moghadam, M.; Soleimani, D.; Pahlavani, N.; Navashenaq, J.G. A comprehensive review of long non-coding RNAs in the pathogenesis and development of non-alcoholic fatty liver disease. Nutr. Metab. 2021, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wan, L.Y.; Liang, J.J.; Zhang, Y.Q.; Ai, W.B.; Wu, J.F. The roles of lncRNA in hepatic fibrosis. Cell Biosci. 2018, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yan, I.; Haga, H.; Patel, T. Long noncoding RNA in liver diseases. Hepatology 2014, 60, 744–753. [Google Scholar] [CrossRef]
- Wang, Y.; Hylemon, P.B.; Zhou, H. Long non-coding RNA H19: A key player in liver diseases. Hepatology 2021. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circRNAs. Embo. J. 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Chen, Q.; Fu, L.; Guo, J. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol. Res. 2017, 47, 497–504. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE 2016, 11, e0148407. [Google Scholar] [CrossRef] [Green Version]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e498. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Harries, L.W. Circular RNAs (circRNAs) in Health and Disease. Genes 2017, 8, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.Y.; Wang, S.W.; Hu, M.Y.; Jiang, Z.L.; Shen, L.L.; Zhou, Y.P.; Guo, J.M.; Hu, Y.R. Circular RNAs in liver diseases: Mechanisms and therapeutic targets. Life Sci. 2021, 264, 118707. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Venø, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Xia, L.; Sun, M.; Yang, C.; Wang, F. Circular RNA in Liver: Health and Diseases. Adv. Exp. Med. Biol. 2018, 1087, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Liao, J.; Liang, J.; Chen, X.P.; Zhang, B.; Chu, L. Circular RNA HIPK3: A Key Circular RNA in a Variety of Human Cancers. Front. Oncol. 2020, 10, 773. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Li, J. Role of miRNA sponges in hepatocellular carcinoma. Clin. Chim. Acta 2020, 500, 10–19. [Google Scholar] [CrossRef]
- Shang, W.; Adzika, G.K.; Li, Y.; Huang, Q.; Ding, N.; Chinembiri, B.; Rashid, M.S.; Machuki, J.O. Molecular mechanisms of circular RNAs, transforming growth factor-β, and long noncoding RNAs in hepatocellular carcinoma. Cancer Med. 2019, 8, 6684–6699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, L.; Xu, H.; Ji, M.; Shang, D.; Lu, Z.; Wu, Y.; Tu, Z.; Liu, H. Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms. Life Sci. 2019, 231, 116660. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Li, P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers 2018, 10, 258. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.P.; Wu, Y.H.; Yu, X.F.; Tang, Q.; Chen, L.; Chen, K.P. The Emerging Role of Circular RNAs in Hepatocellular Carcinoma. J. Cancer 2018, 9, 1548–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Li, P.; Song, Y.; Ge, Y.X.; Meng, X.M.; Huang, C.; Li, J.; Xu, T. Progress and prospects of circular RNAs in Hepatocellular carcinoma: Novel insights into their function. J. Cell Physiol. 2018, 233, 4408–4422. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Jiang, Z.; Li, T.; Hu, Y.; Guo, J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018, 7, 3101–3109. [Google Scholar] [CrossRef]
- Tanaka, A.; Leung, P.S.C.; Gershwin, M.E. The genetics of primary biliary cholangitis. Curr. Opin Gastroenterol. 2019, 35, 93–98. [Google Scholar] [CrossRef]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef]
- Chien, Y.; Tsai, P.H.; Lai, Y.H.; Lu, K.H.; Liu, C.Y.; Lin, H.F.; Huang, C.S.; Wu, W.W.; Wang, C.Y. CircularRNA as novel biomarkers in liver diseases. J. Chin. Med. Assoc. 2020, 83, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic liver disease. Nat. Rev. Dis. Primers 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, T.S.; Liu, M.; Shah, V.H. The knowns and unknowns of treatment for alcoholic hepatitis. Lancet Gastroenterol. Hepatol. 2020, 5, 494–506. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Xuan, W.; Ye, J.; Yao, H.; Huang, C.; Li, J. Circ_1639 induces cells inflammation responses by sponging miR-122 and regulating TNFRSF13C expression in alcoholic liver disease. Toxicol. Lett. 2019, 314, 89–97. [Google Scholar] [CrossRef]
- Bajaj, J.S. Alcohol, liver disease and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 235–246. [Google Scholar] [CrossRef]
- Szabo, G.; Satishchandran, A. MicroRNAs in alcoholic liver disease. Semin. Liver Dis. 2015, 35, 36–42. [Google Scholar] [CrossRef] [Green Version]
- Satishchandran, A.; Ambade, A.; Rao, S.; Hsueh, Y.C.; Iracheta-Vellve, A.; Tornai, D.; Lowe, P.; Gyongyosi, B.; Li, J.; Catalano, D.; et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology 2018, 154, 238–252.e237. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.; Wang, L.; You, H.; Huang, C.; Li, J. Circular RNA expression profile of liver tissues in an EtOH-induced mouse model of alcoholic hepatitis. Eur. J. Pharmacol. 2019, 862, 172642. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Feng, L.; Ying, N.; Ding, Q.; Song, Q.; Jiang, F.; Wang, C.; Li, S. RNA Sequencing Reveals a Comprehensive Circular RNA Expression Profile in a Mouse Model of Alcoholic Liver Disease. Alcohol. Clin. Exp. Res. 2020, 44, 415–422. [Google Scholar] [CrossRef]
- Francis, H.; McDaniel, K.; Han, Y.; Liu, X.; Kennedy, L.; Yang, F.; McCarra, J.; Zhou, T.; Glaser, S.; Venter, J.; et al. Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J. Biol. Chem. 2014, 289, 27526–27539. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Ren, T.; Zhu, Z.; Cheng, M.; Mou, Q.; Mu, M.; Liu, Y.; Yao, Y.; Cheng, Y.; Zhang, B.; et al. Thymosin-β4 Mediates Hepatic Stellate Cell Activation by Interfering with CircRNA-0067835/miR-155/FoxO3 Signaling Pathway. Cell Physiol. Biochem. 2018, 51, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Huang, J.; Li, X.; Xing, J.; Chen, Q.; Liu, R.; Hua, F.; Qiu, Z.; Song, Y.; Bai, C.; et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes 2020, 12, 1788891. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J. Past, present and future perspectives in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 377–386. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Non-alcoholic fatty liver disease. BMC Med. 2017, 15, 45. [Google Scholar] [CrossRef] [Green Version]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Yu, G.; Yang, Z.; Peng, T.; Lv, Y. Circular RNAs: Rising stars in lipid metabolism and lipid disorders. J. Cell Physiol. 2021, 236, 4797–4806. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, X.; Deng, Y.; Cui, Q.; Zhu, J.; Ren, H.; Liu, Y.; Hu, X.; Zuo, J.; Peng, Y. Regulatory roles of circRNAs in adipogenesis and lipid metabolism: Emerging insights into lipid-related diseases. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.Y.; He, C.X.; Wang, Y.Q.; Sun, C.; Li, G.M.; Su, Q.; Pan, Q.; Fan, J.G. Circular RNA Profiling and Bioinformatic Modeling Identify Its Regulatory Role in Hepatic Steatosis. Biomed. Res. Int. 2017, 2017, 5936171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Shan, K.; Liu, Y.; Zhang, Y.; Xu, L.; Xu, L. CircScd1 Promotes Fatty Liver Disease via the Janus Kinase 2/Signal Transducer and Activator of Transcription 5 Pathway. Dig. Dis. Sci. 2019, 64, 113–122. [Google Scholar] [CrossRef]
- Yuan, X.; Diao, J.; Du, A.; Wen, S.; Zhou, L.; Pan, Y. Circular RNA expression profiles and features in NAFLD mice: A study using RNA-seq data. J. Transl. Med. 2020, 18, 476. [Google Scholar] [CrossRef]
- Guo, X.Y.; Sun, F.; Chen, J.N.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J. Gastroenterol. 2018, 24, 323–337. [Google Scholar] [CrossRef]
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid Med. Cell Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef]
- Jin, X.; Gao, J.; Zheng, R.; Yu, M.; Ren, Y.; Yan, T.; Huang, Y.; Li, Y. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020, 11, 123. [Google Scholar] [CrossRef]
- Ou, Q.; Zhao, Y.; Zhou, J.; Wu, X. Comprehensive circular RNA expression profiles in a mouse model of nonalcoholic steatohepatitis. Mol. Med. Rep. 2019, 19, 2636–2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Zhou, Y.; Cheng, Y.; Fang, W.; Hu, G.; Wei, J.; Lin, Y.; Man, Y.; Guo, L.; Sun, M.; et al. Metformin-Induced Changes of the Coding Transcriptome and Non-Coding RNAs in the Livers of Non-Alcoholic Fatty Liver Disease Mice. Cell Physiol. Biochem. 2018, 45, 1487–1505. [Google Scholar] [CrossRef]
- Zhu, M.; Li, M.; Zhou, W.; Yang, Y.; Li, F.; Zhang, L.; Ji, G. Qianggan extract improved nonalcoholic steatohepatitis by modulating lncRNA/circRNA immune ceRNA networks. BMC Complement. Altern Med. 2019, 19, 156. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; McDaniel, K.; Wu, N.; Ramos-Lorenzo, S.; Glaser, T.; Venter, J.; Francis, H.; Kennedy, L.; Sato, K.; Zhou, T.; et al. Regulation of Cellular Senescence by miR-34a in Alcoholic Liver Injury. Am. J. Pathol. 2017, 187, 2788–2798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, M.-F.; Chen, D.-S.; Dusheiko, G.M.; Janssen, H.L.A.; Lau, D.T.Y.; Locarnini, S.A.; Peters, M.G.; Lai, C.-L. Hepatitis B virus infection. Nat. Rev. Dis. Primers 2018, 4, 18035. [Google Scholar] [CrossRef]
- Seto, W.K.; Lo, Y.R.; Pawlotsky, J.M.; Yuen, M.F. Chronic hepatitis B virus infection. Lancet 2018, 392, 2313–2324. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z. Circular RNA hsa_circ_0004812 impairs IFN-induced immune response by sponging miR-1287-5p to regulate FSTL1 in chronic hepatitis B. Virol. J. 2020, 17, 40. [Google Scholar] [CrossRef] [Green Version]
- Honda, M.; Kaneko, S.; Kawai, H.; Shirota, Y.; Kobayashi, K. Differential gene expression between chronic hepatitis B and C hepatic lesion. Gastroenterology 2001, 120, 955–966. [Google Scholar] [CrossRef]
- Zhou, T.C.; Li, X.; Chen, L.J.; Fan, J.H.; Lai, X.; Tang, Y.; Zhang, L.; Wei, J. Differential expression profile of hepatic circular RNAs in chronic hepatitis B. J. Viral Hepat. 2018, 25, 1341–1351. [Google Scholar] [CrossRef]
- Sarnow, P.; Sagan, S.M. Unraveling the Mysterious Interactions Between Hepatitis C Virus RNA and Liver-Specific MicroRNA-122. Annu. Rev. Virol. 2016, 3, 309–332. [Google Scholar] [CrossRef] [PubMed]
- Kunden, R.D.; Khan, J.Q.; Ghezelbash, S.; Wilson, J.A. The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 2020, 21, 5677. [Google Scholar] [CrossRef] [PubMed]
- Jost, I.; Shalamova, L.A.; Gerresheim, G.K.; Niepmann, M.; Bindereif, A.; Rossbach, O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018, 15, 1032–1039. [Google Scholar] [CrossRef]
- Chen, T.C.; Tallo-Parra, M.; Cao, Q.M.; Kadener, S.; Böttcher, R.; Pérez-Vilaró, G.; Boonchuen, P.; Somboonwiwat, K.; Díez, J.; Sarnow, P. Host-derived circular RNAs display proviral activities in Hepatitis C virus-infected cells. PLoS Pathog. 2020, 16, e1008346. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.K.; Yuen, M.F.; Yuan, H.; Sum, S.S.; Hui, C.K.; Hall, J.; Lai, C.L. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology 2004, 40, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Sekiba, K.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yamagami, M.; Suzuki, T.; Ishibashi, R.; Seimiya, T.; Tanaka, E.; Koike, K. DHX9 regulates production of hepatitis B virus-derived circular RNA and viral protein levels. Oncotarget 2018, 9, 20953–20964. [Google Scholar] [CrossRef] [Green Version]
- Itoh, T. Stem/progenitor cells in liver regeneration. Hepatology 2016, 64, 663–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xu, L.; Wang, P.; Zheng, X.; Hu, Y.; Luo, J.; Zhang, M.; Xu, M. RNA-seq Used to Explore circRNA Expression and Identify Key circRNAs During the DNA Synthesis Phase of Mice Liver Regeneration. DNA Cell Biol. 2020, 39, 2059–2076. [Google Scholar] [CrossRef]
- Li, L.; Guo, J.; Chen, Y.; Chang, C.; Xu, C. Comprehensive CircRNA expression profile and selection of key CircRNAs during priming phase of rat liver regeneration. BMC Genom. 2017, 18, 80. [Google Scholar] [CrossRef] [Green Version]
- Peralta, C.; Jiménez-Castro, M.B.; Gracia-Sancho, J. Hepatic ischemia and reperfusion injury: Effects on the liver sinusoidal milieu. J. Hepatol. 2013, 59, 1094–1106. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Kong, Q.; Han, J.; Deng, J.; Wu, M.; Deng, H. Circular RNAs are differentially expressed in liver ischemia/reperfusion injury model. J. Cell Biochem. 2018, 119, 7397–7405. [Google Scholar] [CrossRef]
- Qu, X.; Zheng, C.; Wang, B.; Wang, F.; Sun, X.; Gao, Y.; Xia, Q.; Kong, X. Comprehensive analysis of circular RNAs from steatotic livers after ischemia and reperfusion injury by next-generation RNA sequencing. FEBS Lett. 2020, 595, 99–109. [Google Scholar] [CrossRef]
- Tian, X.; Hu, Y.; Liu, Y.; Yang, Z.; Xie, H.; Zhou, L.; Zheng, S. Circular RNA Microarray Analyses in Hepatic Ischemia-Reperfusion Injury With Ischemic Preconditioning Prevention. Front. Med. 2021, 8, 626948. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, B.; Chen, G.; Zhang, L.; Zhuang, Y.; Niu, H.; Zeng, Z. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J. Cell Physiol. 2020, 235, 8270–8282. [Google Scholar] [CrossRef]
- Niu, H.; Zhang, L.; Chen, Y.H.; Yuan, B.Y.; Wu, Z.F.; Cheng, J.C.; Lin, Q.; Zeng, Z.C. Circular RNA TUBD1 Acts as the miR-146a-5p Sponge to Affect the Viability and Pro-Inflammatory Cytokine Production of LX-2 Cells through the TLR4 Pathway. Radiat Res. 2020, 193, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y.; Sun, R.; Sun, Y.; Liu, D.; Lin, M.; Chen, Z.; Zhou, J.; Lv, L.; Tian, X.; et al. circ-CBFB upregulates p66Shc to perturb mitochondrial dynamics in APAP-induced liver injury. Cell Death Dis. 2020, 11, 953. [Google Scholar] [CrossRef]
- Li, Y.; Gao, X.; Wang, Z.; Liu, W.; Xu, F.; Hu, Y.; Li, Y.; Shi, L. Circular RNA 4099 aggravates hydrogen peroxide-induced injury by down-regulating microRNA-706 in L02 cells. Life Sci. 2020, 241, 116826. [Google Scholar] [CrossRef]
- Li, B. The Diagnostic Value and Mechanism of Circular RNA circMARS and circITPR1 in Anti-Tuberculosis Drug- Induced Liver Injury. Master’s Thesis, North China University of Science and Technology, Tangshan, China, 2019. (In Chinese). [Google Scholar]
- Zhang, P.; Ming, Y.; Ye, Q.; Niu, Y. Comprehensive circRNA expression profile during ischemic postconditioning attenuating hepatic ischemia/reperfusion injury. Sci. Rep. 2019, 9, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, H.; Wang, H.; Yu, Q. Circular RNA circ_0003420 mediates inflammation in sepsis-induced liver damage by downregulating neuronal PAS domain protein 4. Immunopharmacol. Immunotoxicol. 2021, 43, 271–282. [Google Scholar] [CrossRef]
- Parola, M.; Pinzani, M. Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Aspects Med. 2019, 65, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Harrington, L.; McPhail, T.; Mar, V.; Zhou, W.; Oulton, R.; Bass, M.B.; Arruda, I.; Robinson, M.O. A mammalian telomerase-associated protein. Science 1997, 275, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Barnard, A.; Moch, A.; Saab, S. Relationship between Telomere Maintenance and Liver Disease. Gut Liver 2019, 13, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Song, F.; Lei, X.; Li, J.; Li, F.; Tan, H. hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging 2020, 12, 11517–11529. [Google Scholar] [CrossRef]
- Jin, H.; Li, C.; Dong, P.; Huang, J.; Yu, J.; Zheng, J. Circular RNA cMTO1 Promotes PTEN Expression Through Sponging miR-181b-5p in Liver Fibrosis. Front. Cell Dev. Biol. 2020, 8, 714. [Google Scholar] [CrossRef]
- Wang, W.; Dong, R.; Guo, Y.; He, J.; Shao, C.; Yi, P.; Yu, F.; Gu, D.; Zheng, J. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell Mol. Med. 2019, 23, 5486–5496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, D.; Chen, G.F.; Wang, J.C.; Ji, S.H.; Wu, X.W.; Lu, X.J.; Chen, J.L.; Li, J.T. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 2020, 12, 1643–1655. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lv, X.; Qu, H.; Zhao, K.; Fu, L.; Zhu, L.; Ye, G.; Guo, J. Differential expression of circular RNAs in hepatic tissue in a model of liver fibrosis and functional analysis of their target genes. Hepatol. Res. 2019, 49, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lv, X.; Qu, H.; Zhao, K.; Fu, L.; Zhu, L.; Ye, G.; Guo, J. Preliminary screening and functional analysis of circular RNAs associated with hepatic stellate cell activation. Gene 2018, 677, 317–323. [Google Scholar] [CrossRef]
- Bu, F.T.; Zhu, Y.; Chen, X.; Wang, A.; Zhang, Y.F.; You, H.M.; Yang, Y.; Yang, Y.R.; Huang, C.; Li, J. Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol. Ther. Nucleic Acids 2021, 23, 847–862. [Google Scholar] [CrossRef]
- Liu, W.; Feng, R.; Li, X.; Li, D.; Zhai, W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging 2019, 11, 9569–9580. [Google Scholar] [CrossRef] [PubMed]
- Riaz, F.; Li, D. Non-coding RNA Associated Competitive Endogenous RNA Regulatory Network: Novel Therapeutic Approach in Liver Fibrosis. Curr. Gene Ther. 2019, 19, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Dai, Q.; Liu, Z.; Zhou, L.; Xu, J. Circular RNAs in Organ Fibrosis. Adv. Exp. Med. Biol. 2018, 1087, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, B.; Wu, Z.; Dong, Y.; Zhang, L.; Zeng, Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 2017, 629, 35–42. [Google Scholar] [CrossRef]
- Marrone, G.; Shah, V.H.; Gracia-Sancho, J. Sinusoidal communication in liver fibrosis and regeneration. J. Hepatol. 2016, 65, 608–617. [Google Scholar] [CrossRef] [Green Version]
- Carbone, M.; Neuberger, J.M. Autoimmune liver disease, autoimmunity and liver transplantation. J. Hepatol. 2014, 60, 210–223. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Sun, B.; Huang, S.; Zhao, L. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 2019, 10, 503. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Li, Z.; Wang, T.; Zhao, Y.; Wang, Y. Microarray Expression Profile of Circular RNAs in Plasma from Primary Biliary Cholangitis Patients. Cell Physiol. Biochem. 2017, 44, 1271–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, Z.; Hao, J.; Chen, H.; Hou, T.; Hao, H. Circular RNAs associated with a mouse model of concanavalin A-induced autoimmune hepatitis: Preliminary screening and comprehensive functional analysis. FEBS Open Bio 2020, 10, 2350–2362. [Google Scholar] [CrossRef]
- Virani, S.; Akers, A.; Stephenson, K.; Smith, S.; Kennedy, L.; Alpini, G.; Francis, H. Comprehensive Review of Molecular Mechanisms during Cholestatic Liver Injury and Cholangiocarcinoma. J. Liver 2018, 7, 231. [Google Scholar] [CrossRef]
- Zhao, Z.; Meng, J.; Su, R.; Zhang, J.; Chen, J.; Ma, X.; Xia, Q. Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 2020, 73, 664–679. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.C.; Chen, X.Y.; Zhang, J.; Zhu, J.S. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol. Cancer 2020, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Chi, F.; Cao, Y.; Chen, Y. Analysis and Validation of circRNA-miRNA Network in Regulating m(6)A RNA Methylation Modulators Reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis Involving the Proliferation of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 560506. [Google Scholar] [CrossRef]
- Pirola, C.J.; Sookoian, S. Epigenetics factors in nonalcoholic fatty liver disease. Expert Rev. Gastroenterol. Hepatol. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Sun, Y.; Sheng, B.; Zheng, Y.; Wu, X.; Xu, K. Role of identified RNA N6-methyladenosine methylation in liver. Anal. Biochem. 2019, 578, 45–50. [Google Scholar] [CrossRef]
- Joshita, S.; Umemura, T.; Tanaka, E.; Ota, M. Genetics and epigenetics in the pathogenesis of primary biliary cholangitis. Clin. J. Gastroenterol. 2018, 11, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hou, C.; Chen, C.; Guo, Y.; Yuan, W.; Yin, D.; Liu, J.; Sun, Z. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol. Cancer 2020, 19, 105. [Google Scholar] [CrossRef]

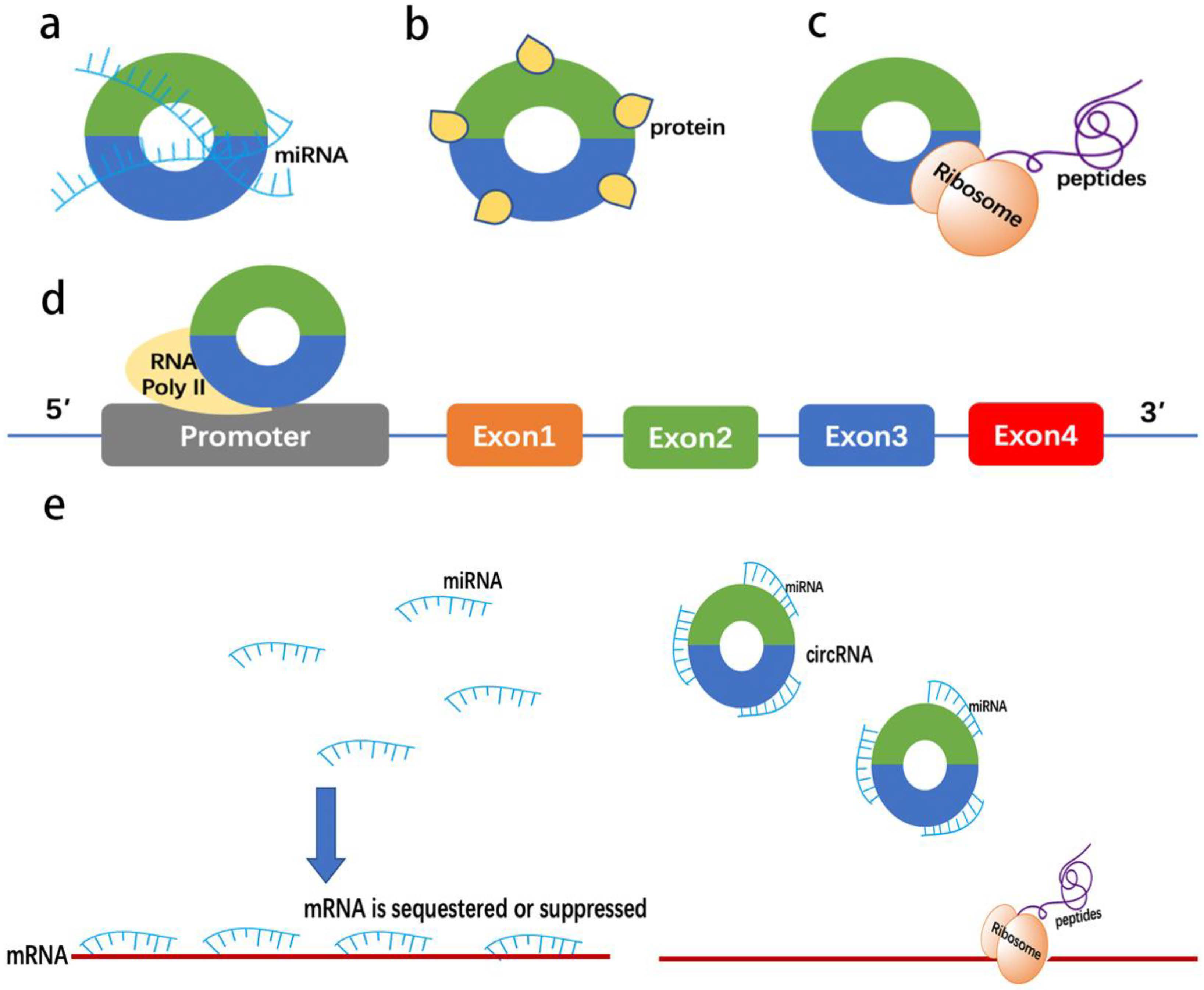
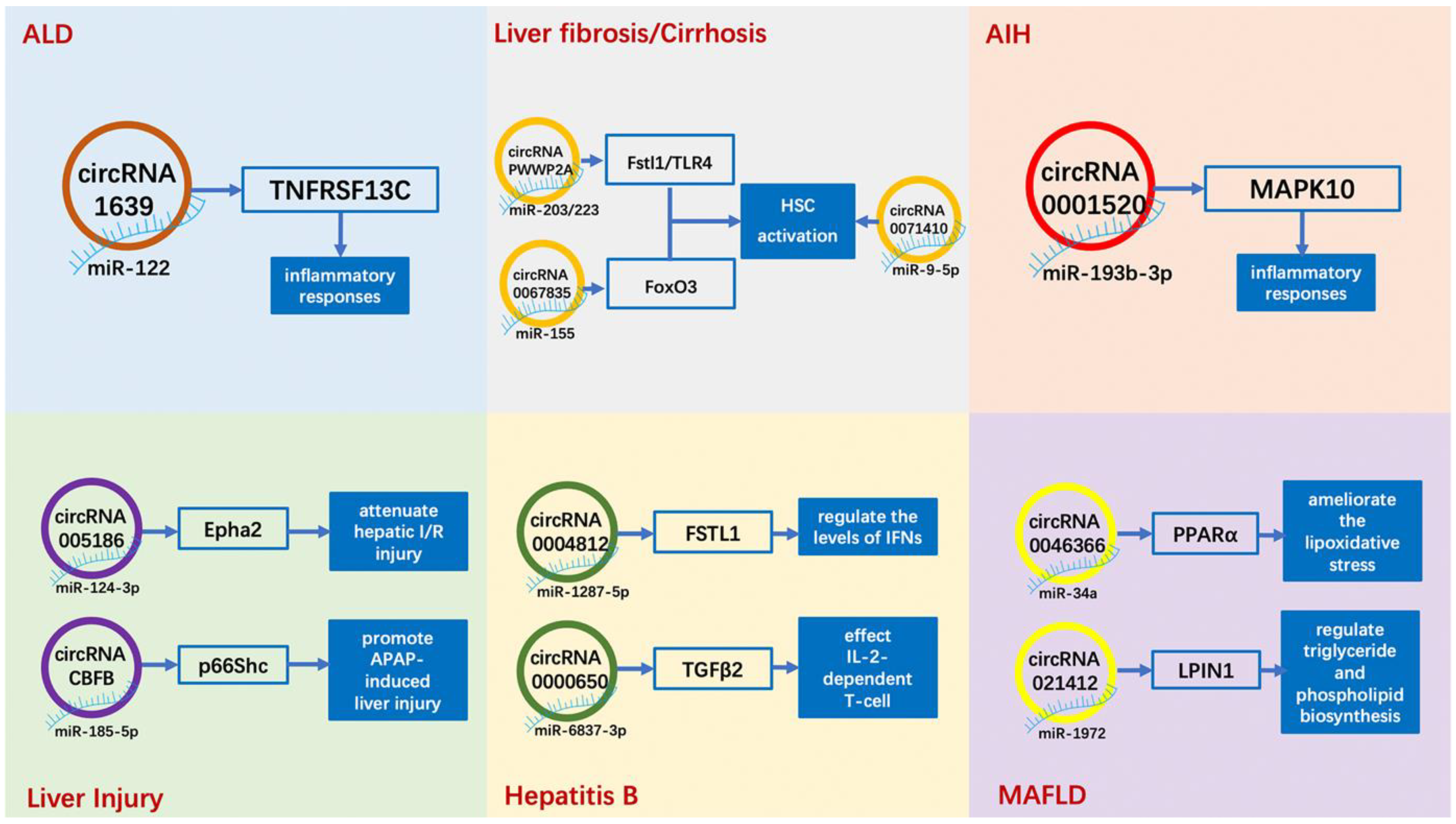
| Diseases | CircRNA or LncRNA | Possible Mechanisms | Expression Change | Refs. |
|---|---|---|---|---|
| ALD | circRNA_1639 | circ_1639/miR-122/TNFRSF13C axis and NF-κB pathway | up-regulation | [49] |
| mou_circ_1657 | sponge miR-96-5p | up-regulation | [54] | |
| MAFLD | circScd1 | JAK2/STAT5 pathway | down-regulation | [66] |
| circRNA_0049392 | target miR-7037-5p and miR-6919-5p | up-regulation | [67] | |
| circRNA_0046366, circRNA_0046367 | circRNA_0046366,7/miR-34a/PPAR α axis | down-regulation | [68,69] | |
| circRNA_021412 | circRNA_021412/miR-1972/LPIN1 axis | down-regulation | [65] | |
| circRNA SCAR | shut down mPTP by binding to ATP5B | down-regulation | [62] | |
| Viral hepatitis | hsa_circ_0004812 | circ_0004812/miR-1287-5p/FSTL1 axis | up-regulation | [77] |
| hsa_circ_0000650 | hsa_circ_0000650/miR-6873-3p and TGF β 2 axis | down-regulation | [79] | |
| circPSD3 | bind factor eIF4A3; nonsense-mediated decay (NMD) pathway | up-regulation | [83] | |
| Liver injury and regeneration | circ432, circ2077, circ1366 and circ15 | controlling the expression level of MAPK14, FN1, TNFRSF21 and GOT1 | up-regulation | [88] |
| circRNA_017753 | circRNA_017753/miR-218-5p, miR-7002-3p, miR-7008-3p/Jade1 | up-regulation | [92] | |
| circRSF1 | promoting inflammatory and fibrotic phenotypes of HSC by sponging miR-146a-5p | up-regulation | [93] | |
| circTUBD1 | circTUBD1/miR-146a-5p/Toll-like receptor 4/NF-κB axis | up-regulation | [94] | |
| circ-CBFB | circ-CBFB/miR-185-5p/p66Shc axis | up-regulation | [95] | |
| circRNA-4099 | circRNA-4099/miR-706/keap1/Nrf2 and p38MAPK axis | down-regulation | [96] | |
| circMARS | circMARS—miR-6808-5p/-6874-3p/-3157-5p—KMT2C—EGFR functional axis | up-regulation | [97] | |
| mmu_circRNA_005186 | mmu_circRNA_005186/miR-124-3p/Epha2 axis | up-regulation | [98] | |
| Liver fibrosis/Cirrhosis | hsa_circ_0004018 | hsa_circ_0004018/hsa-miR-660-3p/TEP1 axis; inhibits liver fibrosis | down-regulation | [104] |
| hsa_circ_0007874 (cMTO1) | inhibit HSC activation through sponging miR-181b-5p and miR-17-5p | down-regulation | [105,106] | |
| hsa_circ_0070963 | inhibits liver fibrosis via regulation of miR-223-3p that related to HSC activation | down-regulation | [107] | |
| circFBXW4 | circFBXW4/miR-18b-3p/FBXW7 axis; inhibit fibrosis | down-regulation | [108] | |
| mmu_circ_34116 | mmu_circ_34116/miR-22-3P/BMP7 signal axis; inhibit HSC activation | down-regulation | [109,110] | |
| circPSD3 | circPSD3/miR-92b-3p/Smad7 | down-regulation | [111] | |
| circ-PWWP2A | circ-PWWP2A/miR-203 and miR-223/Fstl 1 and TLR4 axis; promote HSC activation | up-regulation | [112,113] | |
| circRNA-0067835 | circRNA-0067835/miR-155/FoxO3 axis; promote HSC activation | up-regulation | [56] | |
| hsa_circ_0071410 | sponge miR-9-5p; promote irradiation-induced HSC activation | up-regulation | [114,115] | |
| Autoimmune liver disease | hsa_circ_402458 | sponge hsa-miR-522-3p and hsa-miR-943 | up-regulation | [119] |
| mmu_circ_0001520 | mmu_circ_0001520/mmu-miR-193b-3p/MAPK10 | up-regulation | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Yuan, X.; Cai, Q.; Tang, C.; Gao, J. Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases. Cells 2021, 10, 1945. https://doi.org/10.3390/cells10081945
Zeng X, Yuan X, Cai Q, Tang C, Gao J. Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases. Cells. 2021; 10(8):1945. https://doi.org/10.3390/cells10081945
Chicago/Turabian StyleZeng, Xianhui, Xianglei Yuan, Qiuyu Cai, Chengwei Tang, and Jinhang Gao. 2021. "Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases" Cells 10, no. 8: 1945. https://doi.org/10.3390/cells10081945
APA StyleZeng, X., Yuan, X., Cai, Q., Tang, C., & Gao, J. (2021). Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases. Cells, 10(8), 1945. https://doi.org/10.3390/cells10081945






