Organellar Introns in Fungi, Algae, and Plants
Abstract
1. Introduction
2. Organellar Introns: Group I and Group II Introns
3. Distribution and Impact of Introns on Organellar Genome Architecture
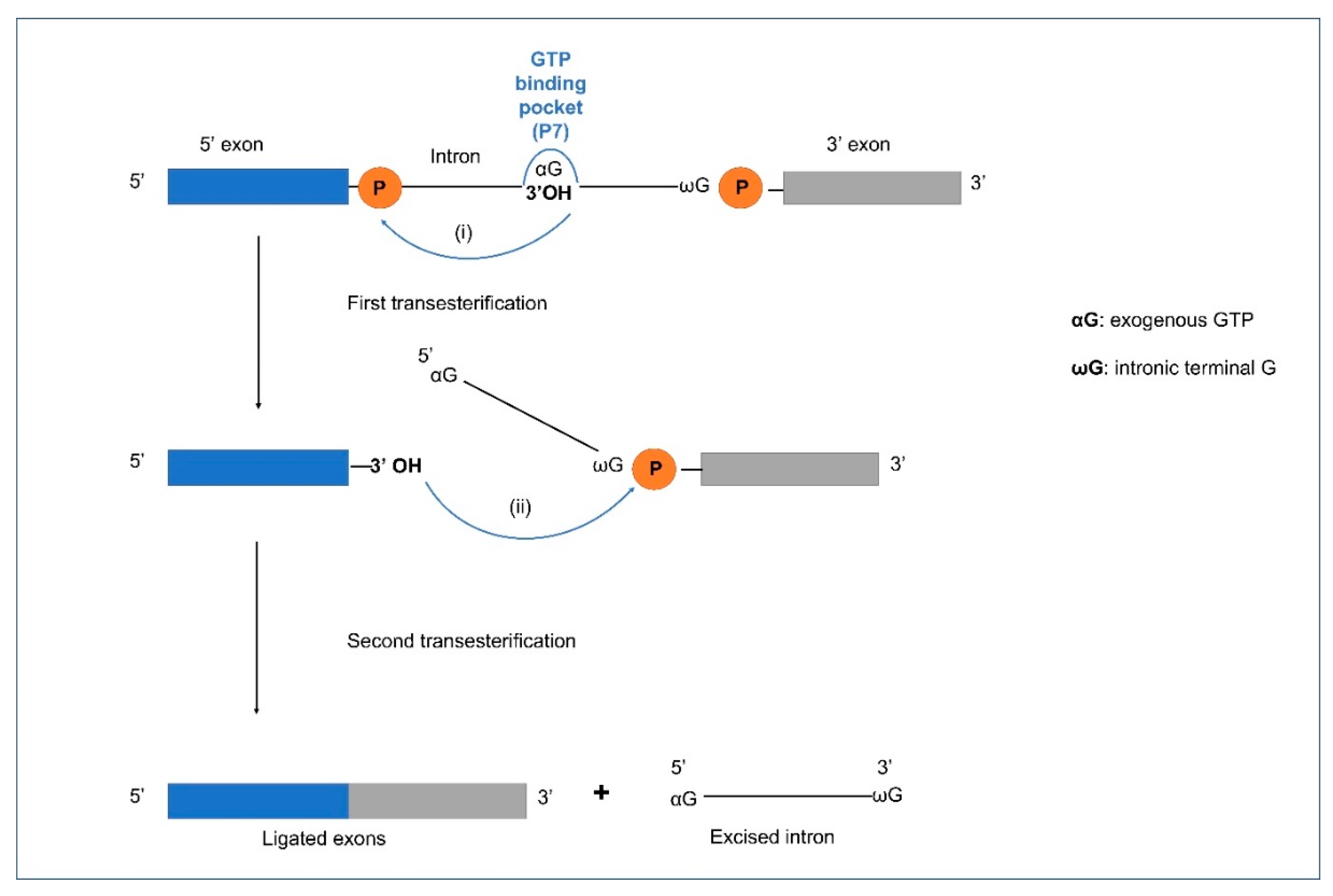
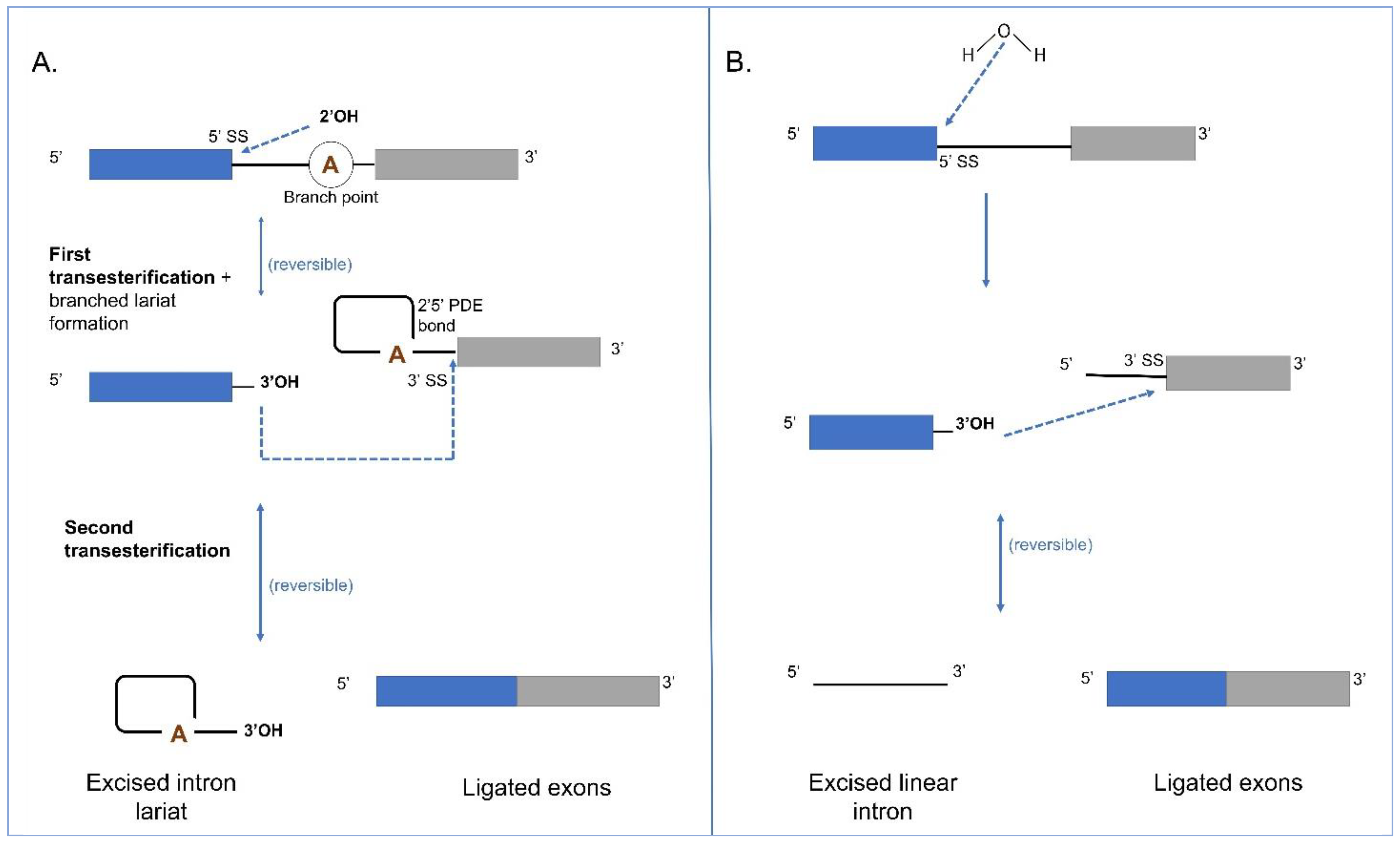
4. Splicing in Group I and II Introns
5. Intron-Encoded Proteins in Guiding Intron Mobility and Multifunctional Roles
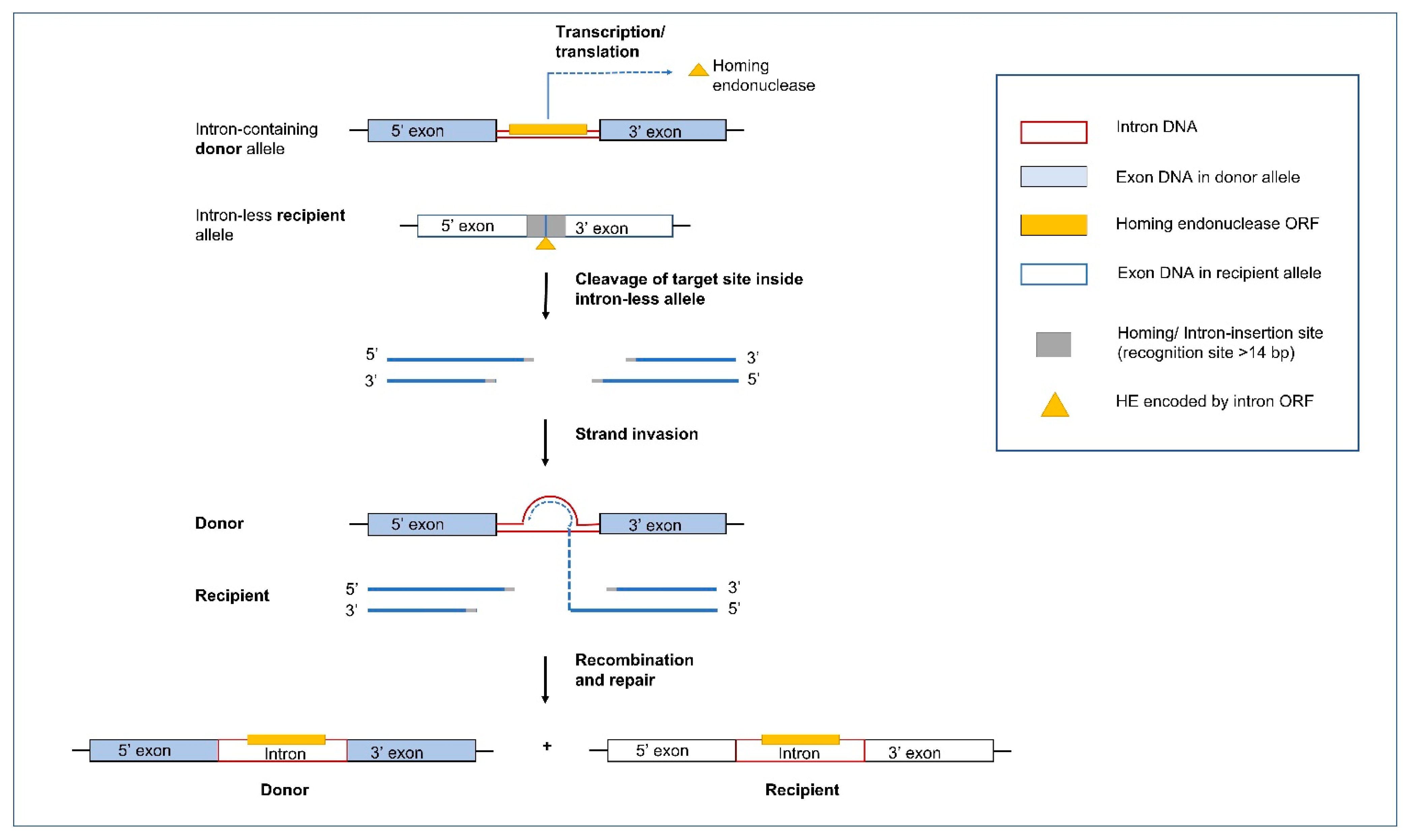
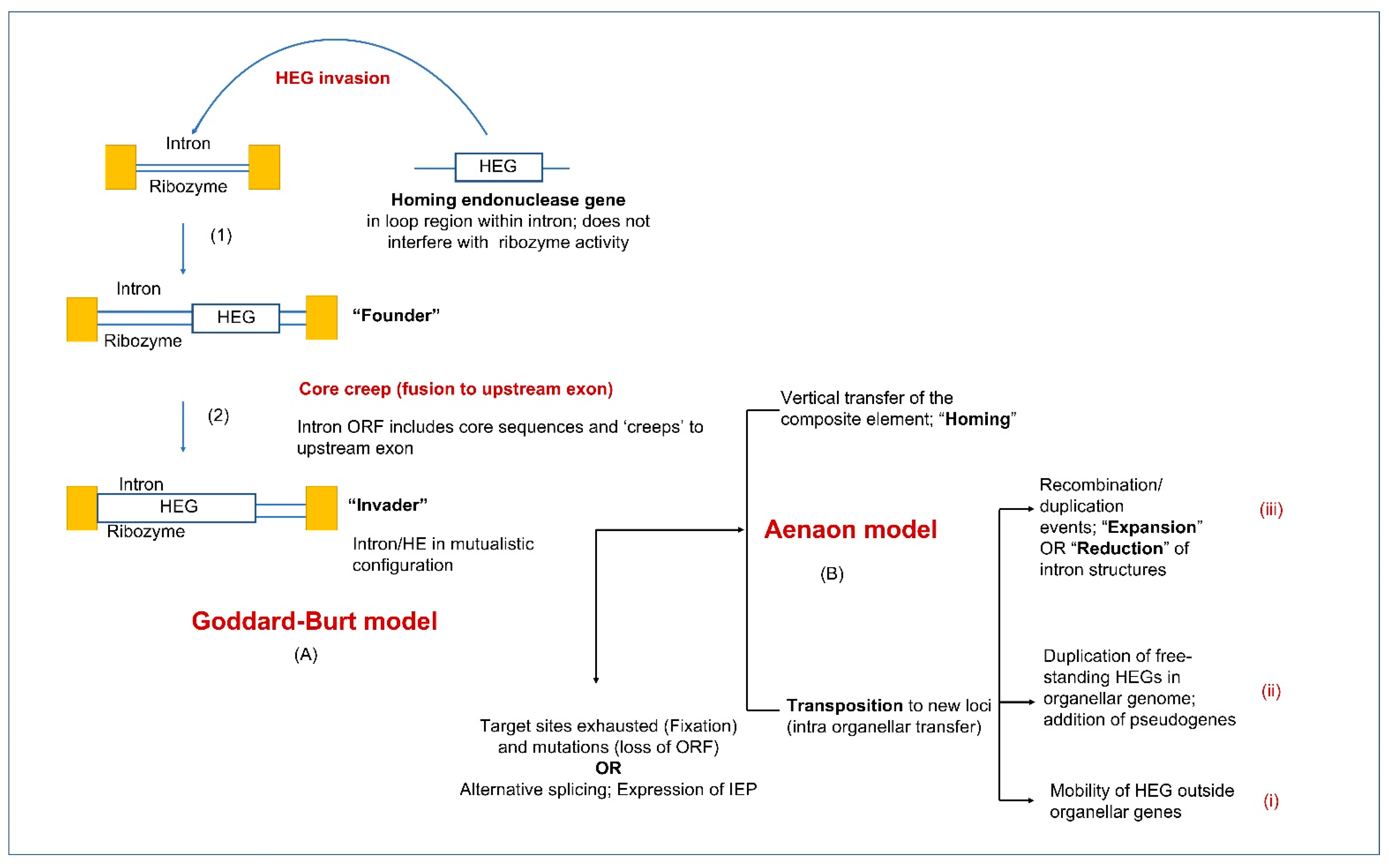
5.1. Group I Introns and Homing Endonucleases
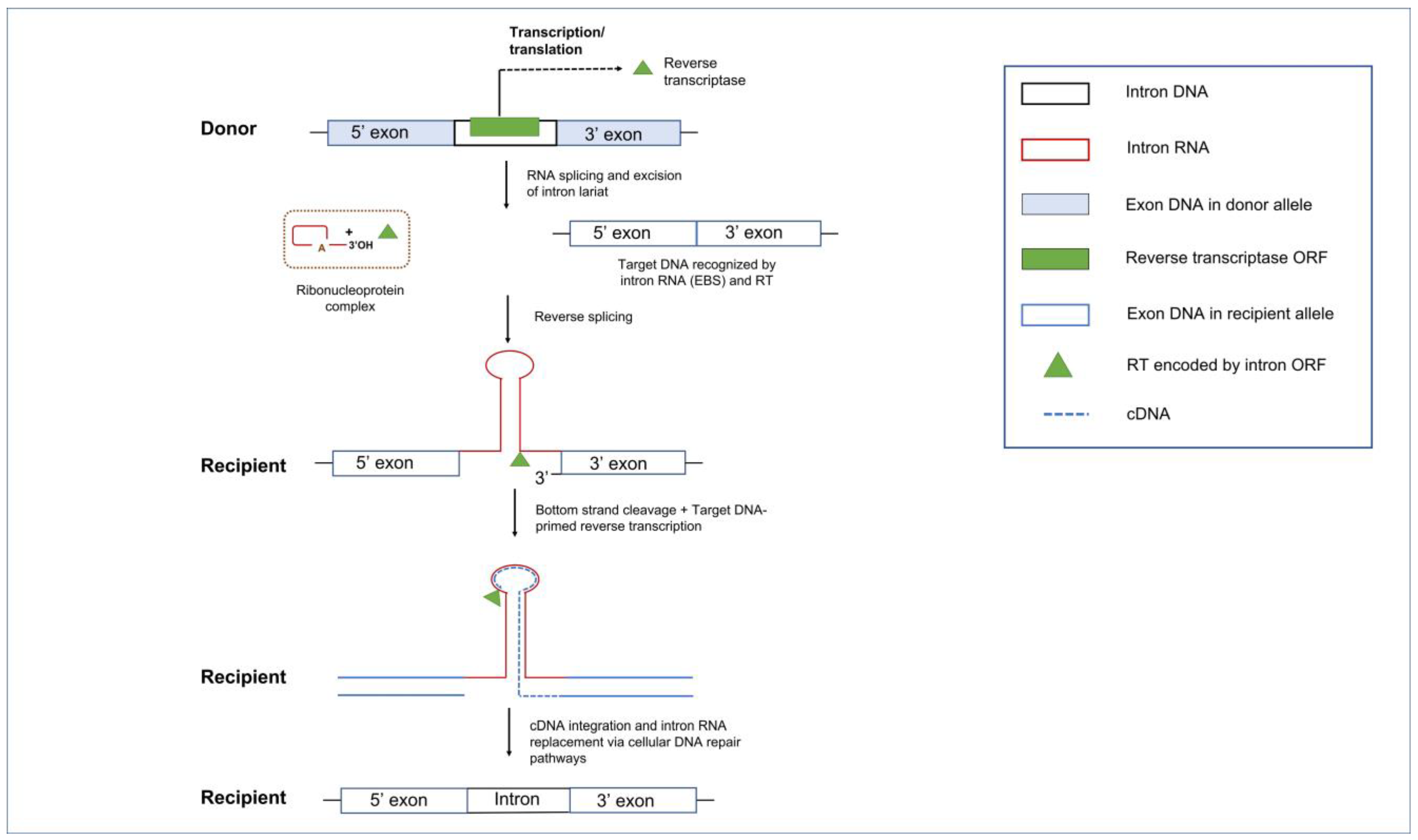
5.2. Group II Introns and Reverse Transcriptases
5.3. Maturase and Splicing Factors
6. Introns: At the Fulcrum of Adaptation and Elimination
7. Introns Impacting Gene and Phenotypic Expression
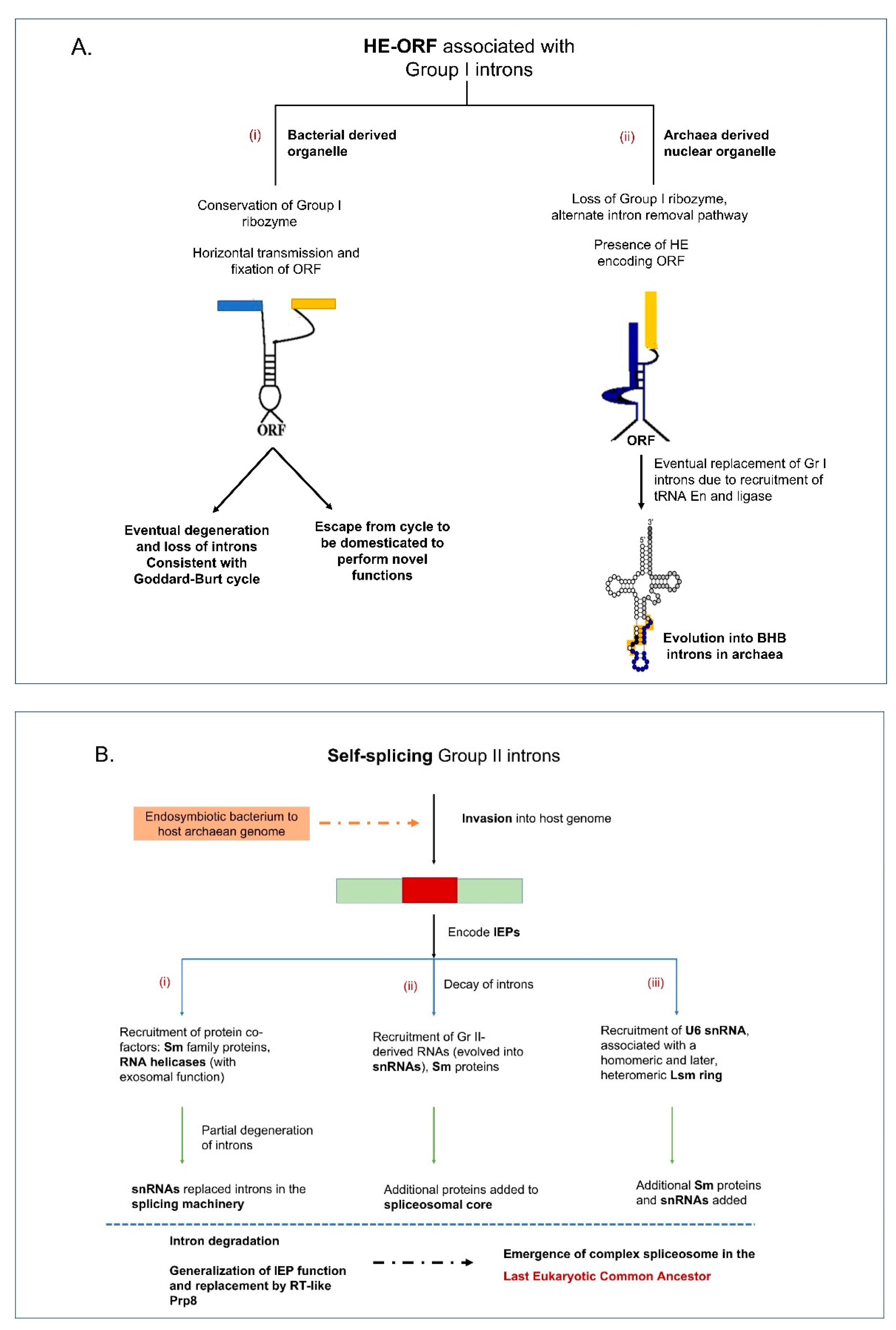
8. Evolutionary Spread of Mobile Organellar Introns across the Three Domains and Organelles
9. Intron Gain and Loss Resulting in Dynamic Organellar Genomes
10. Building Up the Complexity in Introns
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parenteau, J.; Elela, S.A. Introns: Good day junk is bad day treasure. Trends Genet. 2019, 35, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Frumkin, I.; Yofe, I.; Bar-Ziv, R.; Gurvich, Y.; Lu, Y.Y.; Voichek, Y.; Towers, R.; Schirman, D.; Krebber, H.; Pilpel, Y. Evolution of intron splicing towards optimized gene expression is based on various Cis- and Trans-molecular mechanisms. PLoS Bio. 2019, e3000423. [Google Scholar] [CrossRef] [PubMed]
- Zaffagni, M.; Kadener, S. Craving for introns. Mol. Cell 2019, 73, 1095–1096. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Senkevich, T.G.; Dolja, V.V. The ancient virus world and evolution of cells. Biol. Direct 2006, 1, 29. [Google Scholar] [CrossRef]
- Martin, W.; Koonin, E.V. Introns and the origin of nucleus-cytosol compartmentalization. Nature 2006, 440, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.F.; Laforest, M.J.; Burger, G. Mitochondrial introns: A critical view. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef]
- Jo, B.; Choi, S.S. Introns: The functional benefits of introns in genomes. Genom. Inform. 2014, 13, 112–118. [Google Scholar] [CrossRef]
- Dujon, B. Group I introns as mobile genetic elements: Facts and mechanistic speculation-a review. Gene 1989, 82, 91–114. [Google Scholar] [CrossRef]
- Lambowitz, A.M.; Zimmerly, S. Mobile group II introns. Annu Rev. Genet. 2004, 38, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.; Zingler, N. Group II introns: Structure, folding and splicing mechanism. Biol. Chem. 2007, 388, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Anziano, P.Q.; Hanson, D.K.; Mahler, H.R.; Perlman, P.S. Functional domains in introns: Trans-acting and cis-acting regions of intron 4 of the cob gene. Cell 1982, 30, 925–932. [Google Scholar] [CrossRef]
- Lamb, M.R.; Anziano, P.Q.; Glaus, K.R.; Hanson, D.K.; Klappe, H.J.; Perlman, P.S.; Mahler, H.R. Functional domains in introns. RNA processing intermediates in cis- and trans-acting mutants in the penultimate intron of the mitochondrial gene for cytochrome b. J. Biol. Chem. 1983, 258, 1991–1999. [Google Scholar] [CrossRef]
- Copertino, D.W.; Hallick, R.B. Group II twintron: An intron within an intron in a chloroplast cytochrome b-559 gene. EMBO J. 1991, 10, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Copertino, D.W.; Hallic, R.B. Group II and group III introns of twintrons: Potential relationships with nuclear pre-mRNA introns. Trends Biochem. Sci. 1993, 18, 467–471. [Google Scholar] [CrossRef]
- Drager, R.G.; Hallick, R.B. A complex twintron is excised as four individual introns. Nucleic Acids Res. 1993, 21, 2389–2394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hafez, M.; Hausner, G. Convergent evolution of twintron-like configurations: One is never enough. RNA Biol. 2015, 12, 1275–1288. [Google Scholar] [CrossRef]
- Adams, P.L.; Stahley, M.R.; Gill, M.L.; Kosek, A.B.; Wang, J.; Strobel, S.A. Crystal structure of a group I intron splicing intermediate. RNA 2004, 10, 1867–1887. [Google Scholar] [CrossRef]
- Michel, F.; Costa, M.; Westhof, E. The ribozyme core of group II introns: A structure in want of partners. Trends Biochem. Sci. 2009, 34, 189–199. [Google Scholar] [CrossRef] [PubMed]
- van der Horst, G.; Inoue, T. Requirements of a Group I Intron for Reactions at the 3′ Splice Site. J. Mol. Bio. 1993, 229, 685–694. [Google Scholar] [CrossRef]
- Sellem, C.H.; Belcour, L. Intron open reading frames as mobile elements and evolution of a group I intron. Mol. Biol. Evol. 1997, 14, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, A.; Dujon, B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 1985, 41, 383–394. [Google Scholar] [CrossRef]
- Ho, Y.; Kim, S.J.; Waring, R.B. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc. Natl. Acad. Sci. USA 1997, 94, 8994–8999. [Google Scholar] [CrossRef] [PubMed]
- Caprara, M.G.; Waring, R.B. Group I introns and their maturases: Uninvited, but welcome guests. In Homing Endonucleases and Inteins; Belfort, M., Wood, D.W., Stoddard, B.L., Derbyshire, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Matsuura, M.; Saldanha, R.; Ma, H.; Wank, H.; Yang, J.; Mohr, G.; Cavanagh, S.; Dunny, G.M.; Belfort, M.; Lambowitz, A.M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: Biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997, 11, 2910–2924. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Lambowitz, A.M. Group II Intron RNPs and reverse transcriptases: From retroelements to research tools. Cold Spring Harb Perspect Biol. 2019, 11, a032375. [Google Scholar] [CrossRef]
- Burger, G.; Yan, Y.; Javadi, P.; Lang, B.F. Group I-intron trans-splicing and mRNA editing in the mitochondria of placozoan animals. Trends Genet. 2009, 25, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Glanz, S.; Kück, U. Trans-splicing of organelle introns-a detour to continuous RNAs. Bioessays 2009, 31, 921–934. [Google Scholar] [CrossRef]
- Haugen, P.; Simon, D.M.; Bhattacharya, D. The natural history of group I introns. Trends Genet. 2005, 21, 111–119. [Google Scholar] [CrossRef]
- Bonocora, R.P.; Shub, D.A. A likely pathway for formation of mobile group I introns. Curr. Biol. 2009, 19, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Vicens, Q.; Cech, T.R. Atomic level architecture of Group I introns revealed. Trends Biochem. Sci. 2006, 31, 41–51. [Google Scholar] [CrossRef]
- Michel, F.; Westhof, E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990, 216, 585–610. [Google Scholar] [CrossRef]
- Suh, S.O.; Jones, K.G.; Blackwell, M. A group I intron in the nuclear small subunit rRNA gene of Cryptendoxyla hypophloia, an ascomycetous fungus: Evidence for a new major class of Group I introns. J. Mol. Evol. 1999, 48, 493–500. [Google Scholar] [CrossRef]
- Jackson, S.A.; Koduvayur, S.; Woodson, S.A. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA 2006, 12, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, A.; Rosbash, M. Efficient trans-splicing of a yeast mitochondrial RNA group II intron implicates a strong 5′ exon-intron interaction. Science 1986, 234, 1099–1104. [Google Scholar] [CrossRef][Green Version]
- Jacquier, A.; Michel, F. Multiple exon-binding sites in class II self-splicing introns. Cell 1987, 50, 17–29. [Google Scholar] [CrossRef]
- Peebles, C.L.; Perlman, P.S.; Mecklenburg, K.L.; Petrillo, M.L.; Tabor, J.H.; Jarrell, K.A.; Cheng, H.L. A self-splicing RNA excises an intron lariat. Cell 1986, 44, 213–223. [Google Scholar] [CrossRef]
- Peebles, C.L.; Benatan, E.J.; Jarrell, K.A.; Perlman, P.S. Group II intron self-splicing: Development of alternative reaction conditions and identification of a predicted intermediate. Cold Spring Harb Symp Quant. Biol. 1987, 52, 223–232. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, R.; Kwakman, J.H.; Grivell, L.A. Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J. 1987, 6, 3827–3831. [Google Scholar] [CrossRef]
- Daniels, D.L.; Michel, W.J., Jr.; Pyle, A.M. Two competing pathways for self-splicing by group II introns: A quantitative analysis of in vitro reaction rates and products. J. Mol. Biol. 1996, 256, 31–49. [Google Scholar] [CrossRef]
- Pyle, A.M. The tertiary structure of group II introns: Implications for biological function and evolution. Crit Rev. Biochem Mol. Biol. 2010, 45, 215–232. [Google Scholar] [CrossRef]
- Costa, M.; Michel, F.; Westhof, E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000, 19, 5007–5018. [Google Scholar] [CrossRef]
- Michel, F.; Ferat, J.L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995, 64, 435–446. [Google Scholar] [CrossRef]
- Nagy, V.; Pirakitikulr, N.; Zhou, K.I.; Chillón, I.; Luo, J.; Pyle, A.M. Predicted group II intron lineages E and F comprise catalytically active ribozymes. RNA 2013, 19, 1266–1278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smathers, C.M.; Robart, A.R. The mechanism of splicing as told by group II introns: Ancestors of the spliceosome. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2019, 1862, 194390. [Google Scholar] [CrossRef]
- Liu, N.; Dong, X.; Hu, C.; Zeng, J.; Wang, J.; Wang, J.; Wang, H.W.; Belfort, M. Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing. Nucleic Acids Res. 2020, 48, 11185–11198. [Google Scholar] [CrossRef]
- Toor, N.; Hausner, G.; Zimmerly, S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 2001, 7, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Robart, A.R.; Zimmerly, S. Group II intron retroelements: Function and diversity. Cytogenet. Genome Res. 2005, 110, 589–597. [Google Scholar] [CrossRef]
- Dabbagh, N.; Preisfeld, A. The chloroplast genome of Euglena mutabilis —Cluster arrangement, intron analysis, and intrageneric trends. J. Eukaryot. MicroBiol. 2016, 64, 31–44. [Google Scholar] [CrossRef]
- DePriest, P.T.; Been, M.D. Numerous Group I introns with variable distributions in the ribosomal DNA of a lichen fungus. J. Mol. Biol. 1992, 228, 315–321. [Google Scholar] [CrossRef]
- Lasker, B.A.; Smith, G.W.; Kobayashi, G.S.; Whitney, A.M.; Mayer, L.W. Characterization of a single group I intron in the 18S rRNA gene of the pathogenic fungus Histoplasma capsulatum. Med Mycol. 1998, 36, 205–212. [Google Scholar] [CrossRef]
- Beagley, C.T.; Okada, N.A.; Wolstenholme, D.R. Two mitochondrial group I introns in a metazoan, the sea anemone Metridium senile: One intron contains genes for subunits 1 and 3 of NADH dehydrogenase. Proc. Natl. Acad. Sci. USA 1996, 93, 5619–5623. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, C.; Sun, X.; Zhang, R.; Gleason, M.L.; Eiji, T.; Sun, G. Multiple Group I introns in the small-subunit rDNA of Botryosphaeria dothidea: Implication for intraspecific genetic diversity. PLoS ONE 2013, 8, e67808. [Google Scholar]
- Schuster, A.; Lopez, J.V.; Becking, L.E.; Kelly, M.; Pomponi, S.A.; Wörheide, G.; Erpenbeck, D.; Cárdenas, P. Evolution of Group I introns in Porifera: New evidence for intron mobility and implications for DNA barcoding. BMC Evol. Biol. 2017, 17, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Huchon, D.; Szitenberg, A.; Shefer, S.; Ilan, M.; Feldstein, T. Mitochondrial group I and group II introns in the sponge orders Agelasida and Axinellida. BMC Evol. Biol. 2015, 15, 278. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, A.F.; Li, Y.; Smith, C.R.; Halanych, K.M. Multiple introns in a deep-sea Annelid (Decemunciger: Ampharetidae) mitochondrial genome. Sci. Rep. 2017, 7, 4295. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, D.; Köhsler, M.; Venditti, D.; Rott, M.B.; Walochnik, J. Recovery of an acanthamoeba strain with two Group I introns in the nuclear 18S rRNA Gene. Eur. J. Protistol. 2019, 68, 88098. [Google Scholar] [CrossRef]
- Johansen, S.D.; Chi, S.I.; Dubin, A.; Jørgensen, T.E. The mitochondrial genome of the sea anemone Stichodactyla haddoni reveals catalytic introns, insertion-like element, and unexpected phylogeny. Life 2021, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.; Böhm, M.; Grebenjuk, V.A.; Skorokhod, A.; Müller, I.M.; Gamulin, V. Conservation of the positions of metazoan introns from sponges to humans. Gene 2002, 295, 299–309. [Google Scholar] [CrossRef]
- Hausner, G. Introns, mobile elements, and plasmids. In Organelle Genetics; Bullerwell, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar] [CrossRef]
- Corsaro, D.; Venditti, D. Putative Group I introns in the eukaryote nuclear internal transcribed spacers. Curr. Genet. 2020, 66, 373–384. [Google Scholar] [CrossRef]
- Goryunov, D.V.; Sotnikova, E.A.; Goryunova, S.V.; Kuznetsova, O.I.; Logacheva, M.D.; Milyutina, I.A.; Fedorova, A.V.; Fedosov, V.E.; Troitsky, A.V. The mitochondrial genome of nematodontous moss Polytrichum commune and Analysis of intergenic repeats distribution among bryophyta. Diversity 2021, 13, 54. [Google Scholar] [CrossRef]
- Shivji, M.S.; Stanhope, M.J.; Rogers, S.O. Group I introns (“spintrons”) are present in shark nuclear ribosomal DNA internal transcribed spacers. In Proceedings of the American Elasmobranch Society, 13th Annual Meeting, Seattle, WA, USA, 26 June–2 July 1997. [Google Scholar]
- Bonen, L.; Vogel, J. The ins and outs of group II introns. Trends Genet. 2001, 17, 322–331. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Xu, A.; Sagasser, S.; Jakob, W.; Moreno, M.A.; Buss, L.W.; Schierwater, B. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc. Nat. Acad. Sci. USA 2006, 103, 8751–8756. [Google Scholar] [CrossRef]
- Palmer, J.D.; Logsdon, J.M., Jr. The recent origins of introns. Curr Opin Genet. Dev. 1991, 1, 470–477. [Google Scholar] [CrossRef]
- Vallès, Y.; Halanych, K.M.; Boore, J.L. Group II introns break new boundaries: Presence in a bilaterian’s genome. PLoS ONE 2008, 3, e1488. [Google Scholar] [CrossRef]
- Simon, D.M.; Clarke, N.A.; McNeil, B.A.; Johnson, I.; Pantuso, D.; Dai, L.; Chai, D.; Zimmerly, S. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 2008, 14, 1704–1713. [Google Scholar] [CrossRef]
- Bonen, L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 2008, 8, 26–34. [Google Scholar] [CrossRef]
- Brown, G.G.; Colas des Francs-Small, C.; Ostersetzer-Biran, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.A.; Matera, A.G. tRNA introns: Presence, processing, and purpose. Wiley Interdiscip Rev. RNA 2020, 11, e1583. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Jones, T.A.; Eddy, S.R. Group I introns are widespread in archaea. Nucleic Acids Res. 2018, 46, 7970–7976. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wu, X.; Wen, D.; Huang, H.; Chen, Y.; Mukhtar, I.; Yue, L.; Wang, L.; Wen, Z. Intraspecific mitochondrial DNA comparison of mycopathogen Mycogone perniciosa provides insight into mitochondrial transfer RNA introns. Phytopathology 2021, 111, 639–648. [Google Scholar] [CrossRef]
- Stern, D.B.; Goldschmidt-Clermont, M.; Hanson, M.R. Chloroplast RNA metabolism. Annu Rev. Plant Biol. 2010, 61, 125–155. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Koonin, E.V.; Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002, 30, 1427–1464. [Google Scholar] [CrossRef]
- Veretnik, S.; Wills, C.; Youkharibache, P.; Valas, R.E.; Bourne, P.E. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput. Biol. 2009, 5, e1000315. [Google Scholar] [CrossRef] [PubMed]
- Del Hoyo, A.; Álvarez, R.; Gasulla, F.; Casano, L.M.; Del Campo, E.M. Origin and evolution of chloroplast Group I introns in lichen algae. J. Phycol. 2018, 54, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Zimmerly, S.; Hausner, G.; Wu, X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001, 29, 1238–1250. [Google Scholar] [CrossRef][Green Version]
- Lambowitz, A.M.; Zimmerly, S. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011, 3, a003616. [Google Scholar] [CrossRef]
- Kroeger, T.S.; Watkins, K.P.; Friso, G.; van Wijk, K.J.; Barkan, A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 4537–4542. [Google Scholar] [CrossRef]
- Wu, B.; Hao, W. Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns. G3 (Bethesda) 2014, 4, 605–612. [Google Scholar] [CrossRef]
- Chen, C.; Li, Q.; Fu, R.; Wang, J.; Deng, G.; Chen, X.; Lu, D. Comparative mitochondrial genome analysis reveals intron dynamics and gene rearrangements in two Trametes species. Sci. Rep. 2021, 11, 2569. [Google Scholar] [CrossRef]
- Cheng, J.; Luo, Q.; Ren, Y.; Luo, Z.; Liao, W.; Wang, X.; Li, Q. Panorama of intron dynamics and gene rearrangements in the phylum Basidiomycota as revealed by the complete mitochondrial genome of Turbinellus floccosus. Appl Microbiol. Biotechnol 2021, 105, 2017–2032. [Google Scholar] [CrossRef]
- Simpson, C.L.; Stern, D.B. The treasure trove of algal chloroplast genomes: Surprises in architecture and gene content, and their functional implications. Plant Physiol. 2020, 129, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Otis, C.; Lemieux, C. The mitochondrial genome of Chara vulgaris: Insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell 2003, 15, 1888–1903. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, E.V.; Hallick, R.B. Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 2004, 32, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Otis, C.; Lemieux, C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, C.; Otis, C.; Turmel, M. Comparative Chloroplast Genome Analyses of Streptophyte Green Algae Uncover Major Structural Alterations in the Klebsormidiophyceae, Coleochaetophyceae and Zygnematophyceae. Front. Plant Sci. 2016, 7, 697. [Google Scholar] [CrossRef] [PubMed]
- Brouard, J.S.; Turmel, M.; Otis, C.; Lemieux, C. Proliferation of group II introns in the chloroplast genome of the green alga Oedocladium carolinianum (Chlorophyceae). PeerJ 2016, 4, e2627. [Google Scholar] [CrossRef][Green Version]
- Kim, J.I.; Yoon, H.S.; Yi, G.; Shin, W.; Archibald, J.M. Comparative mitochondrial genomics of cryptophyte algae: Gene shuffling and dynamic mobile genetic elements. BMC Genom. 2018, 19, 275. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Lee, H.J.; Lee, J.; Lee, H.O.; Kim, J.H.; Yang, T.J.; Kim, N.S. Plastid genomes of the early vascular plant genus Selaginella have unusual direct repeat structures and drastically reduced gene numbers. Int. J. Mol. Sci. 2021, 22, 641. [Google Scholar] [CrossRef]
- Kwon, E.C.; Kim, J.H.; Kim, N.S. Comprehensive genomic analyses with 115 plastomes from algae to seed plants: Structure, gene contents, GC contents, and introns. Genes Genom. 2020, 42, 553–570. [Google Scholar] [CrossRef]
- Pombert, J.F.; Otis, C.; Lemieux, C.; Turmel, M. The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol. Biol. Evol. 2004, 21, 922–935. [Google Scholar] [CrossRef][Green Version]
- Liu, F.; Melton, J.T. Chloroplast Genomes of the Green-Tide Forming Alga Ulva compressa: Comparative Chloroplast genomics in the genus ulva (Ulvophyceae, Chlorophyta). Front. Mar. Sci. 2021, 8, 387. [Google Scholar] [CrossRef]
- Martínez-Alberola, F.; Barreno, E.; Casano, L.M.; Gasulla, F.; Molins, A.; Del Campo, E.M. Dynamic evolution of mitochondrial genomes in Trebouxiophyceae, including the first completely assembled mtDNA from a lichen-symbiont microalga (Trebouxia sp. TR9). Sci. Rep. 2019, 9, 8209. [Google Scholar] [CrossRef] [PubMed]
- Repetti, S.I.; Jackson, C.J.; Judd, L.M.; Wick, R.R.; Holt, K.E.; Verbruggen, H. The inflated mitochondrial genomes of siphonous green algae reflect processes driving expansion of noncoding DNA and proliferation of introns. PeerJ 2020, 8, e8273. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Verbruggen, H.; Li, T.; Zhu, J.; Chen, Z.; He, H.; Bao, S.; Sun, J. Identification of polycistronic transcriptional units and non-canonical introns in green algal chloroplasts based on long-read RNA sequencing data. BMC Genom. 2021, 22, 298. [Google Scholar] [CrossRef]
- Birgisdottir, A.B.; Johansen, S. Site-specific reverse splicing of a HEG-containing group I intron in ribosomal RNA. Nucleic Acids Res. 2005, 33, 2042–2051. [Google Scholar] [CrossRef] [PubMed]
- Podar, M.; Chu, V.T.; Pyle, A.M.; Perlman, P.S. Group II intron splicing in vivo by first-step hydrolysis. Nature 1998, 391, 915–918. [Google Scholar] [CrossRef] [PubMed]
- de Lencastre, A.; Hamill, S.; Pyle, A.M. A single active-site region for a group II intron. Nat. Struct. Mol. Biol. 2005, 12, 626–627. [Google Scholar] [CrossRef]
- Galej, W.P.; Wilkinson, M.E.; Fica, S.M.; Oubridge, C.; Newman, A.J.; Nagai, K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016, 537, 197–201. [Google Scholar] [CrossRef]
- Robart, A.R.; Chan, R.T.; Peters, J.K.; Rajashankar, K.R.; Toor, N. Crystal structure of a eukaryotic group II intron lariat. Nature 2014, 514, 193–197. [Google Scholar] [CrossRef]
- Zhao, C.; Pyle, A.M. The group II intron maturase: A reverse transcriptase and splicing factor go hand in hand. Curr. Opin. Struct. Biol. 2017, 47, 30–39. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Li, X.; Raabe, C.A.; Cui, D.; Vollmar, J.F.; Rozhdestvensky, T.S.; Skryabin, B.V.; Brosius, J. A universal approach to investigate circRNA protein coding function. Sci. Rep. 2019, 9, 11684. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.I.; Dahl, M.; Emblem, Å.; Johansen, S.D. Giant group I intron in a mitochondrial genome is removed by RNA back-splicing. BMC Mol. Biol. 2019, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Nadimi, M.; Beaudet, D.; Forget, L.; Hijri, M.; Lang, B.F. Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol. Biol. Evol. 2012, 29, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Dolan, G.F.; Müller, U.F. Trans-splicing with the group I intron ribozyme from Azoarcus. RNA 2014, 20, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Kamikawa, R.; Shiratori, T.; Ishida, K.; Miyashita, H.; Roger, A.J. Group II Intron-Mediated Trans-Splicing in the Gene-Rich Mitochondrial Genome of an Enigmatic Eukaryote, Diphylleia rotans. Genome Biol. Evol. 2016, 8, 458–466. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Adams, R.P.; Mower, J.P. Extensive Shifts from Cis- to Trans-splicing of Gymnosperm Mitochondrial Introns. Mol. Biol. Evol. 2020, 37, 1615–1620. [Google Scholar] [CrossRef]
- Mitsuhashi, H.; Homma, S.; Beermann, M.L.; Ishimaru, S.; Takeda, H.; Yu, B.K.; Liu, K.; Duraiswamy, S.; Boyce, F.M.; Miller, J.B. Efficient system for upstream mRNA trans-splicing to generate covalent, head-to-tail, protein multimers. Sci. Rep. 2019, 9, 2274. [Google Scholar] [CrossRef]
- Farré, J.C.; Aknin, C.; Araya, A.; Castandet, B. RNA editing in mitochondrial trans-introns is required for splicing. PLoS ONE 2012, 7, e52644. [Google Scholar] [CrossRef]
- Kück, U.; Schmitt, O. The chloroplast Trans-Splicing RNA-Protein supercomplex from the green alga Chlamydomonas reinhardtii. Cells 2021, 10, 290. [Google Scholar] [CrossRef]
- LaRoche-Johnston, F.; Monat, C.; Coulombe, S.; Cousineau, B. Bacterial group II introns generate genetic diversity by circularization and trans-splicing from a population of intron-invaded mRNAs. PLoS Gen. 2018, 14, e1007792. [Google Scholar] [CrossRef]
- Zlotorynski, E. Intron definition, exon definition and back-splicing revisited. Nat. Rev. Mol. Cell Biol. 2019, 20, 661. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.V.; Mecklenburg, K.L.; Sass, P.; Belcher, S.M.; Mahnke, D.; Lewin, A.; Perlman, P. Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: Initial definition of the maturase domain of the group II intron aI2. Nucleic Acids Res. 1994, 22, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Gramespacher, J.A.; Burton, A.J.; Guerra, L.F.; Muir, T.W. Proximity induced splicing utilizing caged split inteins. J. Am. Chem. Soc. 2019, 141, 13708–13712. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, R.; Minnick, M.F. Group I introns and inteins: Disparate origins but convergent parasitic strategies. J. Bacteriol. 2009, 191, 6193–6202. [Google Scholar] [CrossRef]
- Dujon, B.; Bolotin-Fukuhara, M.; Coen, D.; Deutsch, J.; Netter, P.; Slonimski, P.P.; Weill, L. Mitochondrial genetics. XI. Mutations at the mitochondrial locus omega affecting the recombination of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 1976, 143, 131–165. [Google Scholar] [CrossRef]
- Dujon, B.; Colleaux, L.; Jacquier, A.; Michel, F.; Monteilhet, C. Mitochondrial introns as mobile genetic elements: The role of intron-encoded proteins. Basic Life Sci. 1986, 40, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Colleaux, L.; d’Auriol, L.; Galibert, F.; Dujon, B. Recognition and cleavage site of the intron encoded omega transposase. Proc. Natl. Acad. Sci. USA 1988, 85, 6022–6026. [Google Scholar] [CrossRef]
- Dujon, B. Homing Endonucleases and the Yeast Mitochondrial ω Locus—A Historical Perspective. In Homing Endonucleases and Inteins; Belfort, M., Wood, D.W., Stoddard, B.L., Derbyshire, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 16. [Google Scholar] [CrossRef]
- Edgell, D.R.; Derbyshire, V.; Van Roey, P.; LaBonne, S.; Stanger, M.J.; Li, Z.; Boyd, T.M.; Shub, D.A.; Belfort, M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat. Struct. Mol. Biol. 2004, 11, 936–944. [Google Scholar] [CrossRef]
- Scalley-Kim, M.; McConnell-Smith, A.; Stoddard, B.L. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007, 372, 1305–1319. [Google Scholar] [CrossRef]
- Barzel, A.; Naor, A.; Privman, E.; Kupiec, M.; Gophna, U. Homing endonucleases residing within inteins: Evolutionary puzzles awaiting genetic solutions. Biochem. Soc. Trans. 2011, 39, 169–173. [Google Scholar] [CrossRef]
- Chevalier, B.S.; Stoddard, B.L. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001, 29, 3757–3774. [Google Scholar] [CrossRef]
- Stoddard, B.L. Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob. DNA 2014, 5, 7. [Google Scholar] [CrossRef]
- Fang, X.; Jiang, Y.; Li, K.; Zeng, Q. F-CphI represents a new homing endonuclease family using the Endo VII catalytic motif. Mob. DNA 2018, 9, 27. [Google Scholar] [CrossRef]
- Coughlan, A.Y.; Lombardi, L.; Braun-Galleani, S.; Martos, A.A.; Galeote, V.; Bigey, F.; Dequin, S.; Byrne, K.P.; Wolfe, K.H. The yeast mating-type switching endonuclease HO is a domesticated member of an unorthodox homing genetic element family. eLife 2020, 9, e55336. [Google Scholar] [CrossRef]
- Belfort, M. Two for the price of one: A bifunctional intron-encoded DNA endonuclease-RNA maturase. Genes Dev. 2003, 17, 2860–2863. [Google Scholar] [CrossRef] [PubMed]
- Mota, E.M.; Collins, R.A. Independent evolution of structural and coding regions in a Neurospora mitochondrial intron. Nature 1988, 332, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Megarioti, A.H.; Kouvelis, V.N. The coevolution of fungal mitochondrial introns and their homing endonucleases (giy-yig and laglidadg). Genome Biol. Evolution. 2020, 12, 1337–1354. [Google Scholar] [CrossRef] [PubMed]
- Gimble, F.S. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 2000, 185, 99–107. [Google Scholar] [CrossRef]
- Chevalier, B.; Monnat, R.J.; Stoddard, B.L. The laglidadg homing endonuclease family. In Homing Endonucleases and Inteins; Belfort, M., Wood, D.W., Stoddard, B.L., Derbyshire, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 16. [Google Scholar] [CrossRef]
- Kowalski, J.C.; Belfort, M.; Stapleton, M.A.; Holpert, M.; Dansereau, J.T.; Pietrokovski, S.; Baxter, S.M.; Derbyshire, V. Configuration of the catalytic GIY–YIG domain of intron endonucleases I-Tev I: Coincidence of computational and molecular findings. Nucleic Acids Res. 1999, 27, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L. Homing endonuclease structure and function. Q Rev. Biophys. 2005, 38, 49–95. [Google Scholar] [CrossRef] [PubMed]
- Guha, T.K.; Wai, A.; Mullineux, S.T.; Hausner, G. The intron landscape of the mtDNA cytb gene among the Ascomycota: Introns and intron-encoded open reading frames. Mitochondrial DNA Part A 2018, 29, 1015–1024. [Google Scholar] [CrossRef]
- Edgell, D.R.; Gibb, E.A.; Belfort, M. Mobile DNA elements in T4 and related phages. Virol. J. 2010, 7, 290. [Google Scholar] [CrossRef] [PubMed]
- Brankovics, B.; Kulik, T.; Sawicki, J.; Bilska, K.; Zhang, H.; de Hoog, G.S.; van der Lee, T.A.; Waalwijk, C.; van Diepeningen, A.D. First steps towards mitochondrial pan-genomics: Detailed analysis of Fusarium graminearum mitogenomes. PeerJ 2018, 6, e5963. [Google Scholar] [CrossRef]
- Zubaer, A.; Wai, A.; Hausner, G. The mitochondrial genome of Endoconidiophora resinifera is intron rich. Sci. Rep. 2018, 8, 17591. [Google Scholar] [CrossRef]
- Zubaer, A.; Wai, A.; Patel, N.; Perillo, J.; Hausner, G. The Mitogenomes of Ophiostoma minus and Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales. Front. Microbiol. 2021, 12, 618649. [Google Scholar] [CrossRef] [PubMed]
- Hausner, G.; Hafez, M.; Edgell, D.R. Bacterial group I introns: Mobile RNA catalysts. Mobile DNA 2014, 5, 8. [Google Scholar] [CrossRef]
- Lang, B.F.; O’Kelly, C.; Nerad, T.; Gray, M.W.; Burger, G. The closest unicellular relatives of animals. Curr. Biol. 2002, 12, 1773–1778. [Google Scholar] [CrossRef]
- Mullineux, S.T.; Costa, M.; Bassi, G.S.; Michel, F.; Hausner, G. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA 2010, 16, 1818–1831. [Google Scholar] [CrossRef]
- Pfeifer, A.; Martin, B.; Kämper, J.; Basse, C.W. The mitochondrial LSU rRNA group II intron of Ustilago maydis encodes an active homing endonuclease likely involved in intron mobility. PLoS ONE 2012, 7, e49551. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Li, Z.; Yan, L.; Yu, Y.; Cheng, Y.; Chen, J.; Liu, Y.; Gao, C.; Zeng, L.; Sun, X.; et al. Deletion of the sex-determining gene SXI1α enhances the spread of mitochondrial introns in Cryptococcus neoformans. Mobile DNA 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Kaer, K.; Speek, M. Intronic retroelements. Mobile Gen. El. 2012, 2, 154–157. [Google Scholar] [CrossRef]
- Zimmerly, S.; Guo, H.; Perlman, P.S.; Lambowitz, A.M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 1995, 82, 545–554. [Google Scholar] [CrossRef]
- Zimmerly, S.; Guo, H.; Eskes, R.; Yang, J.; Perlman, P.S.; Lambowitz, A.M. A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell 1995, 83, 529–538. [Google Scholar] [CrossRef]
- Yang, J.; Zimmerly, S.; Perlman, P.S.; Lambowitz, A.M. Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature 1996, 381, 332–335. [Google Scholar] [CrossRef]
- Cousineau, B.; Smith, D.; Lawrence-Cavanagh, S.; Mueller, J.E.; Yang, J.; Mills, D.; Manias, D.; Dunny, G.; Lambowitz, A.M.; Belfort, M. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 1998, 94, 451–462. [Google Scholar] [CrossRef]
- Zhang, F.; Mastroianni, M.; White, T.B.; Lambowitz, A.M. Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation. Proc. Natl. Acad. Sci. USA 2009, 106, 18189–18194. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Zhong, J.; Matsuura, M.; Lambowitz, A.M.; Belfort, M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev. 2005, 19, 2477–2487. [Google Scholar] [CrossRef]
- Coros, C.J.; Piazza, C.L.; Chalamcharla, V.R.; Smith, D.; Belfort, M. Global regulators orchestrate group II intron retromobility. Mol. Cell 2009, 34, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Contreras, L.M.; Huang, T.; Piazza, C.L.; Smith, D.; Qu, G.; Gelderman, G.; Potratz, J.P.; Russell, R.; Belfort, M. Group II intron-ribosome association protects intron RNA from degradation. RNA 2013, 19, 1497–1509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- García-Rodríguez, F.M.; Neira, J.L.; Marcia, M.; Molina-Sánchez, M.D.; Toro, N. A group II intron-encoded protein interacts with the cellular replicative machinery through the β-sliding clamp. Nucleic Acids Res. 2019, 47, 7605–7617. [Google Scholar] [CrossRef]
- Qu, G.; Kaushal, P.S.; Wang, J.; Shigematsu, H.; Piazza, C.L.; Agrawal, R.K.; Belfort, M.; Wang, H.W. Structure of a group II intron in complex with its reverse transcriptase. Nat. Struct. Mol. Biol. 2016, 23, 549–557. [Google Scholar] [CrossRef]
- Novikova, O.; Belfort, M. Mobile Group II Introns as ancestral eukaryotic elements. Trends Genet. 2017, 33, 773–783. [Google Scholar] [CrossRef]
- Chan, R.T.; Peters, J.K.; Robart, A.R.; Wiryaman, T.; Rajashankar, K.R.; Toor, N. Structural basis for the second step of group II intron splicing. Nat. Commun. 2018, 9, 4676. [Google Scholar] [CrossRef] [PubMed]
- Haack, D.B.; Toor, N. Retroelement origins of pre-mRNA splicing. Wiley Interdiscip Rev. RNA. 2020, 11, e1589. [Google Scholar] [CrossRef]
- Gupta, K.; Contreras, L.M.; Smith, D.; Qu, G.; Huang, T.; Spruce, L.A.; Seeholzer, S.H.; Belfort, M.; Van Duyne, G.D. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res. 2014, 42, 5347–5360. [Google Scholar] [CrossRef]
- Vosseberg, J.; Snel, B. Domestication of self-splicing introns during eukaryogenesis: The rise of the complex spliceosomal machinery. Biol. Direct. 2017, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Galej, W.P.; Oubridge, C.; Newman, A.J.; Nagai, K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 2013, 493, 638–643. [Google Scholar] [CrossRef]
- Dlakić, M.; Mushegian, A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 2011, 17, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Mohr, G.; Lambowitz, A.M. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res. 2003, 31, 647–652. [Google Scholar] [CrossRef]
- Malik, S.; Upadhyaya, K.C.; Khurana, S.P. Phylogenetic Analysis of Nuclear-Encoded RNA Maturases. Evol. Bioinform. Online 2017, 13, 1176934317710945. [Google Scholar] [CrossRef] [PubMed]
- Hausner, G.; Olson, R.; Simon, D.; Johnson, I.; Sanders, E.R.; Karol, K.G.; McCourt, R.M.; Zimmerly, S. Origin and evolution of the chloroplast trnK (matK) intron: A model for evolution of group II intron RNA structures. Mol. Biol. Evol. 2006, 23, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Lazowska, J.; Jacq, C.; Slonimski, P.P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell 1980, 22, 333–348. [Google Scholar] [CrossRef]
- Lambowitz, A.M.; Caprara, M.G.; Zimmerly, S.; Perlman, P.S. Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In The RNA World; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1999; pp. 451–485. [Google Scholar]
- Contamine, V.; Picard, M. Maintenance and integrity of the mitochondrial genome: A plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol Rev. 2000, 64, 281–315. [Google Scholar] [CrossRef]
- Wenzlau, J.M.; Saldanha, R.J.; Butow, R.A.; Perlman, P.S. A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell 1989, 56, 421–430. [Google Scholar] [CrossRef]
- Bolduc, J.M.; Spiegel, P.C.; Chatterjee, P.; Brady, K.L.; Downing, M.E.; Caprara, M.G.; Waring, R.B.; Stoddard, B.L. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003, 17, 2875–2888. [Google Scholar] [CrossRef]
- Bertrand, H.; Bridge, P.; Collins, R.A.; Garriga, G.; Lambowitz, A.M. RNA splicing in Neurospora mitochondria. Characterization of new nuclear mutants with defects in splicing the mitochondrial large rRNA. Cell 1982, 29, 517–526. [Google Scholar] [PubMed]
- Dujardin, G.; Pajot, P.; Groudinsky, O.; Slonimski, P.P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol. Gen. Genet. 1980, 179, 469–482. [Google Scholar] [CrossRef]
- Garriga, G.; Bertrand, H.; Lambowitz, A.M. RNA splicing in Neurospora mitochondria: Nuclear mutants defective in both splicing and 3′ end synthesis of the large rRNA. Cell 1984, 36, 623–634. [Google Scholar] [CrossRef]
- Mohr, G.; Rennard, R.; Cherniack, A.D.; Stryker, J.; Lambowitz, A.M. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J. Mol. Biol. 2001, 307, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Stryker, J.M.; Lambowitz, A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 2002, 109, 769–779. [Google Scholar] [CrossRef]
- Huang, H.R.; Rowe, C.E.; Mohr, S.; Jiang, Y.; Lambowitz, A.M.; Perlman, P.S. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. USA 2005, 102, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Jarmoskaite, I.; Bhaskaran, H.; Seifert, S.; Russell, R. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc. Natl. Acad. Sci. USA 2014, 111, E2928–E2936. [Google Scholar] [CrossRef] [PubMed]
- Halls, C.; Mohr, S.; Del Campo, M.; Yang, Q.; Jankowsky, E.; Lambowitz, A.M. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007, 365, 835–855. [Google Scholar] [CrossRef]
- Dobinson, K.F.; Henderson, M.; Kelley, R.L.; Collins, R.A.; Lambowitz, A.M. Mutations in nuclear gene cyt-4 of Neurospora crassa result in pleiotropic defects in processing and splicing of mitochondrial RNAs. Genetics 1989, 123, 97–108. [Google Scholar] [CrossRef]
- Turcq, B.; Dobinson, K.F.; Serizawa, N.; Lambowitz, A.M. A protein required for RNA processing and splicing in Neurospora mitochondria is related to gene products involved in cell cycle protein phosphatase functions. Proc. Natl. Acad. Sci. USA 1992, 89, 1676–1680. [Google Scholar] [CrossRef]
- Labouesse, M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990, 224, 209–221. [Google Scholar] [CrossRef]
- Dmochowska, A.; Golik, P.; Stepien, P.P. The novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr. Genet. 1995, 28, 108–112. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.; Poliquin, S.; Zeng, R.; Zamudio-Ochoa, A.; Marrero, N.; Perez-Martinez, X.; Fontanesi, F.; Barrientos, A. The DEAD-box helicase Mss116 plays distinct roles in mitochondrial ribogenesis and mRNA-specific translation. Nucleic Acids Res. 2017, 45, 6628–6643. [Google Scholar] [CrossRef]
- Zimmerly, S.; Semper, C. Evolution of group II introns. Mob. DNA 2015, 6, 7. [Google Scholar] [CrossRef]
- Zoschke, R.; Nakamura, M.; Liere, K.; Sugiura, M.; Börner, T.; Schmitz-Linneweber, C. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 2010, 107, 3245–3250. [Google Scholar] [CrossRef]
- Barthet, M.M.; Pierpont, C.L.; Tavernier, E.K. Unraveling the role of the enigmatic MatK maturase in chloroplast group IIA intron excision. Plant Direct. 2020, 4, e00208. [Google Scholar] [CrossRef]
- Shevtsov-Tal, S.; Best, C.; Matan, R.; Chandran, S.A.; Brown, G.G.; Ostersetzer-Biran, O. nMAT3 is an essential maturase splicing factor required for holo-complex I biogenesis and embryo development in Arabidopsis thaliana plants. Plant J. 2021, 106, 1128–1147. [Google Scholar] [CrossRef]
- de Longevialle, A.F.; Hendrickson, L.; Taylor, N.L.; Delannoy, E.; Lurin, C.; Badger, M.; Millar, A.H.; Small, I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008, 56, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhu, H.; Xing, Y.; Tan, J.; Chen, X.; Zhang, J.; Peng, H.; Xie, Q.; Zhang, Z. Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 2016, 67, 5339–5347. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Fan, K.; Fang, T.; Zhang, J.; Yang, L.; Wang, J.; Wang, G.; Liu, Y. Maize Empty Pericarp602 Encodes a P-Type PPR Protein That Is Essential for Seed Development. Plant Cell Physiol. 2019, 60, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Li, S.; Sun, F.; Sun, Q.; Zhao, H.; Ren, X.; Zhao, Y.; Tan, B.C.; Zhang, Z.; Qiu, F. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 2017, 91, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Front. Plant Sci. 2021, 12, 1. [Google Scholar] [CrossRef]
- Bobik, K.; McCray, T.N.; Ernest, B.; Fernandez, J.C.; Howell, K.A.; Lane, T.; Staton, M.; Burch-Smith, T.M. The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. Plant J. 2017, 91, 114–131. [Google Scholar] [CrossRef]
- Lee, K.; Park, S.J.; Han, J.H.; Jeon, Y.; Pai, H.S.; Kang, H. A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development. BMC Plant Biol. 2019, 19, 244. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, X.; Shen, Y.; Wang, H.; Liu, R.; Wang, X.; Gao, D.; Yang, Y.Z.; Liu, Y.; Tan, B.C. The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. Plant J. 2018, 95, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Pan, Z.; Zhao, H.; Zhao, J.; Cai, M.; Li, J.; Zhang, Z.; Qiu, F. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J. Exp. Bot. 2017, 68, 4571–4581. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.R.; Hallinan, J.P.; Shen, B.W.; Chik, J.K.; Bolduc, J.M.; Kulshina, N.; Robins, L.I.; Kaiser, B.K.; Jarjour, J.; Havens, K.; et al. Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity. Structure 2016, 24, 862–873. [Google Scholar] [CrossRef]
- Gogarten, J.P.; Hilario, E. Inteins, introns, and homing endonucleases: Recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 2006, 6, 94. [Google Scholar] [CrossRef]
- Mullineux, S.T.; Willows, K.; Hausner, G. Evolutionary dynamics of the mS952 intron: A novel mitochondrial group II intron encoding a LAGLIDADG homing endonuclease gene. J. Mol. Evol. 2011, 72, 433–449. [Google Scholar] [CrossRef]
- Goddard, M.R.; Burt, A. Recurrent invasion and extinction of a selfish gene. Proc. Natl. Acad. Sci. USA 1999, 96, 13880–13885. [Google Scholar] [CrossRef]
- Edgell, D.R.; Chalamcharla, V.R.; Belfort, M. Learning to live together: Mutualism between self-splicing introns and their hosts. BMC Biol. 2011, 9, 22. [Google Scholar] [CrossRef]
- Zubaer, A.; Wai, A.; Hausner, G. The fungal mitochondrial Nad5 pan-genic intron landscape. Mitochondrial DNA Part A 2019, 30, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Wai, A.; Shen, C.; Carta, A.; Dansen, A.; Crous, P.W.; Hausner, G. Intron-encoded ribosomal proteins and N-acetyltransferases within the mitochondrial genomes of fungi: Here today, gone tomorrow? Mitochondrial DNA Part A 2019, 30, 573–584. [Google Scholar] [CrossRef]
- Cinget, B.; Bélanger, R.R. Discovery of new group I-D introns leads to creation of subtypes and link to an adaptive response of the mitochondrial genome in fungi. RNA Biol. 2020, 17, 1252–1260. [Google Scholar] [CrossRef]
- Grasso, V.; Palermo, S.; Sierotzki, H.; Garibaldi, A.; Gisi, U. Cytochrome b gene structure and consequences tor resistance to Qo inhibitor fungicides in plant pathogens. Pest. Manag. Sci. 2006, 62, 465–472. [Google Scholar] [CrossRef]
- Baidyaroy, D.; Hausner, G.; Hafez, M.; Michel, F.; Fulbright, D.W.; Bertrand, H. A 971-bp insertion in the rns gene is associated with mitochondrial hypovirulence in a strain of Cryphonectria parasitica isolated from nature. Fungal Genet. Biol. 2011, 48, 775–783. [Google Scholar] [CrossRef]
- Rose, A.B. Introns as Gene Regulators: A brick on the accelerator. Front. Genet. 2019, 9, 672. [Google Scholar] [CrossRef]
- Rudan, M.; Bou Dib, P.; Musa, M.; Kanunnikau, M.; Sobočanec, S.; Rueda, D.; Warnecke, T.; Kriško, A. Normal mitochondrial function in Saccharomyces cerevisiae has become dependent on inefficient splicing. eLife 2018, 7, e35330. [Google Scholar] [CrossRef]
- Johansen, S.; Emblem, Å. Mitochondrial Group I introns in hexacorals are regulatory genetic elements. In Advances in the Study of Benthic Zone; Soto, L.A., Ed.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Belfort, M. Mobile self-splicing introns and inteins as environmental sensors. Curr. Opin. Microbiol. 2017, 38, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Qu, G.; Piazza, C.L.; Belfort, M. Group II intron as cold sensor for self-preservation and bacterial conjugation. Nucleic Acids Res. 2020, 48, 6198–6209. [Google Scholar] [CrossRef]
- Lee, E.R.; Baker, J.L.; Weinberg, Z.; Sudarsan, N.; Breaker, R.R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 2010, 329, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.M.; Sidote, D.J.; Russell, R.; Lambowitz, A.M. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core. Proc. Natl. Acad. Sci. USA 2013, 110, E3800–E3809. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Random drift and the shifting balance theory of evolution. Biomathematics 1970, 1. [Google Scholar] [CrossRef]
- Lynch, M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. USA 2007, 104, 8597–8604. [Google Scholar] [CrossRef]
- Lynch, M.; Ackerman, M.; Gout, J.F.; Long, H.; Sung, W.; Thomas, W.K.; Foster, P.L. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 2016, 17, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.; Ackerman, M.S.; Miller, S.F.; Doak, T.G.; Lynch, M. The drift-barrier hypothesis and mutation-rate evolution. Proc. Natl Acad. Sci. USA 2012, 109, 18488–18492. [Google Scholar] [CrossRef]
- Koonin, E.V. Splendor and misery of adaptation, or the importance of neutral null for understanding evolution. BMC Biol. 2016, 14, 114. [Google Scholar] [CrossRef]
- Grainger, R.J.; Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA 2005, 11, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Ruck, E.C.; Linard, S.R.; Nakov, T.; Theriot, E.C.; Alverson, A.J. Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr. Genet. 2017, 63, 499–507. [Google Scholar] [CrossRef]
- Ruck, E.C.; Nakov, T.; Jansen, R.K.; Theriot, E.C.; Alverson, A.J. Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 2014, 6, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, P.; Moreira, D. Reevaluating the green contribution to diatom genomes. Genome Biol. Evol. 2012, 4, 683–688. [Google Scholar] [CrossRef]
- Kamikawa, R.; Masuda, I.; Demura, M.; Oyama, K.; Yoshimatsu, S.; Kawachi, M.; Sako, Y. Mitochondrial group II introns in the raphidophycean flagellate Chattonella spp. suggest a diatom-to-Chattonella lateral group II intron transfer. Protist 2009, 160, 364–375. [Google Scholar] [CrossRef]
- Medlin, L.K. Evolution of the diatoms: Major steps in their evolution and a review of the supporting molecular and morphological evidence. Phycologia 2016, 55, 79–103. [Google Scholar] [CrossRef]
- Parks, M.B.; Wickett, N.J.; Alverson, A.J. Signal, uncertainty, and conflict in phylogenomic data for a diverse lineage of microbial eukaryotes (Diatoms, Bacillariophyta). Mol. Biol. Evol. 2018, 35, 80–93. [Google Scholar] [CrossRef]
- Guillory, W.X.; Onyshchenko, A.; Ruck, E.C.; Parks, M.; Nakov, T.; Wickett, N.J.; Alverson, A.J. Recurrent Loss, Horizontal Transfer, and the Obscure Origins of Mitochondrial Introns in Diatoms (Bacillariophyta). Genome Biol. Evol. 2018, 10, 1504–1515. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. The puzzle of plastid evolution. Curr. Biology. 2009, 19, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Woehle, C.; Dagan, T.; Martin, W.F.; Gould, S.B. Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol. Evol. 2011, 3, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Beszteri, B.; Maier, U.G.; Bowler, C.; Valentin, K.; Bhattacharya, D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 2009, 324, 1724–1726. [Google Scholar] [CrossRef] [PubMed]
- Brembu, T.; Winge, P.; Tooming-Klunderud, A.; Nederbragt, A.J.; Jakobsen, K.S.; Bones, A.M. The chloroplast genome of the diatom Seminavis robusta: New features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genom. 2014, 16, 17–27. [Google Scholar] [CrossRef]
- Nishimura, Y.; Shiratori, T.; Ishida, K.I.; Hashimoto, T.; Ohkuma, M.; Inagaki, Y. Horizontally-acquired genetic elements in the mitochondrial genome of a centrohelid Marophrys sp. SRT127. Sci. Rep. 2019, 9, 4850. [Google Scholar] [CrossRef]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Abbona, C.C.; Zhuo, C.; Tepe, E.J.; Bohs, L.; Olmstead, R.G.; Palmer, J.D. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended co-conversion of flanking exons. BMC Evol. Biol. 2011, 11, 277. [Google Scholar] [CrossRef]
- Mower, J.P.; Stefanović, S.; Hao, W.; Gummow, J.S.; Jain, K.; Ahmed, D.; Palmer, J.D. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010, 8, 150. [Google Scholar] [CrossRef]
- Hao, W.; Richardson, A.O.; Zheng, Y.; Palmer, J.D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA 2010, 107, 21576–21581. [Google Scholar] [CrossRef] [PubMed]
- Filip, E.; Skuza, L. Horizontal Gene Transfer Involving Chloroplasts. Int. J. Mol. Sci. 2021, 22, 4484. [Google Scholar] [CrossRef]
- Vaughn, J.C.; Mason, M.T.; Sper-Witis, G.L.; Kuhlman, P.; Palmer, J.D. Fungal Origin by Horizontal Transfer of a Plant Mitochondrial Group I Intron in the Chimeric Coxl Gene of Peperomia. J. Mol. Evol. 1995, 41, 563–572. [Google Scholar] [CrossRef]
- Sinn, B.T.; Barrett, C.G. Ancient Mitochondrial Gene Transfer between Fungi and the Orchids. Mol. Biol. Evol. 2020, 37, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, X.; Xie, B.; Lin, L.; Hsiang, T.; Lin, X.; Lin, Y.; Zhang, X.; Ma, Y.; Miao, W.; et al. Intra-specific comparison of mitochondrial genomes reveals host gene fragment exchange via intron mobility in Tremella fuciformis. BMC Genom. 2020, 21, 426. [Google Scholar] [CrossRef]
- Rot, C.; Goldfarb, I.; Ilan, M.; Huchon, D. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol. Biol. 2006, 6, 71. [Google Scholar] [CrossRef]
- Ye, J.; Cheng, J.; Ren, Y.; Liao, W.; Li, Q. The First Mitochondrial Genome for Geastrales (Sphaerobolus stellatus) Reveals Intron Dynamics and Large-Scale Gene Rearrangements of Basidiomycota. Front. Microbiol. 2020, 11, 1970. [Google Scholar] [CrossRef]
- Li, Q.; Wu, P.; Li, L.; Feng, H.; Tu, W.; Bao, Z.; Xiong, C.; Gui, M.; Huang, W. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int. J. Biol Macromol. 2021, 172, 560–572. [Google Scholar] [CrossRef]
- Cuenca, A.; Ross, T.G.; Graham, S.W.; Barrett, C.F.; Davis, J.I.; Seberg, O.; Petersen, G. Localized Retroprocessing as a Model of Intron Loss in the Plant Mitochondrial Genome. Genome Biol. Evol. 2016, 8, 2176–2189. [Google Scholar] [CrossRef] [PubMed]
- Hepburn, N.J.; Schmidt, D.W.; Mower, J.P. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol. Biol. Evol. 2012, 29, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Pogoda, C.S.; Keepers, K.G.; Nadiadi, A.Y.; Bailey, D.W.; Lendemer, J.C.; Tripp, E.A.; Kane, N.C. Genome streamlining via complete loss of introns has occurred multiple times in lichenized fungal mitochondria. Ecol Evol. 2019, 9, 4245–4263. [Google Scholar] [CrossRef]
- Beaudet, D.; Nadimi, M.; Iffis, B.; Hijri, M. Rapid Mitochondrial Genome Evolution through Invasion of Mobile Elements in Two Closely Related Species of Arbuscular Mycorrhizal Fungi. PLoS ONE 2013, 8, e60768. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.; Kitahara, M.; Fukami, H.; Tracey, D.; Miller, D.J.; Chen, C.A. Loss and Gain of Group I introns in the Mitochondrial Cox1 Gene of the Scleractinia (Cnidaria; Anthozoa). Zool Stud. 2017, 56, e9. [Google Scholar] [CrossRef]
- Suzuki, H.; Kameyama, T.; Ohe, K.; Tsukahara, T.; Mayeda, A. Nested introns in an intron: Evidence of multi-step splicing in a large intron of the human dystrophin pre-mRNA. FEBS Lett. 2013, 587, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, Q.; Ming, R.; Lin, L.; Lin, X.; Lin, Y.; Li, X.; Xie, B.; Wen, Z. Analysis of the mitochondrial genome in Hypomyces aurantius reveals a novel twintron complex in fungi. Int. J. Mol. Sci. 2016, 17, 1049. [Google Scholar] [CrossRef] [PubMed]
- Hafez, M.; Majer, A.; Sethuraman, J.; Rudski, S.M.; Michel, F.; Hausner, G. The mtDNA rns gene landscape in the Ophiostomatales and other fungal taxa: Twintrons, introns, and intron-encoded proteins. Fungal Genet. Biol. 2013, 53, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Guha, T.K.; Hausner, G. Using group II introns for attenuating the in vitro and in vivo expression of a homing endonuclease. PLoS ONE 2016, 11, e0150097. [Google Scholar] [CrossRef]
- Guha, T.K.; Wai, A.; Hausner, G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput. Struct. Biotechnol. J. 2017, 15, 146–160. [Google Scholar] [CrossRef]
- Zumkeller, S.; Philipp Gerke, P.; Knoop, V. A functional twintron, ‘zombie’ twintrons and a hypermobile group II intron invading itself in plant mitochondria. Nucleic Acids Res. 2020, 48, 2661–2675. [Google Scholar] [CrossRef]
- Takahara, K.; Schwarze, U.; Imamura, Y.; Hoffman, G.G.; Toriello, H.; Smith, L.T.; Byers, P.H.; Greenspan, D.S. Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a col5a1 acceptor-site mutation that results in abnormal pro-α1(v) n-propeptides and ehlers-danlos syndrome type I. AJHJ 2002, 71, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hsiang, T.; Li, S.; Lin, L.; Wang, Q.; Chen, Q.; Xie, B.; Ming, R. Comparison of the mitochondrial genome sequences of six annulohypoxylon stygium isolates suggests short fragment insertions as a potential factor leading to larger genomic size. Front. Microbiol. 2018, 10, 2079. [Google Scholar] [CrossRef] [PubMed]
- Pombert, J.F.; James, E.R.; Janouškovec, J.; Keeling, P.J. Evidence for Transitional stages in the evolution of euglenid Group II introns and twintrons in the Monomorphina aenigmatica plastid genome. PLoS ONE 2012, 7, e53433. [Google Scholar] [CrossRef] [PubMed]
- Pfreundt, U.; Hess, W.R. Sequential splicing of a group II twintron in the marine cyanobacterium Trichodesmium. Sci. Rep. 2015, 5, 16829. [Google Scholar] [CrossRef]
- Guha, T.K.; Hausner, G. A homing endonuclease with a switch: Characterization of a twintron encoded homing endonuclease. Fungal Genet. Biol. 2014, 65, 57–68. [Google Scholar] [CrossRef]
- Chen, S.Y.; Li, C.; Jia, X.; Lai, S.J. Sequence and evolutionary features for the alternatively spliced exons of eukaryotic genes. Int. J. Mol. Sci. 2019, 20, 3834. [Google Scholar] [CrossRef]
- Rudski, S.M.; Hausner, G. The mtDNA rps3 locus has been invaded by a group I intron in some species of Grosmannia. Mycoscience 2012, 53, 471–475. [Google Scholar] [CrossRef]
- Hong, L.; Hallick, R.B. A group III intron is formed from domains of two individual group II introns. Genes Dev. 1994, 8, 1589–1599. [Google Scholar] [CrossRef]
- Maier, U.G.; Rensing, S.A.; Igloi, G.L.; Maerz, M. Twintrons are not unique to the Euglena chloroplast genome: Structure and evolution of a plastome cpn60 gene from a cryptomonad. Mol. Gen. Genet. 1995, 246, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Archibald, J.M. Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res. 2008, 36, 3043–3053. [Google Scholar] [CrossRef]
- Perrineau, M.M.; Price, D.C.; Mohr, G.; Bhattacharya, D. Recent mobility of plastid encoded group II introns and twintrons in five strains of the unicellular red alga Porphyridium. PeerJ 2015, 3, e1017. [Google Scholar] [CrossRef]
- Medina, R.; Franco, M.E.E.; Bartel, L.C.; Martinez Alcántara, V.; Saparrat, M.C.N.; Balatti, P.A. Fungal mitogenomes: Relevant features to planning plant disease management. Front. Microbiol. 2020, 11, 978. [Google Scholar] [CrossRef]
- Gomes, F.E.E.S.; Arantes, T.D.; Fernandes, J.A.L.; Ferreira, L.C.; Romero, H.; Bosco, S.M.G.; Oliveira, M.T.B.; Del Negro, G.M.B.; Theodoro, R.C. Polymorphism in mitochondrial Group I introns among Cryptococcus neoformans and Cryptococcus gattii Genotypes and its association with drug susceptibility. Front. Microbiol. 2018, 9, 86. [Google Scholar] [CrossRef]
- Lee, C.H.; Han, S.R.; Lee, S.W. Group I intron-based therapeutics through trans-splicing reaction. Prog. Mol. Biol Transl. Sci. 2018, 159, 79–100. [Google Scholar] [CrossRef]
- Jayaguru, P.; Raghunathan, M. Group I intron renders differential susceptibility of Candida albicans to bleomycin. Mol. Biol Rep. 2007, 34, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.M. RNA as a drug target: Recent patents on the catalytic activity of trans-splicing ribozymes derived from group I intron RNA. Recent Pat. DNA Gene Seq. 2010, 4, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.; Jagdmann, G.E., Jr.; Adams, R.L.; Yuan, L.; Van Zandt, M.C.; Pyle, A.M. Small molecules that target group II introns are potent antifungal agents. Nat. Chem Biol. 2018, 14, 1073–1078. [Google Scholar] [CrossRef]
- Cui, X.; Davis, G. Mobile group II intron targeting: Applications in prokaryotes and perspectives in eukaryotes. Front. Biosci. 2007, 12, 4972–4985. [Google Scholar] [CrossRef]
- Enyeart, P.J.; Mohr, G.; Ellington, A.D.; Lambowitz, A.M. Biotechnological applications of mobile group II introns and their reverse transcriptases: Gene targeting, RNA-seq, and non-coding RNA analysis. Mob. DNA 2014, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, R.; Lambert, A.R.; Mak, A.N.; Jacoby, K.; Dickson, R.J.; Gloor, G.B.; Scharenberg, A.M.; Edgell, D.R.; Stoddard, B.L. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. USA 2011, 108, 13077–13082. [Google Scholar] [CrossRef] [PubMed]
- Hafez, M.; Hausner, G. Homing endonucleases: DNA scissors on a mission. Genome 2012, 55, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, A. On the possibility of constructive neutral evolution. J. Mol. Evol. 1999, 49, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Archibald, J.M.; Keeling, P.J.; Doolittle, W.F.; Gray, M.W. How a neutral evolutionary ratchet can build cellular complexity? IUBMB Life 2011, 63, 528–537. [Google Scholar] [CrossRef] [PubMed]

| Twintron Category | Kingdom | Organism | Host Gene | Subcellular Location | Reference |
|---|---|---|---|---|---|
| GI/GI | Fungi | Cryphonectria parasitica | rns | Mitochondria | [254] |
| GI/GI | Fungi | Annulohypoxylon stygium | nad5, cob, cox1 | Mitochondria | [259] |
| GI/GI | Fungi | Mycogone perniciosa | nad1 | Mitochondria | [72] |
| GI/GI 1 | Fungi | Hypomyces aurantius | cox1 | Mitochondria | [253] |
| GI/GI 1 | Fungi | Endoconidiophora resinifera | rns | Mitochondria | [141] |
| GI/GII | Fungi | Chaetomium thermophilum | rns | Mitochondria | [254] |
| GI/GII | Fungi | Grosmannia piceiperda | rnl | Mitochondria | [264] |
| GI/GII | Fungi | Chaetomium thermophilum | ms1247 (position) | Mitochondria | [262] |
| GI/GII | Fungi | Ascomycota: multiple | cytb | Mitochondria | [138] |
| GI/GII 2 | Plant | Lycopodiaceae: multiple | cox1 | Mitochondria | [257] |
| GI/GII 3 | Fungi | Ophiostoma ips | cox3, cob | Mitochondria | [142] |
| GII/GII | Algae | Euglena gracilis | psbF | Chloroplast | [13] |
| GII/GII | Algae | Euglena gracilis | ycf8 | Chloroplast | [265] |
| GII/GII | Algae | Pyrenomonas salina | cpn60 | Chloroplast | [266] |
| GII/GII | Algae | Rhodomonas sp. | groEL | Chloroplast | [267] |
| GII/GII | Algae | Porphyridium purpureum | atpI, rpoC | Plastid | [268] |
| GII/GII | Algae | Eutreptiella pomquetensis | rpoB, psbD, petG | Plastid | [48] |
| GIII/GIII | Algae | Euglena gracilis | rps18 | Chloroplast | [15] |
| GIII/GII | Algae | Euglena gracilis | rps3 | Chloroplast | [13] |
| GII/GIII, GII/GII, GIII/GIII | Algae | Euglena gracilis, Monomorphina aenigmatica | psb, rpoC1, rps3 | Plastid | [260] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukhopadhyay, J.; Hausner, G. Organellar Introns in Fungi, Algae, and Plants. Cells 2021, 10, 2001. https://doi.org/10.3390/cells10082001
Mukhopadhyay J, Hausner G. Organellar Introns in Fungi, Algae, and Plants. Cells. 2021; 10(8):2001. https://doi.org/10.3390/cells10082001
Chicago/Turabian StyleMukhopadhyay, Jigeesha, and Georg Hausner. 2021. "Organellar Introns in Fungi, Algae, and Plants" Cells 10, no. 8: 2001. https://doi.org/10.3390/cells10082001
APA StyleMukhopadhyay, J., & Hausner, G. (2021). Organellar Introns in Fungi, Algae, and Plants. Cells, 10(8), 2001. https://doi.org/10.3390/cells10082001






