The Genes Encoding Small Leucine-Rich Proteoglycans Undergo Differential Expression Alterations in Colorectal Cancer, Depending on Tumor Location
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Total RNA Isolation and cDNA Synthesis
2.3. qRT-PCR Reactions
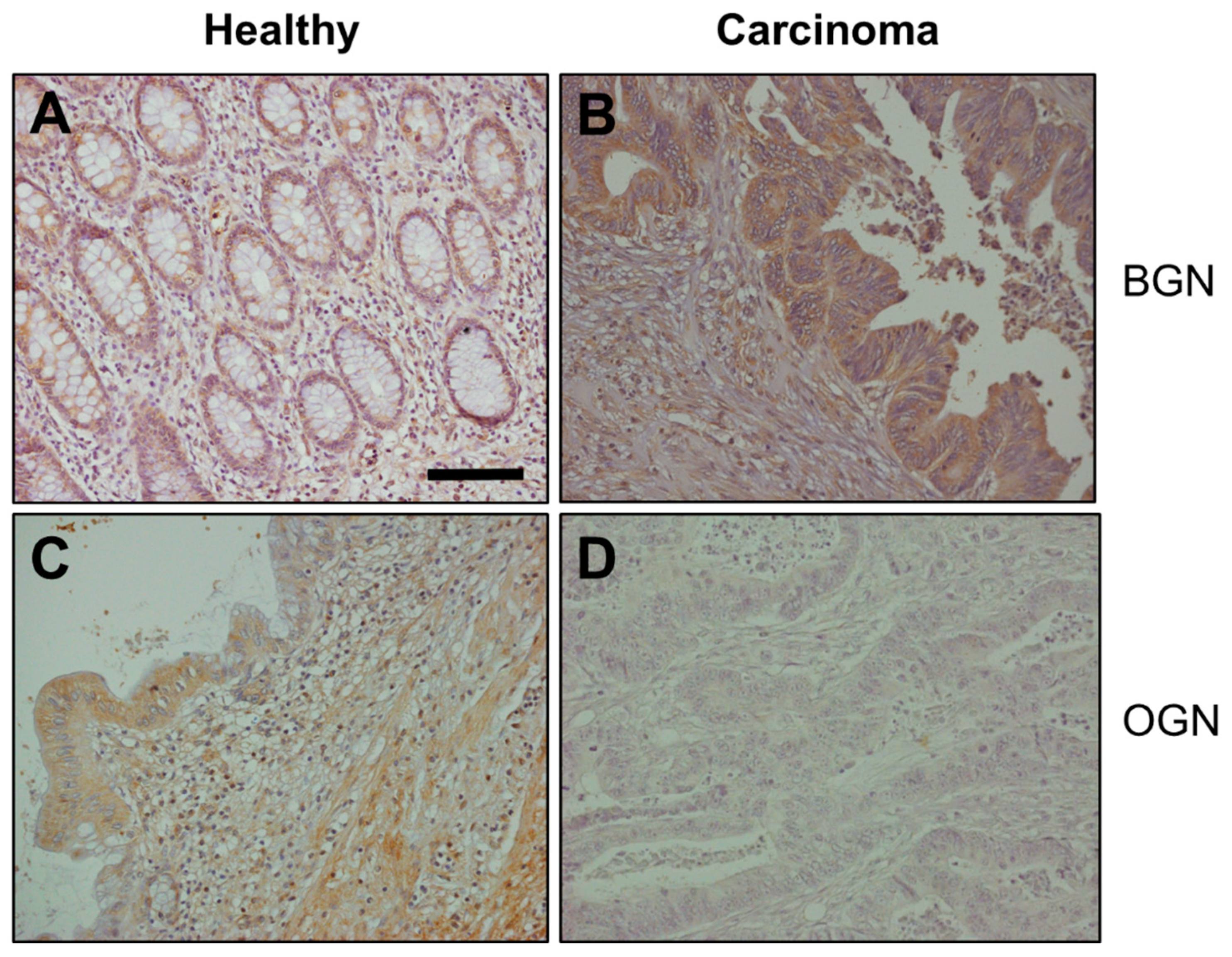
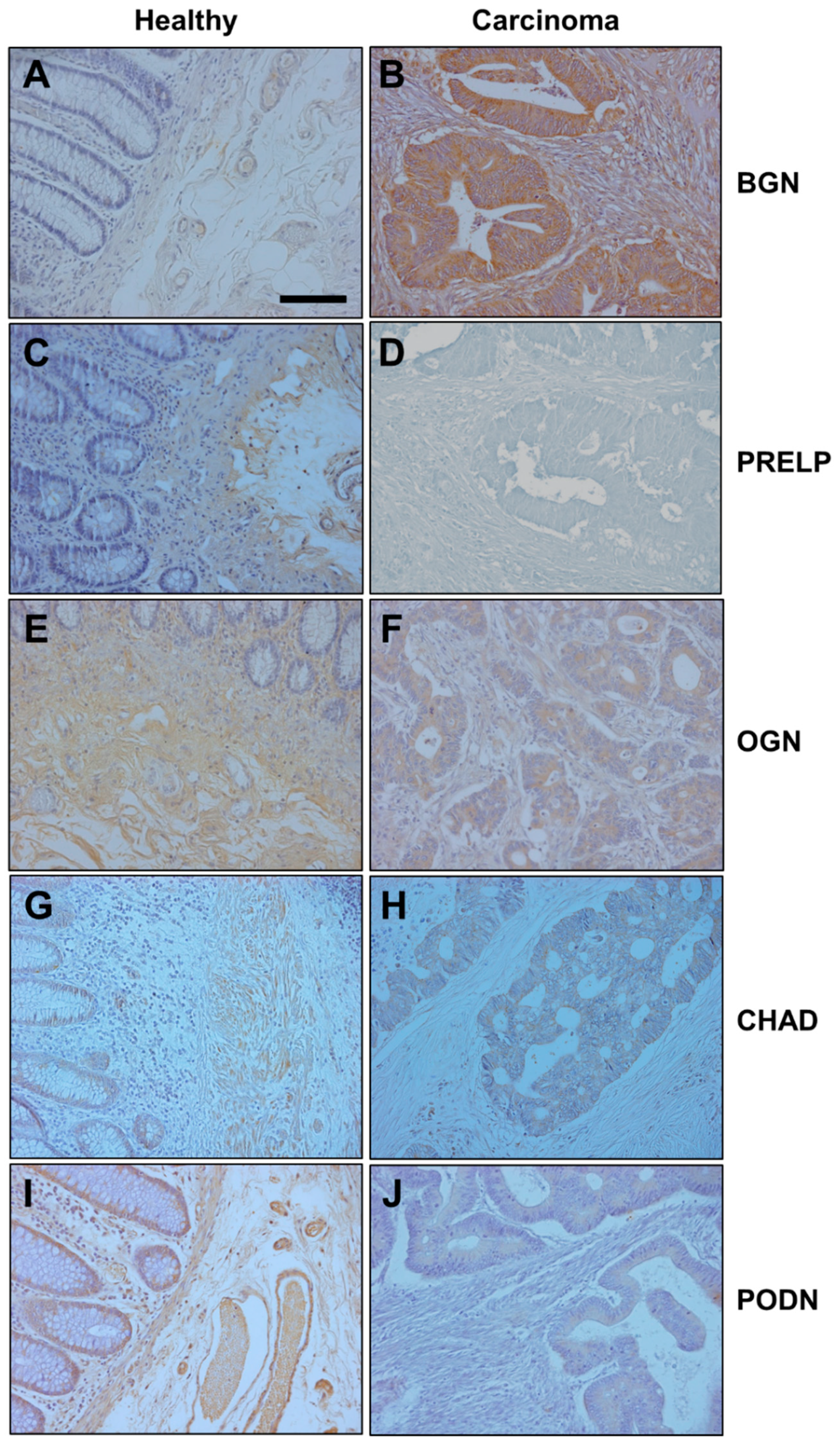
2.4. Immunohistochemistry
2.5. Survival Analysis
2.6. Cell Culture
2.7. Cell Migration Assay
2.8. Cell Proliferation Assay
3. Results
3.1. Analysis of Differential Gene Expression
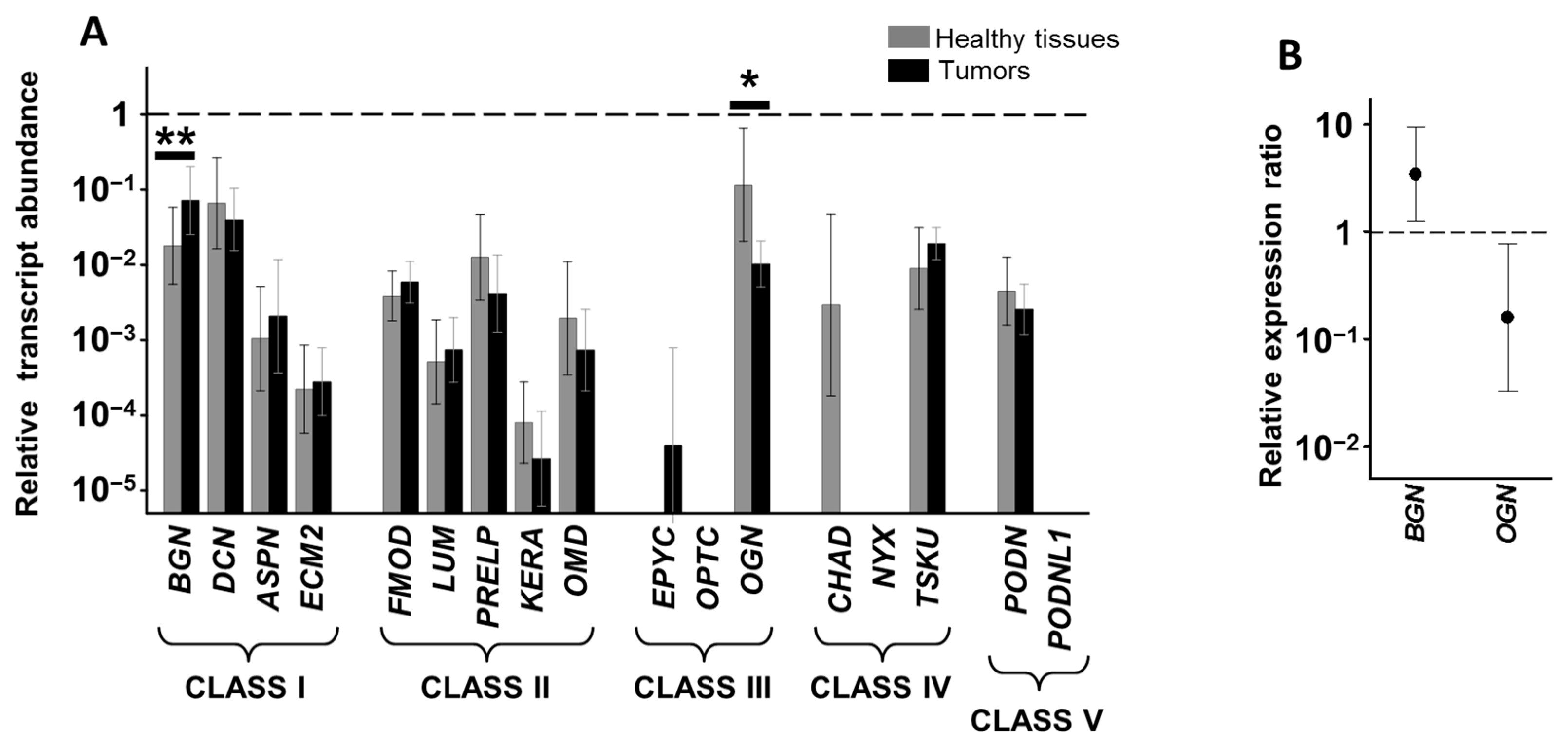
3.2. Differential Expression of Genes Encoding SLRPs in RSCCs
3.3. Differential Expression of Genes Encoding SLRPs in LSCCs
3.4. Relationship between Alteration in Gene Expression and Survival Data
3.5. Effect of Biglycan, Osteoglycine and PRLEP on Cell Migration and Proliferation of the HT29 Cell Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baek, S.K. Laterality: Right-Sided and Left-Sided Colon Cancer. Ann. Coloproctol. 2017, 33, 205–206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stintzing, S.; Tejpar, S.; Gibbs, P.; Thiebach, L.; Lenz, H.J. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur. J. Cancer 2017, 84, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef]
- Shen, H.; Yang, J.; Huang, Q.; Jiang, M.J.; Tan, Y.N.; Fu, J.F.; Zhu, L.Z.; Fang, X.F.; Yuan, Y. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J. Gastroenterol. 2015, 21, 6470–6478. [Google Scholar] [CrossRef]
- Loupakis, F.; Yang, D.; Yau, L.; Feng, S.; Cremolini, C.; Zhang, W.; Maus, M.K.; Antoniotti, C.; Langer, C.; Scherer, S.J.; et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J. Natl. Cancer Inst. 2015, 107, dju427. [Google Scholar] [CrossRef]
- Crotti, S.; Piccoli, M.; Rizzolio, F.; Giordano, A.; Nitti, D.; Agostini, M. Extracellular Matrix and Colorectal Cancer: How Surrounding Microenvironment Affects Cancer Cell Behavior? J. Cell. Physiol. 2017, 232, 967–975. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Naba, A.; Clauser, K.R.; Whittaker, C.A.; Carr, S.A.; Tanabe, K.K.; Hynes, R.O. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 2014, 14, 518. [Google Scholar] [CrossRef]
- Chen, S.; Birk, D.E. The regulatory roles of small leucine-rich proteoglycans in extracellular matrix assembly. FEBS J. 2013, 280, 2120–2137. [Google Scholar] [CrossRef]
- Merline, R.; Schaefer, R.M.; Schaefer, L. The matricellular functions of small leucine-rich proteoglycans (SLRPs). J. Cell Commun. Signal. 2009, 3, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Troup, S.; Njue, C.; Kliewer, E.V.; Parisien, M.; Roskelley, C.; Chakravarti, S.; Roughley, P.J.; Murphy, L.C.; Watson, P.H. Reduced expression of the small leucine-rich proteoglycans, lumican, and decorin is associated with poor outcome in node-negative invasive breast cancer. Clin. Cancer Res. 2003, 9, 207–214. [Google Scholar]
- Naito, Z. Role of the small leucine-rich proteoglycan (SLRP) family in pathological lesions and cancer cell growth. J. Nippon Med. Sch. 2005, 72, 137–145. [Google Scholar] [CrossRef]
- Schaefer, L.; Iozzo, R.V. Small leucine-rich proteoglycans, at the crossroad of cancer growth and inflammation. Curr. Opin. Genet. Dev. 2012, 22, 56–57. [Google Scholar] [CrossRef]
- Appunni, S.; Anand, V.; Khandelwal, M.; Seth, A.; Mathur, S.; Sharma, A. Altered expression of small leucine-rich proteoglycans (Decorin, Biglycan and Lumican): Plausible diagnostic marker in urothelial carcinoma of bladder. Tumour Biol. 2017, 39, 1010428317699112. [Google Scholar] [CrossRef]
- Fernández-Vega, I.; García-Suárez, O.; García, B.; Crespo, A.; Astudillo, A.; Quirós, L.M. Heparan sulfate proteoglycans undergo differential expression alterations in right sided colorectal cancer, depending on their metastatic character. BMC Cancer 2015, 15, 742. [Google Scholar] [CrossRef]
- Crespo, A.; García-Suárez, O.; Fernández-Vega, I.; Solis-Hernandez, M.P.; García, B.; Castañón, S.; Quirós, L.M. Heparan sulfate proteoglycans undergo differential expression alterations in left sided colorectal cancer, depending on their metastatic character. BMC Cancer 2018, 18, 687. [Google Scholar] [CrossRef]
- Dowling, C.M.; Walsh, D.; Coffey, J.C.; Kiely, P.A. The importance of selecting the appropriate reference genes for quantitative real time PCR as illustrated using colon cancer cells and tissue. F1000Research 2016, 5, 99. [Google Scholar] [CrossRef]
- Xu, L.; Luo, H.; Wang, R.; Wu, W.W.; Phue, J.N.; Shen, R.F.; Juhl, H.; Wu, L.; Alterovitz, W.L.; Simonyan, V.; et al. Novel reference genes in colorectal cancer identify a distinct subset of high stage tumors and their associated histologically normal colonic tissues. BMC Med. Genet. 2019, 20, 138. [Google Scholar] [CrossRef] [PubMed]
- Abramoff, M.D.; Magalhaes, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Manne, U.; Shanmugam, C.; Katkoori, V.R.; Bumpers, H.L.; Grizzle, W.E. Development and progression of colorectal neoplasia. Cancer Biomark. 2010, 9, 235–265. [Google Scholar] [CrossRef]
- Ameye, L.; Young, M.F. Mice deficient in small leucine-rich proteoglycans: Novel in vivo models for osteoporosis, osteoarthritis, Ehlers-Danlos syndrome, muscular dystrophy, and corneal diseases. Glycobiology 2008, 12, 107R–116R. [Google Scholar] [CrossRef]
- Schaefer, L.; Iozzo, R.V. Biological functions of the small leucine-rich proteoglycans: From genetics to signal transduction. J. Biol. Chem. 2008, 283, 21305–21309. [Google Scholar] [CrossRef] [PubMed]
- Tasheva, E.S.; Klocke, B.; Conrad, G.W. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol. Vis. 2004, 10, 758–772. [Google Scholar] [PubMed]
- Mager, L.F.; Wasmer, M.H.; Rau, T.T.; Krebs, P. Cytokine-Induced Modulation of Colorectal Cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.G.; Bhattacharyya, I.; Lydiatt, W.M. Aberrant expression and localization of decorin in human oral dysplasia and squamous cell carcinoma. Cancer Res. 2003, 63, 7769–7776. [Google Scholar] [PubMed]
- Liu, Y.; Li, W.; Li, X.; Tai, Y.; Lü, Q.; Yang, N.; Jiang, J. Expression and significance of biglycan in endometrial cancer. Arch. Gynecol. Obstet. 2014, 289, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Aprile, G.; Avellini, C.; Reni, M.; Mazzer, M.; Foltran, L.; Rossi, D.; Cereda, S.; Iaiza, E.; Fasola, G.; Piga, A. Biglycan expression and clinical outcome in patients with pancreatic adenocarcinoma. Tumour Biol. 2013, 34, 131–137. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Yang, F.; Zhang, S.S.; Zeng, T.T.; Xie, X.; Guan, X.Y. High expression of biglycan is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 2497–2505. [Google Scholar] [PubMed]
- Hu, L.; Duan, Y.; Li, J.; Su, L.P.; Yan, M.; Zhu, Z.G.; Liu, B.Y.; Yang, Q.M. Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget 2014, 5, 1885–1896. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, F.; Kraft, J.; Schroeder, C.; Hube-Magg, C.; Kluth, M.; Lang, D.S.; Simon, R.; Sauter, G.; Izbicki, J.R.; Clauditz, T.S.; et al. Up-regulation of Biglycan is Associated with Poor Prognosis and PTEN Deletion in Patients with Prostate Cancer. Neoplasia 2017, 19, 707–715. [Google Scholar] [CrossRef]
- Mikula, M.; Rubel, T.; Karczmarski, J.; Goryca, K.; Dadlez, M.; Ostrowski, J. Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct. Integr. Genom. 2011, 11, 215–224. [Google Scholar] [CrossRef]
- Xing, X.; Gu, X.; Ma, T. Knockdown of biglycan expression by RNA interference inhibits the proliferation and invasion of, and induces apoptosis in, the HCT116 colon cancer cell line. Mol. Med. Rep. 2015, 12, 7538–7544. [Google Scholar] [CrossRef]
- Xing, X.; Gu, X.; Ma, T.; Ye, H. Biglycan up-regulated vascular endothelial growth factor (VEGF) expression and promoted angiogenesis in colon cancer. Tumor Biol. 2015, 36, 1773–1780. [Google Scholar] [CrossRef]
- Lee, J.Y.; Eom, E.M.; Kim, D.S.; Ha-Lee, Y.M.; Lee, D.H. Analysis of gene expression profiles of gastric normal and cancer tissues by SAGE. Genomics 2003, 82, 78–85. [Google Scholar] [CrossRef]
- Röwer, C.; Ziems, B.; Radtke, A.; Schmitt, O.; Reimer, T.; Koy, C.; Thiesen, H.J.; Gerber, B.; Glocker, M.O. Toponostics of invasive ductal breast carcinoma: Combination of spatial protein expression imaging and quantitative proteome signature analysis. Int. J. Clin. Exp. Pathol. 2011, 4, 454–467. [Google Scholar] [PubMed]
- Lomnytska, M.I.; Becker, S.; Hellman, K.; Hellström, A.C.; Souchelnytskyi, S.; Mints, M.; Hellman, U.; Andersson, S.; Auer, G. Diagnostic protein marker patterns in squamous cervical cancer. Proteom. Clin. Appl. 2010, 4, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Z.; Wang, C.; Miao, L.; Zhang, J.; Wang, J.; Jiao, B.; Zhao, S. Quantitative proteomics approach to screening of potential diagnostic and therapeutic targets for laryngeal carcinoma. PLoS ONE 2014, 9, e90181. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, Y.; Lü, B.; Xu, E.; Huang, Q.; Lai, M. Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: A proteomic study. Exp. Biol. Med. 2007, 232, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, Y.Q.; Li, Q.G.; Ma, Y.L.; Peng, J.J.; Cai, S.J. Osteoglycin (OGN) reverses epithelial to mesenchymal transition and invasiveness in colorectal cancer via EGFR/Akt pathway. J. Exp. Clin. Cancer Res. 2018, 37, 41. [Google Scholar] [CrossRef] [PubMed]
- Conrotto, P.; Roesli, C.; Rybak, J.; Kischel, P.; Waltregny, D.; Neri, D.; Castronovo, V. Identification of new accessible tumor antigens in human colon cancer by ex vivo protein biotinylation and comparative mass spectrometry analysis. Int. J. Cancer 2008, 123, 2856–2864. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Wei, W.; Huang, N.; Shi, Y.; Huang, M.; Yan, Y.; Li, D.; Yi, J.; Wang, X. Tumor repressor gene chondroadherin oppose migration and proliferation in hepatocellular carcinoma and predicts a good survival. Oncotarget 2017, 8, 60270–60279. [Google Scholar] [CrossRef]
- Ross, M.D.; Bruggeman, L.A.; Hanss, B.; Sunamoto, M.; Marras, D.; Klotman, M.E.; Klotman, P.E. Podocan, a novel small leucine-rich repeat protein expressed in the sclerotic glomerular lesion of experimental HIV-associated nephropathy. J. Biol. Chem. 2003, 278, 33248–33255. [Google Scholar] [CrossRef]
- Hutter, R.; Huang, L.; Speidl, W.S.; Giannarelli, C.; Trubin, P.; Bauriedel, G.; Klotman, M.E.; Fuster, V.; Badimon, J.J.; Klotman, P.E. Novel small leucine-rich repeat protein podocan is a negative regulator of migration and proliferation of smooth muscle cells, modulates neointima formation, and is expressed in human atheroma. Circulation 2013, 128, 2351–2363. [Google Scholar] [CrossRef]
- Shimizu-Hirota, R.; Sasamura, H.; Kuroda, M.; Kobayashi, E.; Saruta, T. Functional characterization of podocan, a member of a new class in the small leucine-rich repeat protein family. FEBS Lett. 2004, 563, 69–74. [Google Scholar] [CrossRef]


| Primary Tumor Location (PTL) | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| RS-CC | LS-CC | ||||||||
| N | % Total | % RS-CC | N | % Total | % LS-CC | N | % | ||
| Gender | Male | 12 | 48.00 | 100.00 | 13 | 52.00 | 65.00 | 25 | 78.13 |
| Female | 0 | 0.00 | 0.00 | 7 | 100.00 | 35.00 | 7 | 21.88 | |
| Age | ≤70 years old | 5 | 27.78 | 41.67 | 13 | 72.22 | 65.00 | 18 | 56.25 |
| >70 years old | 7 | 50.00 | 58.33 | 7 | 50.00 | 35.00 | 14 | 43.75 | |
| pT | pT3 | 11 | 34.38 | 91.67 | 20 | 62.50 | 100.00 | 31 | 96.88 |
| pT2 | 1 | 3.12 | 8.33 | 0 | 0.00 | 0.00 | 1 | 3.12 | |
| pN | pN0 | 4 | 28.57 | 33.33 | 10 | 71.43 | 50.00 | 14 | 43.75 |
| pN+ | 8 | 44.44 | 66.67 | 10 | 55.56 | 50.00 | 18 | 56.25 | |
| Grade | Low-grade | 8 | 29.63 | 66.67 | 19 | 70.37 | 95.00 | 27 | 84.38 |
| High-grade | 4 | 80.00 | 33.33 | 1 | 20.00 | 5.00 | 5 | 15.63 | |
| Mucinous | No | 11 | 36.67 | 91.67 | 19 | 63.33 | 95.00 | 30 | 93.75 |
| Yes | 1 | 50.00 | 8.33 | 1 | 50.00 | 5.00 | 2 | 6.25 | |
| Perineural invasion | No | 11 | 37.93 | 91.67 | 18 | 62.07 | 90.00 | 29 | 90.63 |
| Yes | 1 | 33.33 | 8.33 | 2 | 66.67 | 10.00 | 3 | 9.38 | |
| Lymphovascular invasion | No | 9 | 33.33 | 75.00 | 18 | 66.67 | 90.00 | 27 | 84.38 |
| Yes | 3 | 60.00 | 25.00 | 2 | 40.00 | 10.00 | 5 | 15.63 | |
| Ulceration | No | 4 | 26.67 | 33.33 | 11 | 73.33 | 55.00 | 15 | 46.88 |
| Yes | 8 | 47.06 | 66.67 | 9 | 52.94 | 45.00 | 17 | 53.13 | |
| Perforation | No | 11 | 39.29 | 91.67 | 17 | 60.71 | 85.00 | 28 | 87.50 |
| Yes | 1 | 25.00 | 8.33 | 3 | 75.00 | 15.00 | 4 | 12.50 | |
| Inflammatory infiltrate | No | 1 | 14.29 | 50.00 | 6 | 85.71 | 54.55 | 7 | 53.85 |
| Yes | 1 | 16.67 | 50.00 | 5 | 83.33 | 45.45 | 6 | 46.15 | |
| Metastatic at diagnosis | No | 8 | 36.36 | 66.67 | 14 | 63.64 | 70.00 | 22 | 68.75 |
| Yes | 4 | 40.00 | 33.33 | 6 | 60.00 | 30.00 | 10 | 31.25 | |
| Metastatic any time | No | 5 | 31.25 | 41.67 | 11 | 68.75 | 55.00 | 16 | 50.00 |
| Yes | 7 | 43.75 | 58.33 | 9 | 56.25 | 45.00 | 16 | 50.00 | |
| CEA at diagnosis | Normal | 4 | 26.67 | 36.36 | 11 | 73.33 | 68.75 | 15 | 55.56 |
| High | 7 | 58.33 | 63.64 | 5 | 41.67 | 31.25 | 12 | 44.44 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solis-Hernandez, M.P.; Martín, C.; García, B.; Pérez-López, N.; García-Mesa, Y.; González-Fernández, S.; García-Suárez, O.; Merayo, J.; Fernández-Vega, I.; Quirós, L.M. The Genes Encoding Small Leucine-Rich Proteoglycans Undergo Differential Expression Alterations in Colorectal Cancer, Depending on Tumor Location. Cells 2021, 10, 2002. https://doi.org/10.3390/cells10082002
Solis-Hernandez MP, Martín C, García B, Pérez-López N, García-Mesa Y, González-Fernández S, García-Suárez O, Merayo J, Fernández-Vega I, Quirós LM. The Genes Encoding Small Leucine-Rich Proteoglycans Undergo Differential Expression Alterations in Colorectal Cancer, Depending on Tumor Location. Cells. 2021; 10(8):2002. https://doi.org/10.3390/cells10082002
Chicago/Turabian StyleSolis-Hernandez, Maria Pilar, Carla Martín, Beatriz García, Natalia Pérez-López, Yolanda García-Mesa, Sara González-Fernández, Olivia García-Suárez, Jesús Merayo, Iván Fernández-Vega, and Luis M. Quirós. 2021. "The Genes Encoding Small Leucine-Rich Proteoglycans Undergo Differential Expression Alterations in Colorectal Cancer, Depending on Tumor Location" Cells 10, no. 8: 2002. https://doi.org/10.3390/cells10082002
APA StyleSolis-Hernandez, M. P., Martín, C., García, B., Pérez-López, N., García-Mesa, Y., González-Fernández, S., García-Suárez, O., Merayo, J., Fernández-Vega, I., & Quirós, L. M. (2021). The Genes Encoding Small Leucine-Rich Proteoglycans Undergo Differential Expression Alterations in Colorectal Cancer, Depending on Tumor Location. Cells, 10(8), 2002. https://doi.org/10.3390/cells10082002







