Transcription Control of Liver Development
Abstract
:1. Introduction
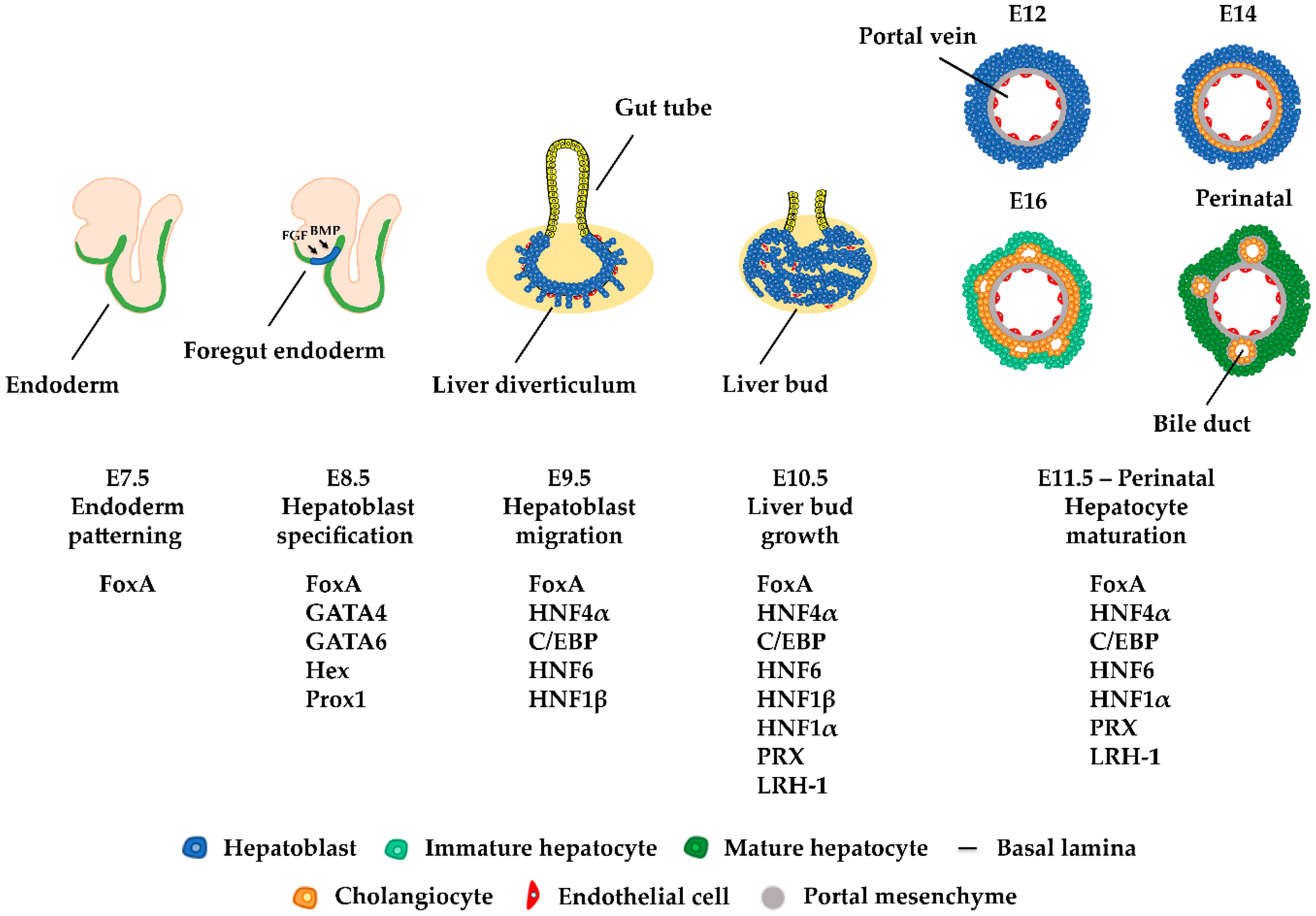
2. Liver Development
3. FoxA Family of Transcription Factors
4. The GATA Family of Transcription Factors
5. Hex and Prox1
6. Hepatocyte Nuclear Factor 4α (HNF4α)
7. The Hepatocyte Nuclear Factor 6 Family (HNF6)
8. C/EBP Family of Transcription Factors
9. Hepatocyte Nuclear Factor 1α and 1β (HNF1α and HNF1β)
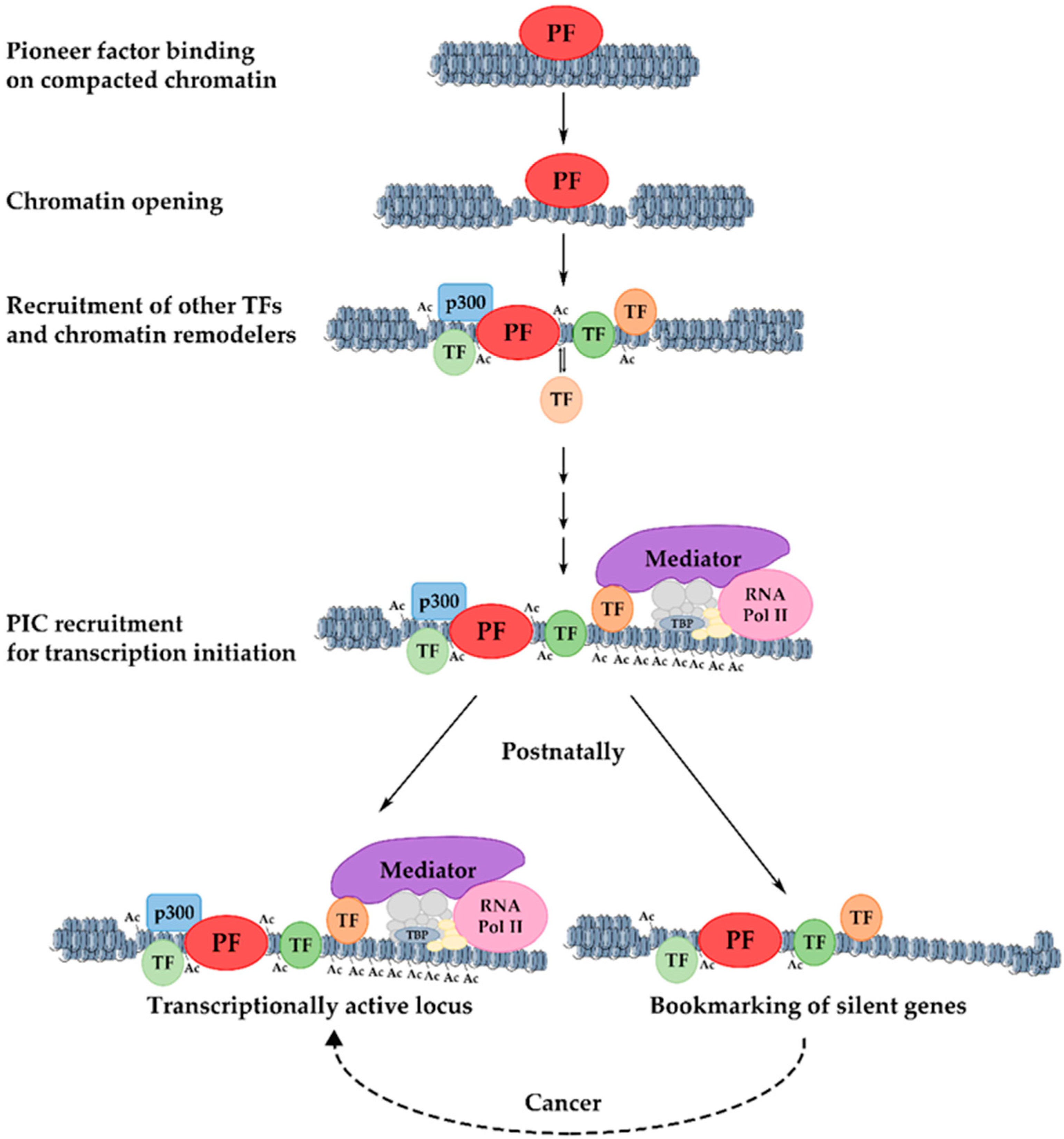
10. Mechanism of Transcriptional Activation of Hepatic Genes during Liver Development
11. Developmental Bookmarking by Pioneer and Non-Pioneer Transcription Factors
12. Significance of Bookmarking in Cancer and Proliferation-Induced Genes
13. Association of Transcription Factors with Their Targets during Mitosis
14. Maintenance of Stable Hepatic Gene Expression Patterns
15. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gordillo, M.; Evans, T.; Gouon-Evans, V. Orchestrating liver development. Development 2015, 142, 2094–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si-Tayeb, K.; Lemaigre, F.P.; Duncan, S.A. Organogenesis and development of the liver. Dev. Cell 2010, 18, 175–189. [Google Scholar] [CrossRef] [Green Version]
- Mu, T.; Xu, L.; Zhong, Y.; Liu, X.; Zhao, Z.; Huang, C.; Lan, X.; Lufei, C.; Zhou, Y.; Su, Y.; et al. Embryonic liver developmental trajectory revealed by single-cell RNA sequencing in the Foxa2eGFP mouse. Commun. Biol. 2020, 3, 642. [Google Scholar] [CrossRef]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011, 475, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011, 475, 390–395. [Google Scholar] [CrossRef]
- Yu, B.; He, Z.Y.; You, P.; Han, Q.W.; Xiang, D.; Chen, F.; Wang, M.J.; Liu, C.C.; Lin, X.W.; Borjigin, U.; et al. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell 2013, 13, 328–340. [Google Scholar] [CrossRef] [Green Version]
- Horisawa, K.; Udono, M.; Ueno, K.; Ohkawa, Y.; Nagasaki, M.; Sekiya, S.; Suzuki, A. The Dynamics of Transcriptional Activation by Hepatic Reprogramming Factors. Mol. Cell 2020, 79, 660–676.e8. [Google Scholar] [CrossRef]
- Costa, R.H.; Grayson, D.R.; Darnell, J.E. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 1989, 9, 1415–1425. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.K.; DiPersio, C.M.; Zaret, K.S. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol. Cell. Biol. 1991, 11, 773–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulweber, B.; Sandhofer, F.; Levy-Wilson, B. The mechanism by which the human apolipoprotein B gene reducer operates involves blocking of transcriptional activation by hepatocyte nuclear factor 3. Mol. Cell. Biol. 1993, 13, 1534–1546. [Google Scholar] [CrossRef] [Green Version]
- Sawaya, P.L.; Stripp, B.R.; Whitsett, J.A.; Luse, D.S. The lung-specific CC10 gene is regulated by transcription factors from the AP-1, octamer, and hepatocyte nuclear factor 3 families. Mol. Cell. Biol. 1993, 13, 3860–3871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippe, J.; Morel, C.; Prezioso, V.R. Glucagon gene expression is negatively regulated by hepatocyte nuclear factor 3 beta. Mol. Cell. Biol. 1994, 14, 3514–3523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, K.L.; Halay, E.D.; Lai, E.; Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993, 364, 412–420. [Google Scholar] [CrossRef]
- Qian, X.; Costa, R.H. Analysis of hepatocyte nuclear factor-3β protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 1995, 23, 1184–1191. [Google Scholar] [CrossRef]
- Lau, H.H.; Ng, N.H.J.; Loo, L.S.W.; Jasmen, J.B.; Teo, A.K.K. The molecular functions of hepatocyte nuclear factors—In and beyond the liver. J. Hepatol. 2018, 68, 1033–1048. [Google Scholar] [CrossRef] [Green Version]
- Lai, E.; Prezioso, V.R.; Tao, W.; Chen, W.S.; Darnell, J.E. Hepatocyte nuclear factor 3α belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991, 5, 416–427. [Google Scholar] [CrossRef] [Green Version]
- Kaestner, K.H.; Knöchel, W.; Martínez, D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000, 14, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, K.H.; Hiemisch, H.; Luckow, B.; Schütz, G. The HNF-3 gene family of transcription factors in mice: Gene structure, cDNA sequence, and mRNA distribution. Genomics 1994, 20, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monaghan, A.P.; Kaestner, K.H.; Grau, E.; Schutz, G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 1993, 119, 567–578. [Google Scholar] [CrossRef]
- Ang, S.L.; Wierda, A.; Wong, D.; Stevens, K.A.; Cascio, S.; Rossant, J.; Zaret, K.S. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 1993, 119, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Hogan, B.L.M. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development 1993, 118, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kaestner, K.H.; Katz, J.; Liu, Y.; Drucker, D.J.; Schütz, G. Inactivation of the winged helix transcription factor HNF3α affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999, 13, 495–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, S.L.; Rossant, J. HNF-3β is essential for node and notochord formation in mouse development. Cell 1994, 78, 561–574. [Google Scholar] [CrossRef]
- Weinstein, D.C.; Ruiz i Altaba, A.; Chen, W.S.; Hoodless, P.; Prezioso, V.R.; Jessell, T.M.; Darnell, J.E. The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell 1994, 78, 575–588. [Google Scholar] [CrossRef]
- Kaestner, K.H.; Hiemisch, H.; Schütz, G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 1998, 18, 4245–4251. [Google Scholar] [CrossRef] [Green Version]
- Kyrmizi, I.; Hatzis, P.; Katrakili, N.; Tronche, F.; Gonzalez, F.J.; Talianidis, I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006, 20, 2293–2305. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Friedman, J.R.; Fulmer, J.T.; Kaestner, K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature 2005, 435, 944–947. [Google Scholar] [CrossRef]
- Li, Z.; White, P.; Tuteja, G.; Rubins, N.; Sackett, S.; Kaestner, K.H. Foxa1 and Foxa2 regulate bile duct development in mice. J. Clin. Investg. 2009, 119, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reizel, Y.; Morgan, A.; Gao, L.; Lan, Y.; Manduchi, E.; Waite, E.L.; Wang, A.W.; Wells, A.; Kaestner, K.H. Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev. 2020, 34, 1039–1050. [Google Scholar] [CrossRef]
- Thakur, A.; Wong, J.C.H.; Wang, E.Y.; Lotto, J.; Kim, D.; Cheng, J.C.; Mingay, M.; Cullum, R.; Moudgil, V.; Ahmed, N.; et al. Hepatocyte nuclear factor 4-alpha is essential for the active epigenetic state at enhancers in mouse liver. Hepatology 2019, 70, 1360–1376. [Google Scholar] [CrossRef]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef]
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Arceci, R.J.; King, A.A.J.; Simon, M.C.; Orkin, S.H.; Wilson, D.B. Mouse GATA-4: A retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 1993, 13, 2235–2246. [Google Scholar] [CrossRef] [Green Version]
- Merika, M.; Orkin, S.H. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 1993, 13, 3999–4010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molkentin, J.D. The zinc finger-containing transcription factors GATA-4, -5, and -6: Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000, 275, 38949–38952. [Google Scholar] [CrossRef] [Green Version]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef] [Green Version]
- Narita, N.; Bielinska, M.; Wilson, D.B. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 1997, 189, 270–274. [Google Scholar] [CrossRef]
- Watt, A.J.; Battle, M.A.; Li, J.; Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 12573–12578. [Google Scholar] [CrossRef] [Green Version]
- Watt, A.J.; Zhao, R.; Li, J.; Duncan, S.A. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev. Biol. 2007, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Crompton, M.R.; Bartlett, T.J.; Macgregor, A.D.; Manfioletti, G.; Buratti, E.; Giancotti, V.; Goodwin, G.H. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 1992, 20, 5661–5667. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.Q.; Brown, A.; Beddington, R.S.P. Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development 1998, 125, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Bogue, C.W.; Ganea, G.R.; Sturm, E.; Ianucci, R.; Jacobs, H.C. Hex expression suggests a role in the development and function of organs derived frown foregut endoderm. Dev. Dyn. 2000, 219, 84–89. [Google Scholar] [CrossRef]
- Martinez Barbera, J.P.; Clements, M.; Thomas, P.; Rodriguez, T.; Meloy, D.; Kioussis, D.; Beddington, R.S.P. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 2000, 127, 2433–2445. [Google Scholar] [CrossRef] [PubMed]
- Keng, V.W.; Yagi, H.; Ikawa, M.; Nagano, T.; Myint, Z.; Yamada, K.; Tanaka, T.; Sato, A.; Muramatsu, I.; Okabe, M.; et al. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem. Biophys. Res. Commun. 2000, 276, 1155–1161. [Google Scholar] [CrossRef]
- Hunter, M.P.; Wilson, C.M.; Jiang, X.; Cong, R.; Vasavada, H.; Kaestner, K.H.; Bogue, C.W. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev. Biol. 2007, 308, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Bort, R.; Martinez-Barbera, J.P.; Beddington, R.S.P.; Zaret, K.S. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 2004, 131, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Bort, R.; Signore, M.; Tremblay, K.; Martinez Barbera, J.P.; Zaret, K.S. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 2006, 290, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Oliver, G.; Sosa-Pineda, B.; Geisendorf, S.; Spana, E.P.; Doe, C.Q.; Gruss, P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech. Dev. 1993, 44, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Wigle, J.T.; Oliver, G. Prox1 function is required for the development of the murine lymphatic system. Cell 1999, 98, 769–778. [Google Scholar] [CrossRef] [Green Version]
- Risebro, C.A.; Searles, R.G.; Melville, A.A.D.; Ehler, E.; Jina, N.; Shah, S.; Pallas, J.; Hubank, M.; Dillard, M.; Harvey, N.L.; et al. Prox1 maintains muscle structure and growth in the developing heart. Development 2009, 136, 495–505. [Google Scholar] [CrossRef] [Green Version]
- Charest-Marcotte, A.; Dufour, C.R.; Wilson, B.J.; Tremblay, A.M.; Eichner, L.J.; Arlow, D.H.; Mootha, V.K.; Giguère, V. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev. 2010, 24, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.H.; Li, T.; Chiang, J.Y.L. A prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4α that regulates the cholesterol 7α-hydroxylase gene. J. Biol. Chem. 2006, 281, 10081–10088. [Google Scholar] [CrossRef] [Green Version]
- Sosa-Pineda, B.; Wigle, J.T.; Oliver, G. Hepatocyte migration during liver development requires Prox1. Nat. Genet. 2000, 25, 254–255. [Google Scholar] [CrossRef] [PubMed]
- Wigle, J.T.; Chowdhury, K.; Gruss, P.; Oliver, G. Prox1 function is crucial for mouse lens-fibre elongation. Nat. Genet. 1999, 21, 318–322. [Google Scholar] [CrossRef]
- Sladek, F.M.; Zhong, W.; Lai, E.; Darnell, J.E. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990, 4, 2353–2365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadzopoulou-Cladaras, M.; Kistanova, E.; Evagelopoulou, C.; Zeng, S.; Cladaras, C.; Ladias, J.A.A. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J. Biol. Chem. 1997, 272, 539–550. [Google Scholar] [CrossRef] [Green Version]
- Duncan, S.A.; Manova, K.; Chen, W.S.; Hoodless, P.; Weinstein, D.C.; Bachvarova, R.F.; Darnell, J.E. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA 1994, 91, 7598–7602. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.S.; Manova, K.; Weinstein, D.C.; Duncan, S.A.; Plump, A.S.; Prezioso, V.R.; Bachvarova, R.F.; Darnell, J.E. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994, 8, 2466–2477. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ning, G.; Duncan, S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000, 14, 464–474. [Google Scholar] [CrossRef]
- Parviz, F.; Matullo, C.; Garrison, W.D.; Savatski, L.; Adamson, J.W.; Ning, G.; Kaestner, K.H.; Rossi, J.M.; Zaret, K.S.; Duncan, S.A. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003, 34, 292–296. [Google Scholar] [CrossRef]
- Taraviras, S.; Paula Monaghan, A.; Schütz, G.; Kelsey, G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech. Dev. 1994, 48, 67–79. [Google Scholar] [CrossRef]
- Nammo, T.; Yamagata, K.; Tanaka, T.; Kodama, T.; Sladek, F.M.; Fukui, K.; Katsube, F.; Sato, Y.; Miyagawa, J.I.; Shimomura, I. Expression of HNF-4α (MODY1), HNF-1β (MODY5), and HNF-1α (MODY3) proteins in the developing mouse pancreas. Gene Expr. Patterns 2008, 8, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.; Tang, J.I.; Seckl, J.R.; Nyirenda, M.J. Developmental and tissue-specific regulation of hepatocyte nuclear factor 4-α (HNF4-α) isoforms in rodents. Gene Expr. 2010, 14, 337–344. [Google Scholar] [CrossRef]
- Hayhurst, G.P.; Lee, Y.-H.; Lambert, G.; Ward, J.M.; Gonzalez, F.J. Hepatocyte nuclear factor 4α (Nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001, 21, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Landry, C.; Clotman, F.; Hioki, T.; Oda, H.; Picard, J.J.; Lemaigre, F.P.; Rousseau, G.G. HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver- enriched transcription factors. Dev. Biol. 1997, 192, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lannoy, V.J.; Bürglin, T.R.; Rousseau, G.G.; Lemaigre, F.P. Isoforms of hepatocyte nuclear factor-6 differ in DNA-binding properties, contain a bifunctional holneodomain, and define the new ONECUT class of homeodomain proteins. J. Biol. Chem. 1998, 273, 13552–13562. [Google Scholar] [CrossRef] [Green Version]
- Lemaigre, F.P.; Durviaux, S.M.; Truong, O.; Lannoy, V.J.; Hsuan, J.J.; Rousseau, G.G. Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc. Natl. Acad. Sci. USA 1996, 93, 9460–9464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausa, F.; Samadani, U.; Ye, H.; Lim, L.; Fletcher, C.F.; Jenkins, N.A.; Copeland, N.G.; Costa, R.H. The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3β in the developing murine liver and pancreas. Dev. Biol. 1997, 192, 228–246. [Google Scholar] [CrossRef] [Green Version]
- Lannoy, V.J.; Rodolosse, A.; Pierreux, C.E.; Rousseau, G.G.; Lemaigre, F.P. Transcriptional stimulation by hepatocyte nuclear factor-6: Target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF). J. Biol. Chem. 2000, 275, 22098–22103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquemin, P.; Lannoy, V.J.; Rousseau, G.G.; Lemaigre, F.P. OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J. Biol. Chem. 1999, 274, 2665–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhorenbeeck, V.; Jacquemin, P.; Lemaigre, F.P.; Rousseau, G.G. OC-3, a novel mammalian member of the ONECUT class of transcription factors. Biochem. Biophys. Res. Commun. 2002, 292, 848–854. [Google Scholar] [CrossRef]
- Rastegar, M.; Szpirer, C.; Rousseau, G.G.; Lemaigre, F.P. Hepatocyte nuclear factor 6: Organization and chromosomal assignment of the rat gene and characterization of its promoter. Biochem. J. 1998, 334, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropp, P.A.; Gannon, M. Onecut transcription factors in development and disease. Trends Dev. Biol. 2016, 9, 43–57. [Google Scholar] [PubMed]
- Jacquemin, P.; Durviaux, S.M.; Jensen, J.; Godfraind, C.; Gradwohl, G.; Guillemot, F.; Madsen, O.D.; Carmeliet, P.; Dewerchin, M.; Collen, D.; et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 2000, 20, 4445–4454. [Google Scholar] [CrossRef] [Green Version]
- Jacquemin, P.; Pierreux, C.E.; Fierens, S.; van Eyll, J.M.; Lemaigre, F.P.; Rousseau, G.G. Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr. Patterns 2003, 3, 639–644. [Google Scholar] [CrossRef]
- Lannoy, V.J.; Decaux, J.F.; Pierreux, C.E.; Lemaigre, F.P.; Rousseau, G.G. Liver Glucokinase Gene Expression Is Controlled by the Onecut Transcription Factor Hepatocyte Nuclear Factor-6. Diabetologia 2002, 45, 1136–1141. [Google Scholar] [CrossRef] [Green Version]
- Maestro, M.A.; Boj, S.F.; Luco, R.F.; Pierreux, C.E.; Cabedo, J.; Servitja, J.M.; German, M.S.; Rousseau, G.G.; Lemaigre, F.P.; Ferrer, J. Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum. Mol. Genet. 2003, 12, 3307–3314. [Google Scholar] [CrossRef] [Green Version]
- Pierreux, C.E.; Poll, A.V.; Kemp, C.R.; Clotman, F.; Maestro, M.A.; Cordi, S.; Ferrer, J.; Leyns, L.; Rousseau, G.G.; Lemaigre, F.P. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology 2006, 130, 532–541. [Google Scholar] [CrossRef]
- Poll, A.V.; Pierreux, C.E.; Lokmane, L.; Haumaitre, C.; Achouri, Y.; Jacquemin, P.; Rousseau, G.G.; Cereghini, S.; Lemaigre, F.P. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes 2006, 55, 61–69. [Google Scholar] [CrossRef]
- Clotman, F.; Lannoy, V.J.; Reber, M.; Cereghini, S.; Cassiman, D.; Jacquemin, P.; Roskams, T.; Rousseau, G.G.; Lemaigre, F.P. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 2002, 129, 1819–1828. [Google Scholar] [CrossRef]
- Clotman, F.; Jacquemin, P.; Plumb-Rudewiez, N.; Pierreux, C.E.; Van Der Smissen, P.; Dietz, H.C.; Courtoy, P.J.; Rousseau, G.G.; Lemaigre, F.P. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev. 2005, 19, 1849–1854. [Google Scholar] [CrossRef] [Green Version]
- Clotman, F.; Lemaigre, F.P. Control of hepatic differentiation by activin/TGFβ signaling. Cell Cycle 2006, 5, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Plumb-Rudewiez, N.; Clotman, F.; Strick-Marchand, H.; Pierreux, C.E.; Weiss, M.C.; Rousseau, G.G.; Lemaigre, F.P. Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3α/Foxa1 gene by TGF-β in mouse liver. Hepatology 2004, 40, 1266–1274. [Google Scholar] [CrossRef]
- Beaudry, J.-B.; Pierreux, C.E.; Hayhurst, G.P.; Plumb-Rudewiez, N.; Weiss, M.C.; Rousseau, G.G.; Lemaigre, F.P. Threshold levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy with HNF-4 and PGC-1α are required for time-specific gene expression during liver development. Mol. Cell. Biol. 2006, 26, 6037–6046. [Google Scholar] [CrossRef] [Green Version]
- Westmacott, A.; Burke, Z.D.; Oliver, G.; Slack, J.M.W.; Tosh, D. C/EBPα and C/EBPβ are markers of early liver development. Int. J. Dev. Biol. 2006, 50, 653–657. [Google Scholar] [CrossRef] [Green Version]
- Landschulz, W.H.; Johnson, P.F.; Adashi, E.Y.; Graves, B.J.; McKnight, S.L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988, 2, 786–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, A.D.; McKnight, S.L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990, 4, 1416–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, D.Q.; Shih, C.H. An “attenuator domain” is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol. Cell. Biol. 1991, 11, 1480–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nerlov, C.; Ziff, E.B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-α on the serum albumin promoter. Genes Dev. 1994, 8, 350–362. [Google Scholar] [CrossRef] [Green Version]
- Hendricks-Taylor, L.R.; Darlington, G.J. Inhibition of cell proliferation by C/EBPα occurs in many cell types, does not require the presence of p53 or Rb, and is not affected by large T-antigen. Nucleic Acids Res. 1995, 23, 4726–4733. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.C.; Baer, M.; Dillner, A.J.; Johnson, P.F. CRP2 (C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995, 14, 3170–3183. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.C.; Cantwell, C.A.; Johnson, P.F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991, 5, 1553–1567. [Google Scholar] [CrossRef] [Green Version]
- Shiojiri, N.; Takeshita, K.; Yamasaki, H.; Iwata, T. Suppression of C/EBP α expression in biliary cell differentiation from hepatoblasts during mouse liver development. J. Hepatol. 2004, 41, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.D.; Finegold, M.J.; Bradley, A.; Ou, C.N.; Abdelsayed, S.V.; Wilde, M.D.; Taylor, L.R.; Wilson, D.R.; Darlington, G.J. Impaired energy homeostasis in C/EBPα knockout mice. Science 1995, 269, 1108–1112. [Google Scholar] [CrossRef]
- Flodby, P.; Barlow, C.; Kylefjord, H.; Ährlund-Richter, L.; Xanthopoulos, K.G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 1996, 271, 24753–24760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, Y.; Inoue, J.; Lambert, G.; Yim, S.H.; Gonzalez, F.J. Disruption of hepatic C/EBPα results in impaired glucose tolerance and age-dependent hepatosteatosis. J. Biol. Chem. 2004, 279, 44740–44748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Croniger, C.M.; Lekstrom-Himes, J.; Zhang, P.; Fenyus, M.; Tenen, D.G.; Darlington, G.J.; Hanson, R.W. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer- binding protein α. J. Biol. Chem. 2005, 280, 38689–38699. [Google Scholar] [CrossRef] [Green Version]
- Diehl, A.M. Roles of CCAAT/enhancer-binding proteins in regulation of liver regenerative growth. J. Biol. Chem. 1998, 273, 30843–30846. [Google Scholar] [CrossRef] [Green Version]
- Frain, M.; Swart, G.; Monaci, P.; Nicosia, A.; Stämpfli, S.; Frank, R.; Cortese, R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell 1989, 59, 145–157. [Google Scholar] [CrossRef]
- Baumhueter, S.; Mendel, D.B.; Conley, P.B.; Kuo, C.J.; Turk, C.; Graves, M.K.; Edwards, C.A.; Courtois, G.; Crabtree, G.R. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 1990, 4, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Blumenfeld, M.; Maury, M.; Chouard, T.; Yaniv, M.; Condamine, H. Hepatic Nuclear Factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development 1991, 113, 589–599. [Google Scholar] [CrossRef]
- Coffinier, C.; Barra, J.; Babinet, C.; Yaniv, M. Expression of the vHNF1/HNF1β homeoprotein gene during mouse organogenesis. Mech. Dev. 1999, 89, 211–213. [Google Scholar] [CrossRef]
- De Simone, V.; De Magistris, L.; Lazzaro, D.; Gerstner, J.; Monaci, P.; Nicosia, A.; Cortese, R. LFB3, a heterodimer-forming homeoprotein of the LFB1 family, is expressed in specialized epithelia. EMBO J. 1991, 10, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Lazzarro, D.; De Simone, V.; De Magistris, L.; Lehtonen, E.; Cortese, R. LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development 1992, 114, 469–479. [Google Scholar] [CrossRef]
- Pontoglio, M.; Barra, J.; Hadchouel, M.; Doyen, A.; Kress, C.; Bach, J.P.; Babinet, C.; Yaniv, M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 1996, 84, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Cereghini, S.; Blumenfeld, M.; Yaniv, M. A liver-specific factor essential for albumin transcription differs between differentiated and dedifferentiated rat hepatoma cells. Genes Dev. 1988, 2, 957–974. [Google Scholar] [CrossRef] [Green Version]
- Baumhueter, S.; Courtois, G.; Crabtree, G.R. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO J. 1988, 7, 2485–2493. [Google Scholar] [CrossRef]
- Rey-Campos, J.; Chouard, T.; Yaniv, M.; Cereghini, S. vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO J. 1991, 10, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.O.; Rey-Campos, J.; Cereghini, S.; Yaniv, M. vHNF1 is expressed in epithelial cells of distinct embryonic origin during development and precedes HNF1 expression. Mech. Dev. 1991, 36, 47–58. [Google Scholar] [CrossRef]
- Nammo, T.; Yamagata, K.; Hamaoka, R.; Zhu, Q.; Akiyama, T.E.; Gonzalez, F.J.; Miyagawa, J.; Matsuzawa, Y. Expression profile of MODY3/HNF-1α protein in the developing mouse pancreas. Diabetologia 2002, 45, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Shih, D.Q.; Stoffel, M. Dissecting the transcriptional network of pancreatic islets during development and differentiation. Proc. Natl. Acad. Sci. USA 2001, 98, 14189–14191. [Google Scholar] [CrossRef] [Green Version]
- Barbacci, E.; Reber, M.; Ott, M.O.; Breillat, C.; Huetz, F.; Cereghini, S. Variant Hepatocyte Nuclear Factor 1 is required for visceral endoderm specification. Development 1999, 126, 4795–4805. [Google Scholar] [CrossRef]
- Coffinier, C.; Gresh, L.; Fiette, L.; Tronche, F.; Schütz, G.; Babinet, C.; Pontoglio, M.; Yaniv, M.; Barra, J. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1β. Development 2002, 129, 1829–1838. [Google Scholar] [CrossRef]
- Gualdi, R.; Bossard, P.; Zheng, M.; Hamada, Y.; Coleman, J.R.; Zaret, K.S. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 1996, 10, 1670–1682. [Google Scholar] [CrossRef] [Green Version]
- Bossard, P.; Zaret, K.S. GATA transcription factors as potentiators of gut endoderm differentiation. Development 1998, 125, 4909–4917. [Google Scholar] [CrossRef]
- Cirillo, L.A.; Lin, F.R.; Cuesta, I.; Friedman, D.; Jarnik, M.; Zaret, K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 2002, 9, 279–289. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Zaret, K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014, 28, 2679–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayran, A.; Drouin, J. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 2018, 293, 13795–13804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, V.; Finch, J.T.; Graziano, V.; Lee, P.L.; Sweet, R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 1993, 362, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, L.A.; McPherson, C.E.; Bossard, P.; Stevens, K.; Cherian, S.; Shim, E.Y.; Clark, K.L.; Burley, S.K.; Zaret, K.S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998, 17, 244–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaya, D.; Hayamizu, T.; Bustin, M.; Zaret, K.S. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 2001, 276, 44385–44389. [Google Scholar] [CrossRef] [Green Version]
- Iwafuchi, M.; Cuesta, I.; Donahue, G.; Takenaka, N.; Osipovich, A.B.; Magnuson, M.A.; Roder, H.; Seeholzer, S.H.; Santisteban, P.; Zaret, K.S. Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat. Genet. 2020, 52, 418–427. [Google Scholar] [CrossRef]
- Sekiya, T.; Muthurajan, U.M.; Luger, K.; Tulin, A.V.; Zaret, K.S. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009, 23, 804–809. [Google Scholar] [CrossRef] [Green Version]
- Caravaca, J.M.; Donahue, G.; Becker, J.S.; He, X.; Vinson, C.; Zaret, K.S. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013, 27, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Lupien, M.; Eeckhoute, J.; Meyer, C.A.; Wang, Q.; Zhang, Y.; Li, W.; Carroll, J.S.; Liu, X.S.; Brown, M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008, 132, 958–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaret, K.S.; Mango, S.E. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 2016, 37, 76–81. [Google Scholar] [CrossRef] [Green Version]
- Hurtado, A.; Holmes, K.A.; Ross-Innes, C.S.; Schmidt, D.; Carroll, J.S. FOXA1 is a critical determinant of Estrogen Receptor function and endocrine response. Nat. Genet. 2011, 43, 27–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, Z.; Li, L.; Chen, B.C.; Revyakin, A.; Hajj, B.; Legant, W.; Dahan, M.; Lionnet, T.; Betzig, E.; et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 2014, 156, 1274–1285. [Google Scholar] [CrossRef] [Green Version]
- Swinstead, E.E.; Miranda, T.B.; Paakinaho, V.; Baek, S.; Goldstein, I.; Hawkins, M.; Karpova, T.S.; Ball, D.; Mazza, D.; Lavis, L.D.; et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 2016, 165, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Kraus, W.L. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 2017, 65, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Franco, H.L.; Nagari, A.; Kraus, W.L. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell 2015, 58, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef] [Green Version]
- Soufi, A.; Fernandez Garcia, M.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Mayran, A.; Khetchoumian, K.; Hariri, F.; Pastinen, T.; Gauthier, Y.; Balsalobre, A.; Drouin, J. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet. 2018, 50, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Iwafuchi-Doi, M.; Zaret, K.S. Cell fate control by pioneer transcription factors. Development 2016, 143, 1833–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soutoglou, E.; Papafotiou, G.; Katrakili, N.; Talianidis, I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem. 2000, 275, 12515–12520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soutoglou, E.; Viollet, B.; Vaxillaire, M.; Yaniv, M.; Pontoglio, M.; Talianidis, I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001, 20, 1984–1992. [Google Scholar] [CrossRef] [Green Version]
- Bevington, S.L.; Cauchy, P.; Piper, J.; Bertrand, E.; Lalli, N.; Jarvis, R.C.; Gilding, L.N.; Ott, S.; Bonifer, C.; Cockerill, P.N. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 2016, 35, 515–535. [Google Scholar] [CrossRef]
- Jacobs, J.; Atkins, M.; Davie, K.; Imrichova, H.; Romanelli, L.; Christiaens, V.; Hulselmans, G.; Potier, D.; Wouters, J.; Taskiran, I.I.; et al. The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet. 2018, 50, 1011–1020. [Google Scholar] [CrossRef]
- Wang, A.; Yue, F.; Li, Y.; Xie, R.; Harper, T.; Patel, N.A.; Muth, K.; Palmer, J.; Qiu, Y.; Wang, J.; et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015, 16, 386–399. [Google Scholar] [CrossRef] [Green Version]
- Karagianni, P.; Moulos, P.; Schmidt, D.; Odom, D.T.; Talianidis, I. Bookmarking by non-pioneer transcription factors during liver development establishes competence for future gene activation. Cell Rep. 2020, 30, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soundararajan, R.; Paranjape, A.N.; Barsan, V.; Chang, J.T.; Mani, S.A. A novel embryonic plasticity gene signature that predicts metastatic competence and clinical outcome. Sci. Rep. 2015, 5, 11766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhart, S.H.; Kretzmer, H.; Holdt, L.M.; Jühling, F.; Ammerpohl, O.; Bergmann, A.K.; Northoff, B.H.; Doose, G.; Siebert, R.; Stadler, P.F.; et al. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Sci. Rep. 2016, 6, 37393. [Google Scholar] [CrossRef]
- Andrisani, O.M.; Studach, L.; Merle, P. Gene signatures in hepatocellular carcinoma (HCC). Semin. Cancer Biol. 2011, 21, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaou, K.C.; Moulos, P.; Chalepakis, G.; Hatzis, P.; Oda, H.; Reinberg, D.; Talianidis, I. Spontaneous development of hepatocellular carcinoma with cancer stem cell properties in PR-SET7-deficient livers. EMBO J. 2015, 34, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Kontaki, H.; Koukaki, M.; Vasilarou, M.; Giakountis, A.; Deligianni, E.; Luo, X.; Kim, Y.; Talianidis, I. Targeting Smyd3 by next-generation antisense oligonucleotides suppresses liver tumor growth. iScience 2021, 24, 102473. [Google Scholar] [CrossRef]
- Wang, Q.; Li, W.; Zhang, Y.; Yuan, X.; Xu, K.; Yu, J.; Chen, Z.; Beroukhim, R.; Wang, H.; Lupien, M.; et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009, 138, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, N.; Ito, E.; Azuma, S.; Honma, R.; Yanagisawa, Y.; Nishikawa, A.; Kawamura, M.; Imai, J.I.; Tatsuta, K.; Inoue, J.I.; et al. FoxA1 as a lineage-specific oncogene in luminal type breast cancer. Biochem. Biophys. Res. Commun. 2008, 365, 711–717. [Google Scholar] [CrossRef]
- Zhou, Y.; Chang, H.; Yang, B. Gata4 is upregulated in nasopharyngeal cancer and facilitates epithelial-mesenchymal transition and metastasis through regulation of slug. Exp. Ther. Med. 2018, 16, 5318–5326. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Semi, K.; Yamamoto, T.; Shimizu, M.; Tanaka, A.; Mitsunaga, K.; Okita, K.; Osafune, K.; Arioka, Y.; Maeda, T.; et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 2014, 156, 663–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, H.; Komura, S.; Yamada, Y.; Sankoda, N.; Tanaka, A.; Ukai, T.; Kabata, M.; Sakurai, S.; Kuze, B.; Woltjen, K.; et al. In vivo reprogramming drives Kras-induced cancer development. Nat. Commun. 2018, 9, 2081. [Google Scholar] [CrossRef] [Green Version]
- So, J.; Kim, A.; Lee, S.H.; Shin, D. Liver progenitor cell-driven liver regeneration. Exp. Mol. Med. 2020, 52, 1230–1238. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.R.; Cole, P.A.; Meyers, D.J.; Kormish, J.; Dent, S.; Zaret, K.S. Chromatin “pre-pattern” and histone modifiers in a fate choice for liver and pancreas. Science 2011, 332, 963–966. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yang, L.; He, Q.; Hu, C.; Zhu, L.; Ma, X.; Ma, X.; Bao, S.; Li, L.; Chen, Y.; et al. A homeostatic Arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated YAP signaling. Cell Stem Cell 2019, 25, 54–68. [Google Scholar] [CrossRef]
- Martínez-Balbás, M.A.; Dey, A.; Rabindran, S.K.; Ozato, K.; Wu, C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 1995, 83, 29–38. [Google Scholar] [CrossRef]
- Hsiung, C.C.S.; Bartman, C.R.; Huang, P.; Ginart, P.; Stonestrom, A.J.; Keller, C.A.; Face, C.; Jahn, K.S.; Evans, P.; Sankaranarayanan, L.; et al. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 2016, 30, 1423–1439. [Google Scholar] [CrossRef] [Green Version]
- Kouskouti, A.; Talianidis, I. Histone modifications defining active genes persist after transcriptional and mitotic in activation. EMBO J. 2005, 24, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Liang, K.; Woodfin, A.R.; Slaughter, B.D.; Unruh, J.R.; Box, A.C.; Rickels, R.A.; Gao, X.; Haug, J.S.; Jaspersen, S.L.; Shilatifard, A. Mitotic transcriptional activation: Clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol. Cell 2015, 60, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Pelham-Webb, B.; Di Giammartino, D.C.; Li, J.; Kim, D.; Kita, K.; Saiz, N.; Garg, V.; Doane, A.; Giannakakou, P.; et al. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 2017, 19, 1283–1293. [Google Scholar] [CrossRef] [Green Version]
- Palozola, K.C.; Lerner, J.; Zaret, K.S. A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol. 2019, 20, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.V.; Sacco-Bubulya, P.A.; Prasanth, S.G.; Spector, D.L. Sequential entry of components of gene expression machinery into daughter cells. Mol. Biol. Cell 2002, 14, 1043–1057. [Google Scholar] [CrossRef] [Green Version]
- Hausnerová, V.V.; Lanctôt, C. Transcriptional output transiently spikes upon mitotic exit. Sci. Rep. 2017, 7, 12607. [Google Scholar] [CrossRef] [PubMed]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 2017, 358, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Michelotti, E.F.; Sanford, S.; Levons, D. Marking of active genes on mitotic chromosomes. Nature 1997, 388, 895–899. [Google Scholar] [CrossRef]
- Burke, L.J.; Zhang, R.; Bartkuhn, M.; Tiwari, V.K.; Tavoosidana, G.; Kurukuti, S.; Weth, C.; Leers, J.; Galjart, N.; Ohlsson, R.; et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005, 24, 3291–3300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egli, D.; Birkhoff, G.; Eggan, K. Mediators of reprogramming: Transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 2008, 9, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Young, D.W.; Hassan, M.Q.; Pratap, J.; Galindo, M.; Zaidi, S.K.; Lee, S.H.; Yang, X.; Xie, R.; Javed, A.; Underwood, J.M.; et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 2007, 445, 442–446. [Google Scholar] [CrossRef]
- Young, D.W.; Hassan, M.Q.; Yang, X.Q.; Galindo, M.; Javed, A.; Zaidi, S.K.; Furcinitti, P.; Lapointe, D.; Montecino, M.; Lian, J.B.; et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. USA 2007, 104, 3189–3194. [Google Scholar] [CrossRef] [Green Version]
- Blobel, G.A.; Kadauke, S.; Wang, E.; Lau, A.W.; Zuber, J.; Chou, M.M.; Vakoc, C.R. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 2009, 36, 970–983. [Google Scholar] [CrossRef] [Green Version]
- Dey, A.; Nishiyama, A.; Karpova, T.; McNally, J.; Ozato, K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 2009, 20, 4899–4909. [Google Scholar] [CrossRef] [Green Version]
- Kadauke, S.; Udugama, M.I.; Pawlicki, J.M.; Achtman, J.C.; Jain, D.P.; Cheng, Y.; Hardison, R.C.; Blobel, G.A. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 2012, 150, 725–737. [Google Scholar] [CrossRef] [Green Version]
- Xing, H.; Wilkerson, D.C.; Mayhew, C.N.; Lubert, E.J.; Skaggs, H.S.; Goodson, M.L.; Hong, Y.; Park-Sarge, O.K.; Sarge, K.D. Mechanism of hsp70i gene bookmarking. Science 2005, 307, 421–423. [Google Scholar] [CrossRef]
- Xing, H.; Vanderford, N.L.; Sarge, K.D. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 2008, 10, 1318–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segil, N.; Guermah, M.; Hoffmann, A.; Roeder, R.G.; Heintz, N. Mitotic regulation of TFIID: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996, 10, 2389–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Hinkley, C.S.; Henry, R.W.; Huang, S. TBP dynamics in living human cells: Constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 2002, 13, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Christova, R.; Oelgeschläger, T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 2002, 4, 79–82. [Google Scholar] [CrossRef]
- Verdeguer, F.; Le Corre, S.; Fischer, E.; Callens, C.; Garbay, S.; Doyen, A.; Igarashi, P.; Terzi, F.; Pontoglio, M. A mitotic transcriptional switch in polycystic kidney disease. Nat. Med. 2010, 16, 106–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, J.; Bagattin, A.; Verdeguer, F.; Makinistoglu, M.P.; Garbay, S.; Felix, T.; Heidet, L.; Pontoglio, M. Human mutations affect the epigenetic/bookmarking function of HNF1B. Nucleic Acids Res. 2016, 44, 8097–8111. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.M.; Byun, J.S.; Sacta, M.; Jin, Q.; Baek, S.J.; Gardner, K. Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLoS ONE 2014, 9, e99989. [Google Scholar] [CrossRef] [Green Version]
- Zhao, R.; Nakamura, T.; Fu, Y.; Lazar, Z.; Spector, D.L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 2011, 13, 1295–1304. [Google Scholar] [CrossRef]
- Festuccia, N.; Dubois, A.; Vandormael-Pournin, S.; Gallego Tejeda, E.; Mouren, A.; Bessonnard, S.; Mueller, F.; Proux, C.; Cohen-Tannoudji, M.; Navarro, P. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol. 2016, 18, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Teves, S.S.; An, L.; Hansen, A.S.; Xie, L.; Darzacq, X.; Tjian, R. A dynamic mode of mitotic bookmarking by transcription factors. Elife 2016, 5, e22280. [Google Scholar] [CrossRef]
- Deluz, C.; Friman, E.T.; Strebinger, D.; Benke, A.; Raccaud, M.; Callegari, A.; Leleu, M.; Manley, S.; Suter, D.M. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 2016, 30, 2538–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaigre, F.P. Mechanisms of Liver Development: Concepts for Understanding Liver Disorders and Design of Novel Therapies. Gastroenterology 2009, 137, 62–79. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tachmatzidi, E.C.; Galanopoulou, O.; Talianidis, I. Transcription Control of Liver Development. Cells 2021, 10, 2026. https://doi.org/10.3390/cells10082026
Tachmatzidi EC, Galanopoulou O, Talianidis I. Transcription Control of Liver Development. Cells. 2021; 10(8):2026. https://doi.org/10.3390/cells10082026
Chicago/Turabian StyleTachmatzidi, Evangelia C., Ourania Galanopoulou, and Iannis Talianidis. 2021. "Transcription Control of Liver Development" Cells 10, no. 8: 2026. https://doi.org/10.3390/cells10082026
APA StyleTachmatzidi, E. C., Galanopoulou, O., & Talianidis, I. (2021). Transcription Control of Liver Development. Cells, 10(8), 2026. https://doi.org/10.3390/cells10082026





