Epigenetic Dysregulation of Mammalian Male Meiosis Caused by Interference of Recombination and Synapsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Mouse Genotypes
2.3. Spermatocyte Preparation
2.4. Immunofluorescence
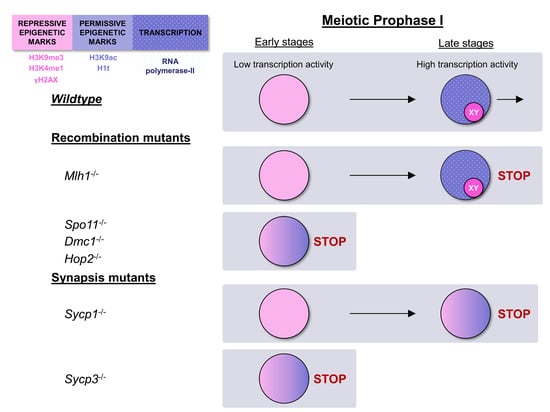
3. Results
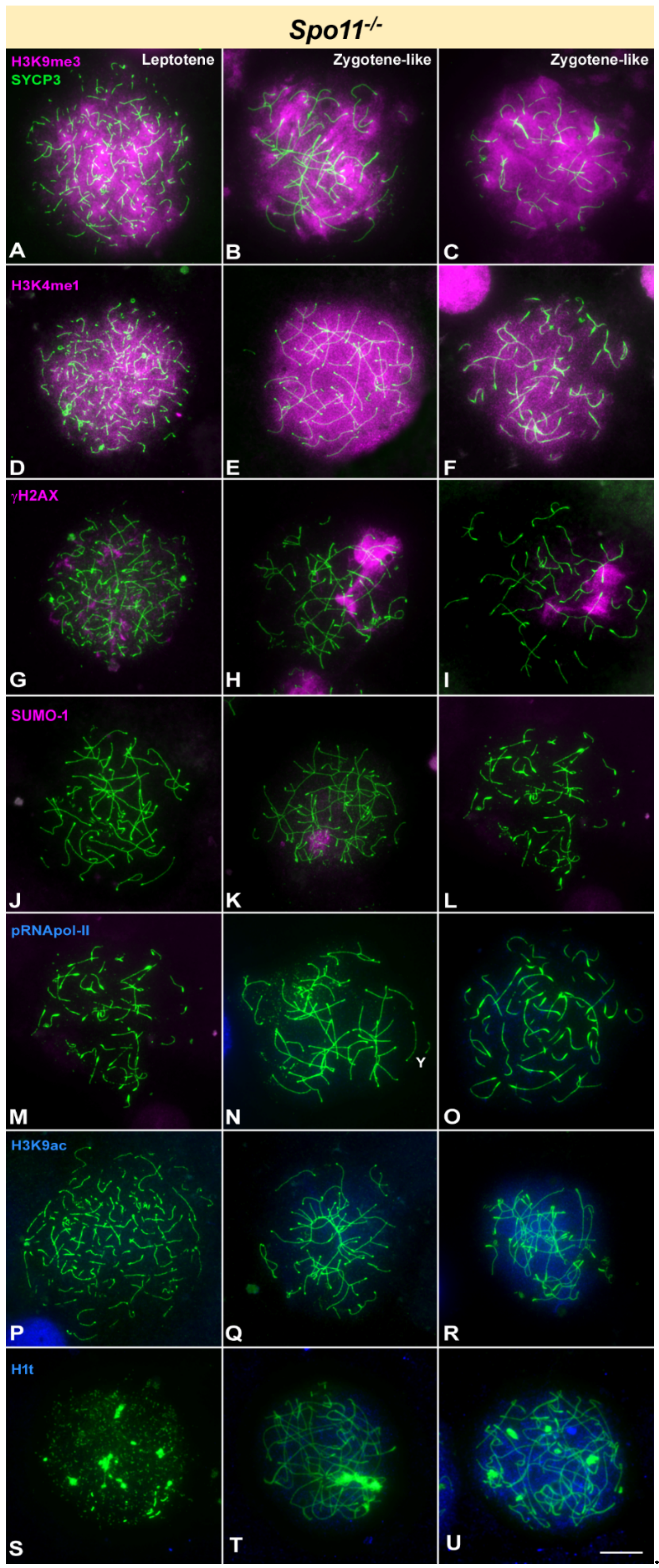
3.1. Epigenetic Markers in Recombination-Defective Mutants
3.2. Epigenetic Markers in Synapsis-Defective Mutants
4. Discussion
4.1. Epigenetic Signatures of Spermatocytes Change along Prophase-I
4.2. Early Epigenetic Signatures Are not Lost in Most Recombination and Synapsis Mutants and Overlap with Late Signatures
4.3. Epigenetic Progression and Inactivation of Sex Chromosomes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016626. [Google Scholar] [CrossRef] [Green Version]
- Fawcett, D.W. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J. Biophys. Biochem. Cytol. 1956, 2, 403–406. [Google Scholar] [CrossRef] [Green Version]
- Moses, M.J. Chromosomal structures in crayfish spermatocytes. J. Biophys. Biochem. Cytol. 1956, 2, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Colaiácovo, M.P. Zipping and Unzipping: Protein Modifications Regulating Synaptonemal Complex Dynamics. Trends Genet. 2018, 34, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Page, S.L.; Hawley, R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004, 20, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Von Wettstein, D.; Rasmussen, S.W.; Holm, P.B. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 1984, 18, 331–413. [Google Scholar] [CrossRef]
- Fraune, J.; Schramm, S.; Alsheimer, M.; Benavente, R. The mammalian synaptonemal complex: Protein components, assembly and role in meiotic recombination. Exp. Cell Res. 2012, 318, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Lammers, J.H.M.; Offenberg, H.H.; van Aalderen, M.; Vink, A.C.G.; Dietrich, A.J.J.; Heyting, C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell Biol. 1994, 14, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Meuwissen, R.L.J.; Offenberg, H.H.; Dietrich, A.J.J.; Riesewijk, A.; van Iersel, M.; Heyting, C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992, 11, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Lam, I.; Keeney, S. Mechanism and Regulation of Meiotic Recombination Initiation. Cold Spring Harb. Perspect. Biol. 2015, 7, a016634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grey, C.; de Massy, B. Chromosome Organization in Early Meiotic Prophase. Front. Cell Dev. Biol. 2021, 9, 688878. [Google Scholar] [CrossRef]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef]
- Shinohara, A.; Ogawa, H.; Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 1992, 69, 457–470. [Google Scholar] [CrossRef]
- Petukhova, G.V.; Pezza, R.J.; Vanevski, F.; Ploquin, M.; Masson, J.Y.; Camerini-Otero, R.D. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005, 12, 449–453. [Google Scholar] [CrossRef]
- Dernburg, A.F.; McDonald, K.; Moulder, G.; Barstead, R.; Dresser, M.; Villeneuve, A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 1998, 94, 387–398. [Google Scholar] [CrossRef] [Green Version]
- McKim, K.S.; Green-Marroquin, B.L.; Sekelsky, J.J.; Chin, G.; Steinberg, C.; Khodosh, R.; Hawley, R.S. Meiotic synapsis in the absence of recombination. Science 1998, 279, 876–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, S.M.; Plug, A.W.; Prolla, T.A.; Bronner, C.E.; Harris, A.C.; Yao, X.; Christie, D.M.; Monell, C.; Arnheim, N.; Bradley, A.; et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996, 13, 336–342. [Google Scholar] [CrossRef]
- Edelmann, W.; Cohen, P.E.; Kane, M.; Lau, K.; Morrow, B.; Bennett, S.; Umar, A.; Kunkel, T.; Cattoretti, G.; Chaganti, R.; et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell 1996, 85, 1125–1134. [Google Scholar] [CrossRef] [Green Version]
- Lipkin, S.M.; Moens, P.B.; Wang, V.; Lenzi, M.; Shanmugarajah, D.; Gilgeous, A.; Thomas, J.; Cheng, J.; Touchman, J.W.; Green, E.D.; et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002, 31, 385. [Google Scholar] [CrossRef]
- Patel, L.; Kang, R.; Rosenberg, S.C.; Qiu, Y.; Raviram, R.; Chee, S.; Hu, R.; Ren, B.; Cole, F.; Corbett, K.D. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct Mol. Biol. 2019, 26, 164–174. [Google Scholar] [CrossRef]
- Vara, C.; Paytuvi-Gallart, A.; Cuartero, Y.; Le Dily, F.; Garcia, F.; Salva-Castro, J.; Gomez, H.L.; Julia, E.; Moutinho, C.; Aiese Cigliano, R.; et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermatogenesis. Cell Rep. 2019, 28, 352–367. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, Y.; Du, Z.; Si, W.; Fan, S.; Qin, D.; Wang, M.; Duan, Y.; Li, L.; et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol. Cell 2019, 73, 547–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrios, S.; Manterola, M.; Prieto, Z.; Lopez-Fenner, J.; Page, J.; Fernandez-Donoso, R. Model of chromosome associations in Mus domesticus spermatocytes. Biol. Res. 2010, 43, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, N.K.; Siegel, E.P.; Alfert, M. Synthetic Activities During Spermatogenesis in the Locust. J. Cell Biol. 1965, 25, 387–395. [Google Scholar] [CrossRef]
- Monesi, V. Ribonucleic Acid Synthesis During Mitosis and Meiosis in the Mouse Testis. J. Cell Biol. 1964, 22, 521–532. [Google Scholar] [CrossRef]
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; Griswold, M.D. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004, 71, 319–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Cruz, I.; Rodríguez-Casuriaga, R.; Santiñaque, F.F.; Farías, J.; Curti, G.; Capoano, C.A.; Folle, G.A.; Benavente, R.; Sotelo-Silveira, J.R.; Geisinger, A. Transcriptome analysis of highly purified mouse spermatogenic cell populations: Gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genom. 2016, 17, 294. [Google Scholar] [CrossRef] [Green Version]
- Geisinger, A.; Rodríguez-Casuriaga, R.; Benavente, R. Transcriptomics of Meiosis in the Male Mouse. Front. Cell Dev. Biol. 2021, 9, 626020. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.-W.G.; Brick, K.; Cheng, G.; Pratto, F.; Camerini-Otero, R.D. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat. Commun. 2019, 10, 3821. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef] [Green Version]
- Page, J.; de la Fuente, R.; Manterola, M.; Parra, M.T.; Viera, A.; Berrios, S.; Fernandez-Donoso, R.; Rufas, J.S. Inactivation or non-reactivation: What accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 2012, 121, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.; Cargile, B.; Handel, M.A. Acquisition of Competence to Condense Metaphase I Chromosomes during Spermatogenesis. Dev. Biol. 1999, 205, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Moens, P.B. Histones H1 and H4 of surface-spread meiotic chromosomes. Chromosoma 1995, 104, 169–174. [Google Scholar] [CrossRef]
- Pradeepa, M.M.; Rao, M.R. Chromatin remodeling during mammalian spermatogenesis: Role of testis specific histone variants and transition proteins. Soc. Reprod. Fertil. Suppl. 2007, 63, 1–10. [Google Scholar]
- Kota, S.K.; Feil, R. Epigenetic Transitions in Germ Cell Development and Meiosis. Dev. Cell 2010, 19, 675–686. [Google Scholar] [CrossRef] [Green Version]
- Tatehana, M.; Kimura, R.; Mochizuki, K.; Inada, H.; Osumi, N. Comprehensive histochemical profiles of histone modification in male germline cells during meiosis and spermiogenesis: Comparison of young and aged testes in mice. PLoS ONE 2020, 15, e0230930. [Google Scholar] [CrossRef] [Green Version]
- Van der Heijden, G.W.; Derijck, A.A.; Posfai, E.; Giele, M.; Pelczar, P.; Ramos, L.; Wansink, D.G.; van der Vlag, J.; Peters, A.H.; de Boer, P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007, 39, 251–258. [Google Scholar] [CrossRef]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; de Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borde, V.; Robine, N.; Lin, W.; Bonfils, S.; Geli, V.; Nicolas, A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009, 28, 99–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, S.; Bowden, R.; Tumian, A.; Bontrop, R.E.; Freeman, C.; MacFie, T.S.; McVean, G.; Donnelly, P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 2010, 327, 876–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parvanov, E.D.; Petkov, P.M.; Paigen, K. Prdm9 controls activation of mammalian recombination hotspots. Science 2010, 327, 835. [Google Scholar] [CrossRef] [Green Version]
- Mahadevaiah, S.K.; Turner, J.M.; Baudat, F.; Rogakou, E.P.; de Boer, P.; Blanco-Rodriguez, J.; Jasin, M.; Keeney, S.; Bonner, W.M.; Burgoyne, P.S. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001, 27, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.C.; Hsieh, C.L.; Liu, N.; Wang, J.; Zhong, M.; Chen, T.; Li, E.; Lin, H. The Essential Function of SETDB1 in Homologous Chromosome Pairing and Synapsis during Meiosis. Cell Rep. 2021, 34, 108575. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Meydan, C.; Lange, J.; Claeys Bouuaert, C.; Lailler, N.; Mason, C.E.; Anderson, K.V.; Keeney, S. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet. 2017, 13, e1006964. [Google Scholar] [CrossRef] [Green Version]
- Lascarez-Lagunas, L.I.; Herruzo, E.; Grishok, A.; San-Segundo, P.A.; Colaiácovo, M.P. DOT-1.1-dependent H3K79 methylation promotes normal meiotic progression and meiotic checkpoint function in C. elegans. PLoS Genet. 2020, 16, e1009171. [Google Scholar] [CrossRef]
- Peters, A.H.; O’Carroll, D.; Scherthan, H.; Mechtler, K.; Sauer, S.; Schofer, C.; Weipoltshammer, K.; Pagani, M.; Lachner, M.; Kohlmaier, A.; et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001, 107, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Hirota, T.; Blakeley, P.; Sangrithi, M.N.; Mahadevaiah, S.K.; Encheva, V.; Snijders, A.P.; ElInati, E.; Ojarikre, O.A.; de Rooij, D.G.; Niakan, K.K.; et al. SETDB1 Links the Meiotic DNA Damage Response to Sex Chromosome Silencing in Mice. Dev. Cell 2018, 47, 646–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, J.A.; Wakefield, M.J.; Toder, R. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum. Mol. Genet. 1998, 7, 1991–1996. [Google Scholar] [CrossRef] [Green Version]
- Solari, A.J. The behavior of the XY pair in mammals. Int. Rev. Cytol. 1974, 38, 273–317. [Google Scholar]
- Handel, M.A. The XY body: A specialized meiotic chromatin domain. Exp. Cell Res. 2004, 296, 57–63. [Google Scholar] [CrossRef]
- Handel, M.A.; Hunt, P.A. Sex-chromosome pairing and activity during mammalian meiosis. Bioessays 1992, 14, 817–822. [Google Scholar] [CrossRef]
- Royo, H.; Polikiewicz, G.; Mahadevaiah, S.K.; Prosser, H.; Mitchell, M.; Bradley, A.; de Rooij, D.G.; Burgoyne, P.S.; Turner, J.M. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 2010, 20, 2117–2123. [Google Scholar] [CrossRef] [Green Version]
- Vernet, N.; Mahadevaiah, S.K.; de Rooij, D.G.; Burgoyne, P.S.; Ellis, P.J.I. Zfy genes are required for efficient meiotic sex chromosome inactivation (MSCI) in spermatocytes. Hum. Mol. Genet. 2016, 25, 5300–5310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchi, M.; Mahadevaiah, S.; Di Giacomo, M.; Baudat, F.; de Rooij, D.G.; Burgoyne, P.S.; Jasin, M.; Keeney, S. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell Biol. 2005, 25, 7203–7215. [Google Scholar] [CrossRef] [Green Version]
- Bellani, M.A.; Romanienko, P.J.; Cairatti, D.A.; Camerini-Otero, R.D. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 2005, 118, 3233–3245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, U.; Wetzker, C.; Lange, J.; Christodoulou, E.G.; Seifert, M.; Beyer, A.; Jessberger, R. Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions. PLoS Genet. 2013, 9, e1003985. [Google Scholar] [CrossRef] [PubMed]
- De Vries, F.A.; de Boer, E.; van den Bosch, M.; Baarends, W.M.; Ooms, M.; Yuan, L.; Liu, J.G.; van Zeeland, A.A.; Heyting, C.; Pastink, A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005, 19, 1376–1389. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Viera, A.; Rufas, J.S.; Martinez, I.; Barbero, J.L.; Ortega, S.; Suja, J.A. CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 2009, 122, 2149–2159. [Google Scholar] [CrossRef] [Green Version]
- Hamer, G.; Novak, I.; Kouznetsova, A.; Höög, C. Disruption of pairing and synapsis of chromosomes causes stage-specific apoptosis of male meiotic cells. Theriogenology 2008, 69, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Handel, M.A.; Schimenti, J.C. Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010, 11, 124–136. [Google Scholar] [CrossRef]
- Fine, A.D.; Ball, R.L.; Fujiwara, Y.; Handel, M.A.; Carter, G.W. Uncoupling of transcriptomic and cytological differentiation in mouse spermatocytes with impaired meiosis. Mol. Biol. Cell 2019, 30, 717–728. [Google Scholar] [CrossRef]
- Baudat, F.; Manova, K.; Yuen, J.P.; Jasin, M.; Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 2000, 6, 989–998. [Google Scholar] [CrossRef]
- Petukhova, G.V.; Romanienko, P.J.; Camerini-Otero, R.D. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 2003, 5, 927–936. [Google Scholar] [CrossRef] [Green Version]
- Romanienko, P.J.; Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 2000, 6, 975–987. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, J.G.; Zhao, J.; Brundell, E.; Daneholt, B.; Hoog, C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 2000, 5, 73–83. [Google Scholar] [CrossRef]
- Pittman, D.L.; Cobb, J.; Schimenti, K.J.; Wilson, L.A.; Cooper, D.M.; Brignull, E.; Handel, M.A.; Schimenti, J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1998, 1, 697–705. [Google Scholar] [CrossRef]
- Peters, A.H.; Plug, A.W.; van Vugt, M.J.; de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997, 5, 66–68. [Google Scholar] [CrossRef]
- Kouznetsova, A.; Novak, I.; Jessberger, R.; Höög, C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J. Cell Sci. 2005, 118, 2271–2278. [Google Scholar] [CrossRef] [PubMed]
- Page, J.; Berrios, S.; Rufas, J.S.; Parra, M.T.; Suja, J.A.; Heyting, C.; Fernandez-Donoso, R. The pairing of X and Y chromosomes during meiotic prophase in the marsupial species Thylamys elegans is maintained by a dense plate developed from their axial elements. J. Cell Sci. 2003, 116, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.M.; Aprelikova, O.; Xu, X.; Wang, R.; Kim, S.; Chandramouli, G.V.; Barrett, J.C.; Burgoyne, P.S.; Deng, C.X. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 2004, 14, 2135–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manterola, M.; Page, J.; Vasco, C.; Berrios, S.; Parra, M.T.; Viera, A.; Rufas, J.S.; Zuccotti, M.; Garagna, S.; Fernandez-Donoso, R. A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet. 2009, 5, e1000625. [Google Scholar] [CrossRef] [Green Version]
- Jenuwein, T.; Allis, C. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowell, I.G.; Aucott, R.; Mahadevaiah, S.K.; Burgoyne, P.S.; Huskisson, N.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; Wu, R.; et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 2002, 111, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Hublitz, P.; Albert, M.; Peters, A. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 2009, 53, 335–354. [Google Scholar] [CrossRef]
- Goetz, P.; Chandley, A.C.; Speed, R.M. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J. Cell Sci. 1984, 65, 249–263. [Google Scholar] [CrossRef]
- Ashley, T.; Gaeth, A.P.; Creemers, L.B.; Hack, A.M.; de Rooij, D.G. Correlation of meiotic events in testis sections and microspreads of mouse spermatocytes relative to the mid-pachytene checkpoint. Chromosoma 2004, 113, 126–136. [Google Scholar] [CrossRef]
- Pelttari, J.; Hoja, M.R.; Yuan, L.; Liu, J.G.; Brundell, E.; Moens, P.; Santucci-Darmanin, S.; Jessberger, R.; Barbero, J.L.; Heyting, C.; et al. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell Biol. 2001, 21, 5667–5677. [Google Scholar] [CrossRef] [Green Version]
- Almstrup, K.; Nielsen, J.E.; Hansen, M.A.; Tanaka, M.; Skakkebaek, N.E.; Leffers, H. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol. Reprod. 2004, 70, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Ontoso, D.; Kauppi, L.; Keeney, S.; San-Segundo, P.A. Dynamics of DOT1L localization and H3K79 methylation during meiotic prophase I in mouse spermatocytes. Chromosoma 2014, 123, 147–164. [Google Scholar] [CrossRef] [Green Version]
- Smirnova, N.A.; Romanienko, P.J.; Khil, P.P.; Camerini-Otero, R.D. Gene expression profiles of Spo11-/- mouse testes with spermatocytes arrested in meiotic prophase I. Reproduction 2006, 132, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Godmann, M.; Auger, V.; Ferraroni-Aguiar, V.; Di Sauro, A.; Sette, C.; Behr, R.; Kimmins, S. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol. Reprod. 2007, 77, 754–764. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, K.; Marley, K.E.; Jeong, B.-r.; Xu, J.; Hesson, J.; Cerny, R.L.; Waterborg, J.H.; Cerutti, H. Monomethyl Histone H3 Lysine 4 as an Epigenetic Mark for Silenced Euchromatin in Chlamydomonas. Plant. Cell Online 2005, 17, 2439–2453. [Google Scholar] [CrossRef] [Green Version]
- Gaysinskaya, V.; Miller, B.F.; De Luca, C.; van der Heijden, G.W.; Hansen, K.D.; Bortvin, A. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenet. Chromatin 2018, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Alavattam, K.G.; Hu, Y.-C.; Pang, Q.; Andreassen, P.R.; Hegde, R.S.; Namekawa, S.H. The Initiation of Meiotic Sex Chromosome Inactivation Sequesters DNA Damage Signaling from Autosomes in Mouse Spermatogenesis. Curr. Biol. 2020, 30, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Baarends, W.M.; Grootegoed, J.A. Chromatin dynamics in the male meiotic prophase. Cytogenet Genome Res. 2003, 103, 225–234. [Google Scholar] [CrossRef]
- Baarends, W.M.; Wassenaar, E.; van der Laan, R.; Hoogerbrugge, J.; Sleddens-Linkels, E.; Hoeijmakers, J.H.; de Boer, P.; Grootegoed, J.A. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell Biol. 2005, 25, 1041–1053. [Google Scholar] [CrossRef] [Green Version]
- Burgoyne, P.S.; Mahadevaiah, S.K.; Turner, J.M. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 2009, 10, 207–216. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Mahadevaiah, S.K.; Celeste, A.; Romanienko, P.J.; Camerini-Otero, R.D.; Bonner, W.M.; Manova, K.; Burgoyne, P.; Nussenzweig, A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 2003, 4, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Ichijima, Y.; Ichijima, M.; Lou, Z.; Nussenzweig, A.; Camerini-Otero, R.D.; Chen, J.; Andreassen, P.R.; Namekawa, S.H. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011, 25, 959–971. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, A.; Schoenmakers, S.; Baarends, W.M. DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 2010, 5, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.M. Meiotic sex chromosome inactivation. Development 2007, 134, 1823–1831. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.M. Meiotic Silencing in Mammals. Annu. Rev. Genet. 2015, 49, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Sosa, E.; Flores, L.; Yan, W.; McCarrey, J.R. Escape of X-linked miRNA genes from meiotic sex chromosome inactivation. Development 2015, 142, 3791–3800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acquaviva, L.; Boekhout, M.; Karasu, M.E.; Brick, K.; Pratto, F.; Li, T.; van Overbeek, M.; Kauppi, L.; Camerini-Otero, R.D.; Jasin, M.; et al. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 2020, 582, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Kauppi, L.; Barchi, M.; Baudat, F.; Romanienko, P.J.; Keeney, S.; Jasin, M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 2011, 331, 916–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Fuente, R.; Pratto, F.; Hernández-Hernández, A.; Manterola, M.; López-Jiménez, P.; Gómez, R.; Viera, A.; Parra, M.T.; Kouznetsova, A.; Camerini-Otero, R.D.; et al. Epigenetic Dysregulation of Mammalian Male Meiosis Caused by Interference of Recombination and Synapsis. Cells 2021, 10, 2311. https://doi.org/10.3390/cells10092311
de la Fuente R, Pratto F, Hernández-Hernández A, Manterola M, López-Jiménez P, Gómez R, Viera A, Parra MT, Kouznetsova A, Camerini-Otero RD, et al. Epigenetic Dysregulation of Mammalian Male Meiosis Caused by Interference of Recombination and Synapsis. Cells. 2021; 10(9):2311. https://doi.org/10.3390/cells10092311
Chicago/Turabian Stylede la Fuente, Roberto, Florencia Pratto, Abrahan Hernández-Hernández, Marcia Manterola, Pablo López-Jiménez, Rocío Gómez, Alberto Viera, María Teresa Parra, Anna Kouznetsova, R. Daniel Camerini-Otero, and et al. 2021. "Epigenetic Dysregulation of Mammalian Male Meiosis Caused by Interference of Recombination and Synapsis" Cells 10, no. 9: 2311. https://doi.org/10.3390/cells10092311