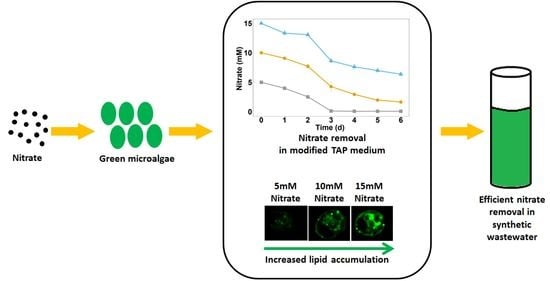
Assessment of Nitrate Removal Capacity of Two Selected Eukaryotic Green Microalgae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microalgae Strains and Growth Media
2.2. Growth Parameters
2.3. Nitrate Determination by the Salicylic Acid Method
2.4. Determination of Reactive Oxygen Species (ROS)
2.5. Total Pigments Extraction and Quantification
2.6. Total Carbohydrates Extraction and Quantification
2.7. Total Proteins Extraction and Quantification
2.8. Total Lipids Extraction and Quantification
2.9. Confocal Microscopy with BODIPY Dye
2.10. Synthetic Wastewater Treatment
2.11. Statistical Analyses
3. Results
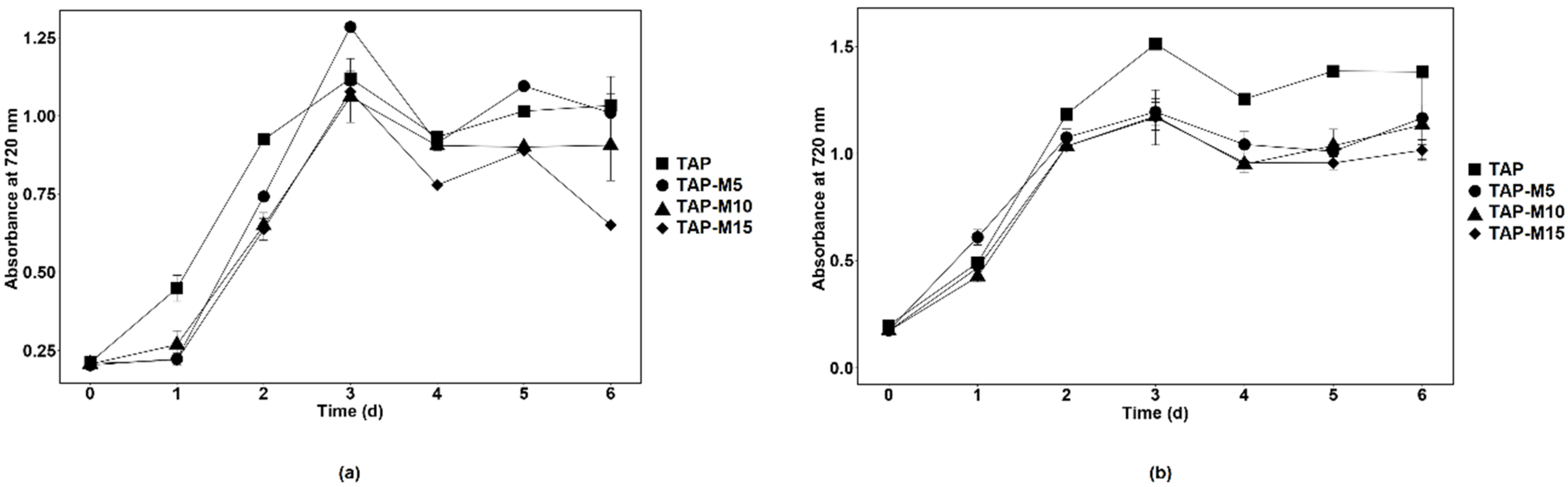
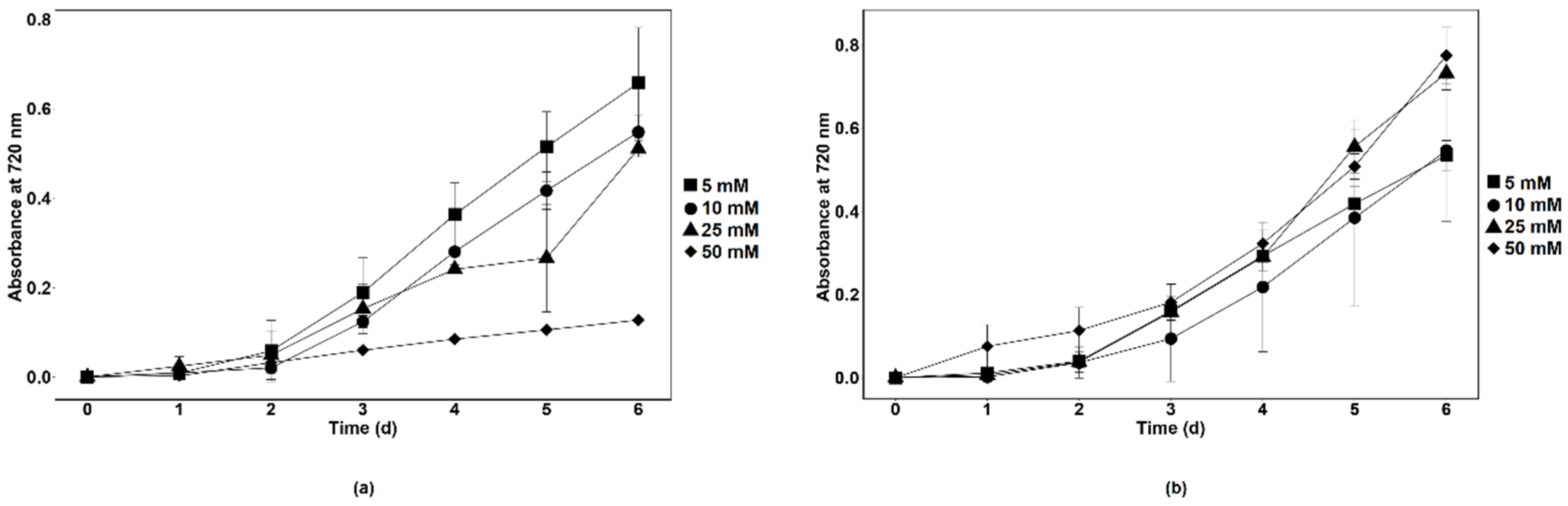
3.1. Influence of Nitrate on the Growth Parameters of Chlamydomonas sp. MACC-216 and Chlorella sp. MACC-360
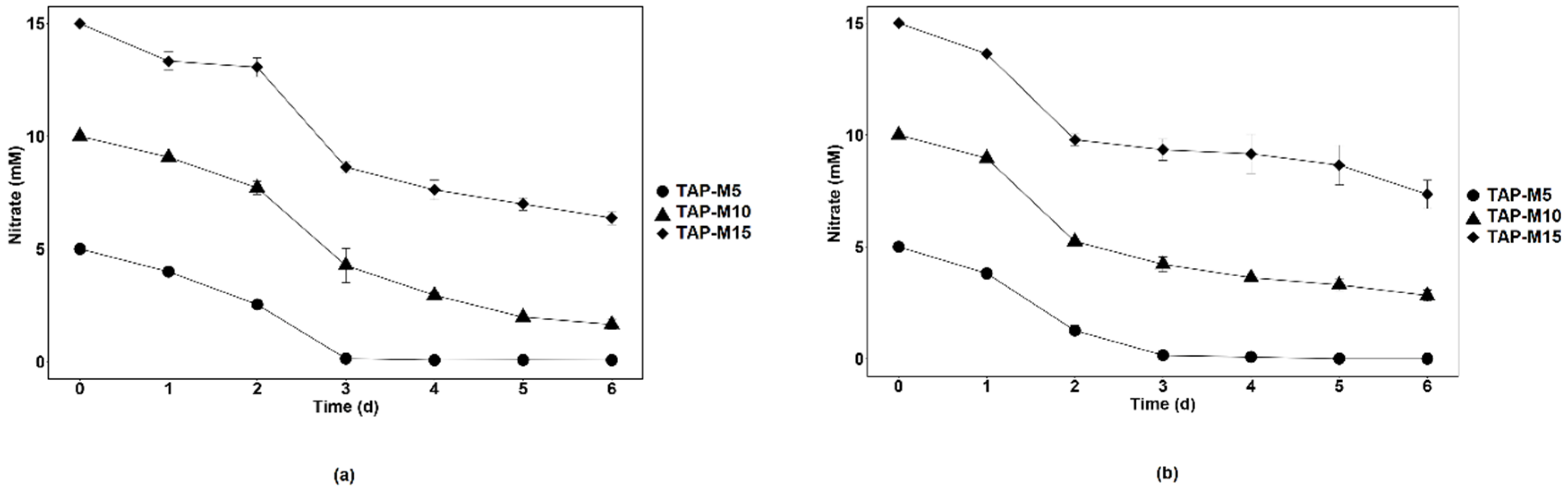
3.2. Nitrate Removal by Chlamydomonas sp. MACC-216 and Chlorella sp. MACC-360
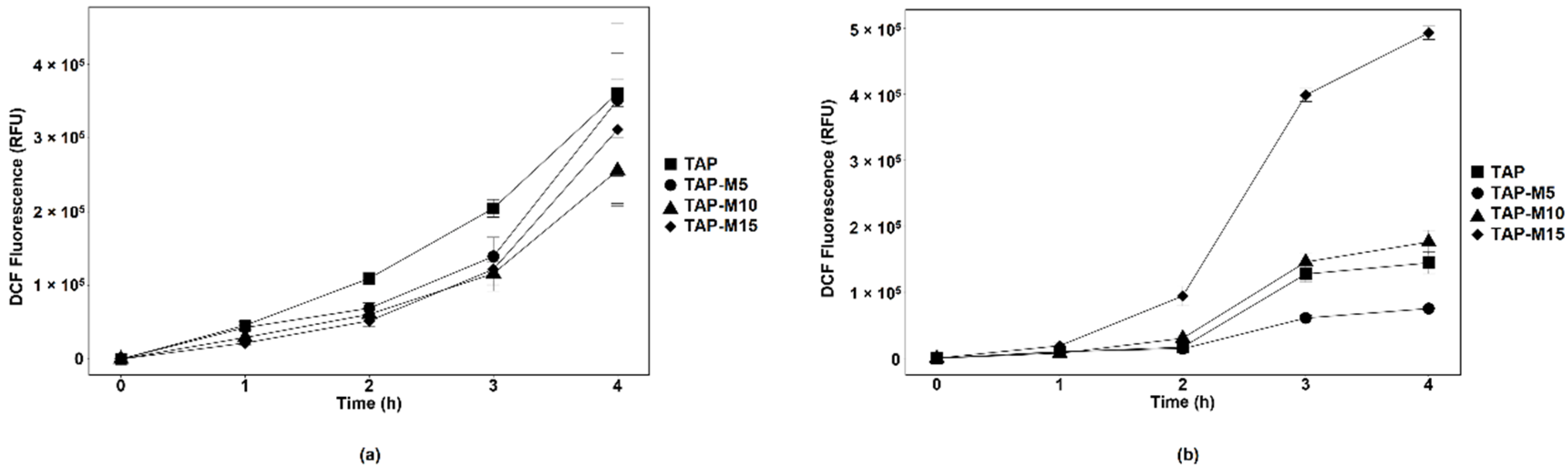
3.3. Nitrate Led to ROS Production in Chlorella sp. MACC-360
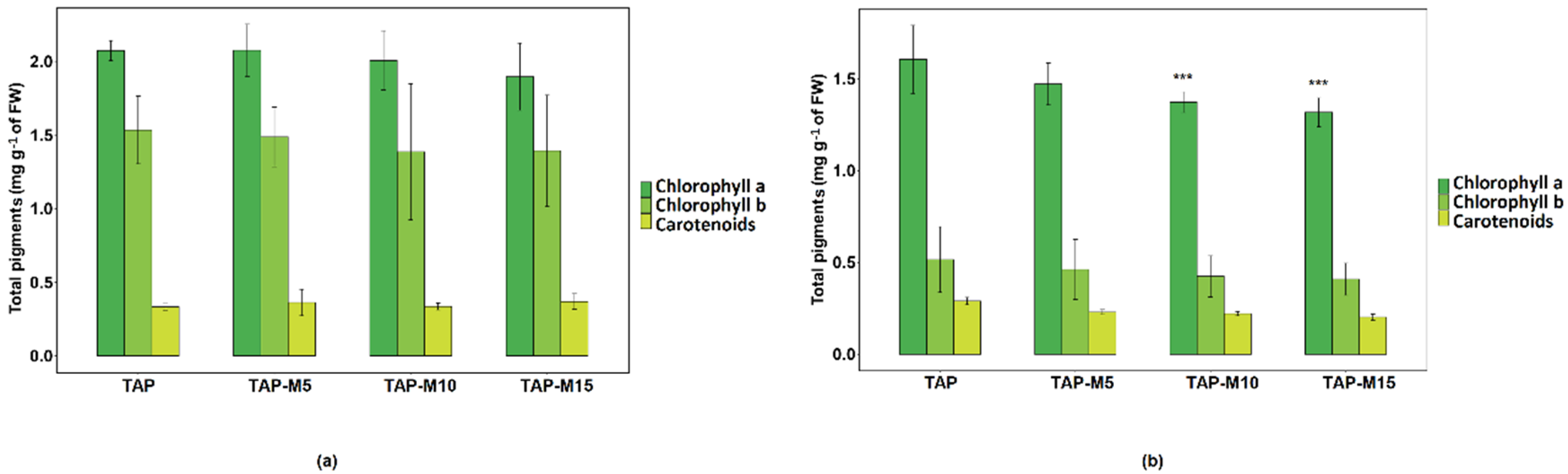
3.4. Nitrate Affected Total Pigments Production
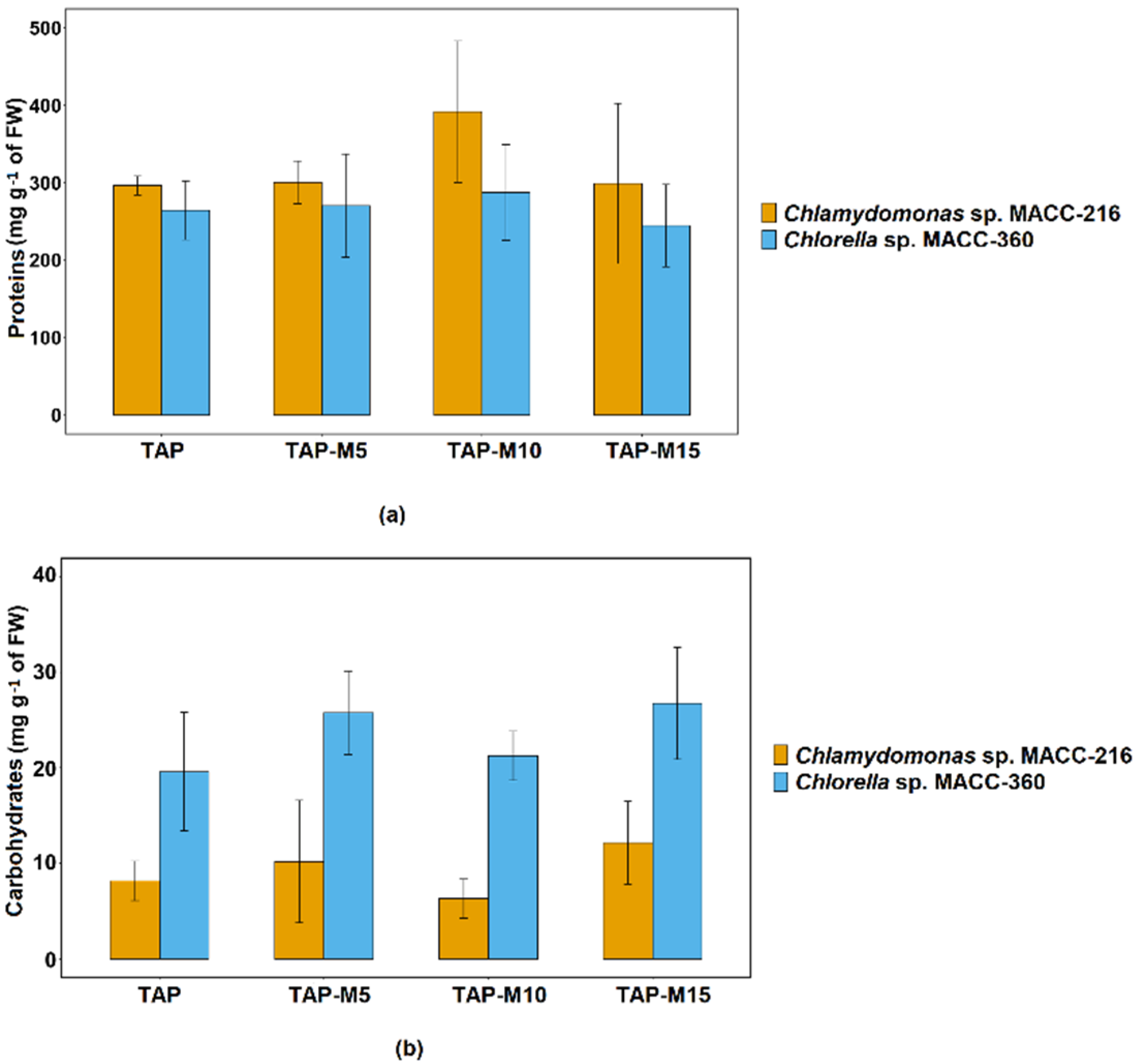
3.5. Effects of Nitrate on Total Protein and Carbohydrate Contents
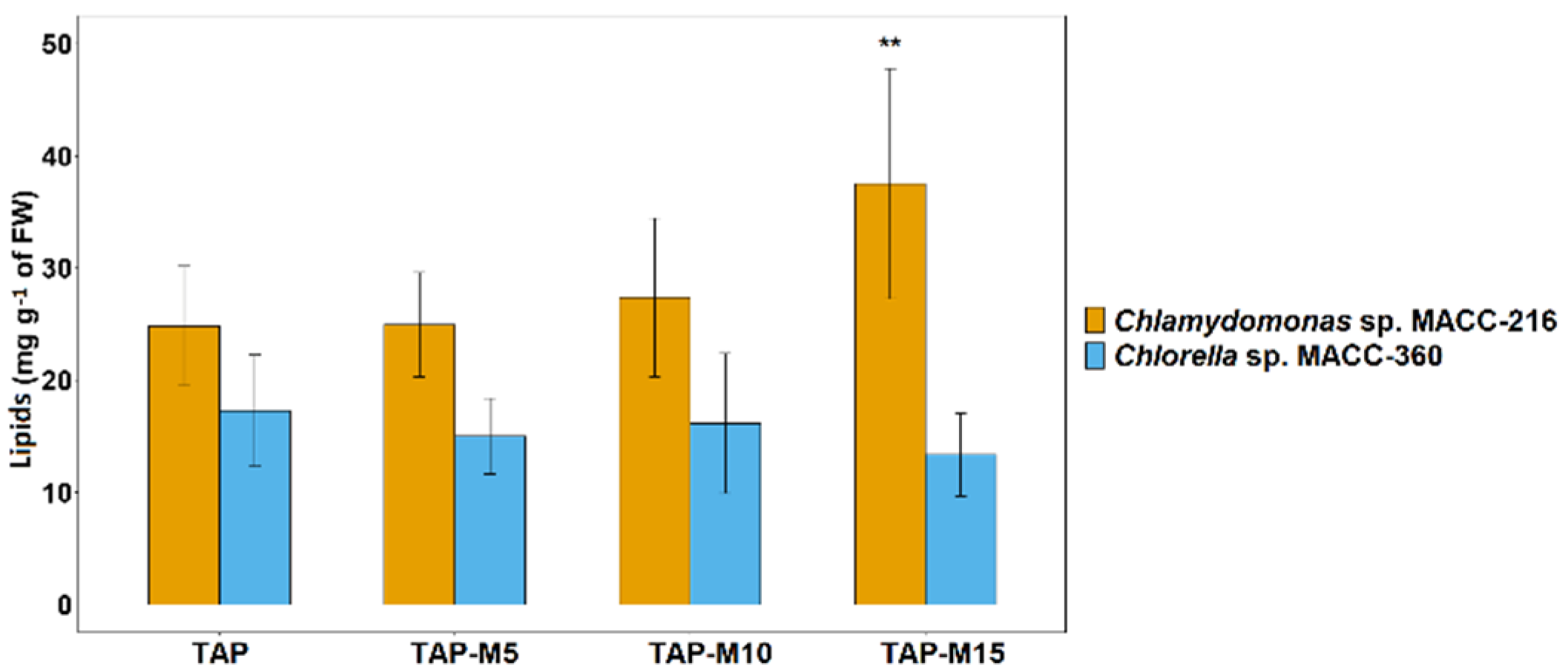
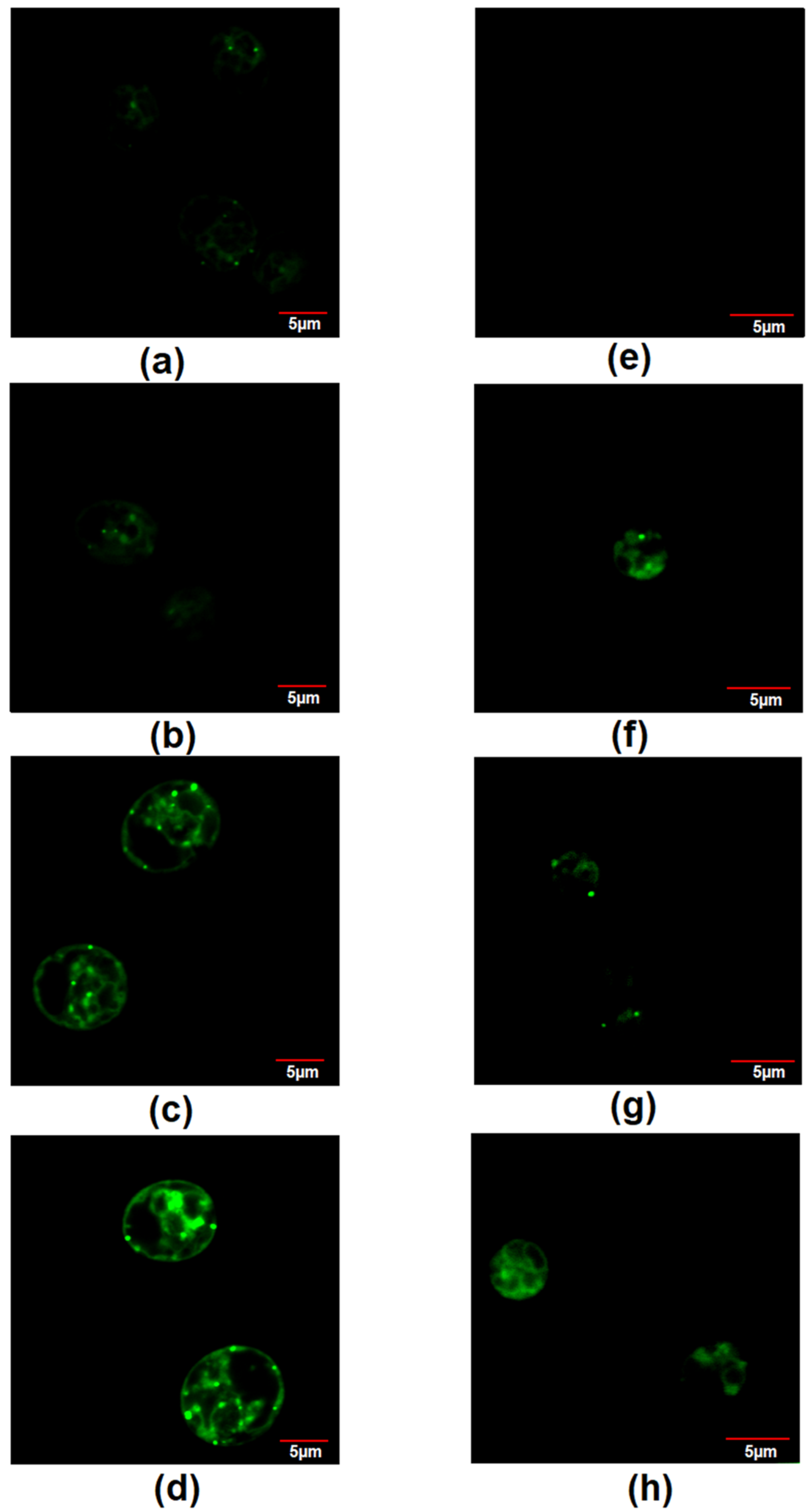
3.6. Lipid Content Increased by Nitrate in Chlamydomonas sp. MACC-216
3.7. Chlorella sp. MACC-360 and Chlamydomonas sp. MACC-216 Efficiently Removed Nitrate from Synthetic Wastewater
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohensi-Bandpi, A.; Elliot, D.J.; Zazouli, M.A. Biological nitrate removal processes from drinking water supply—A review. J. Environ. Health Sci. Eng. 2013, 11, 35. [Google Scholar] [CrossRef]
- Moss, B.; Kosten, S.; Meerhoff, M.; Battarbee, R.W.; Jeppesen, E.; Mazzeo, N.; Havens, K.; Lacerot, G.; Liu, Z.; de Meester, L. Allied attack: Climate change and eutrophication. Inland Waters 2011, 1, 101–105. [Google Scholar] [CrossRef] [Green Version]
- Maberly, S.C.; Pitt, J.-A.; Davies, P.S.; Carvalho, L. Nitrogen and phosphorus limitation and the management of small productive lakes. Inland Waters 2020, 10, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Grizzetti, B.; Bouraoui, F.; Billen, G.; Grinsven, H.V.; Cardoso, A.C.; Thieu, V.; Garnier, J.; Curtis, C.; Howarth, R.; Johnes, P. Nitrogen as a threat to European water quality. In The European Nitrogen Assessment; Sutton, M.A., Howard, C.M., Erisman, J.W., Billen, G., Bleeker, A., Grennfelt, P., Grinsven, H.V., Grizzetti, B., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 379–404. [Google Scholar] [CrossRef] [Green Version]
- Lewis, L.A.; Lewis, P.O. Unearthing the molecular phylodiversity of desert soil green algae (Chlorophyta). Syst. Biol. 2005, 54, 936–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foflonker, F.; Price, D.C.; Qiu, H.; Palenik, B.; Wang, S.; Bhattacharya, D. Genome of halotolerant green alga Picochlorum sp. reveals strategies for thriving under fluctuating environmental conditions. Environ. Microbiol. 2015, 17, 412–426. [Google Scholar] [CrossRef]
- Su, Y.; Mennerich, A.; Urban, B. Comparison of nutrient removal capacity and biomass settleability of four high-potential microalgal species. Bioresour. Technol. 2012, 124, 157–162. [Google Scholar] [CrossRef]
- Prasad, M.S.V.; Varma, A.K.; Kumari, P.; Mondal, P. Production of lipid containing microalgal biomass and simultaneous removal of nitrate and phosphate from synthetic wastewater. Environ. Technol. 2017, 39, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Munoz, R.; Guieysse, B. Algal–bacterial processes for the treatment of hazardous contaminants: A review. Water Res. 2006, 40, 2799–2815. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, A.C.; Mulbry, W.W. Recovery of dairy manure nutrients by benthic freshwater algae. Bioresour. Technol. 2002, 84, 81–91. [Google Scholar] [CrossRef]
- Ogbonna, J.C.; Yoshizawa, H.; Tanaka, H. Treatment of high strength organic wastewater by a mixed culture of photosynthetic microorganisms. J. Appl. Phycol. 2000, 12, 277–284. [Google Scholar] [CrossRef]
- Xin, L.; Hong-ying, H.; Ke, G.; Ying-xue, S. Effects of different nitrogen and phosphorus concentrations on the growth, nutrient uptake, and lipid accumulation of a freshwater microalga Scenedesmus sp. Bioresour. Technol. 2010, 101, 5494–5500. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Bai, F.W.; Zhao, X.Q. Effects of Nitrogen Concentration and Media Replacement on Cell Growth and Lipid Production of Oleaginous Marine Microalga Nannochloropsis oceanica DUT01. Biochem. Eng. J. 2013, 78, 32–38. [Google Scholar] [CrossRef]
- Paes, C.R.P.S.; Faria, G.R.; Tinoco, N.A.B.; Castro, D.J.F.A.; Barbarino, E.; Lourenço, S.O. Growth, nutrient uptake and chemical composition of Chlorella sp. and Nannochloropsis oculata under nitrogen starvation. Lat. Am. J. Aquat. Res. 2016, 44, 275–292. [Google Scholar] [CrossRef]
- Kong, Q.X.; Li, L.; Martinez, B.; Chen, P.; Ruan, R. Culture of microalgae Chlamydomonas reinhardtii in wastewater for biomass feedstock production. Appl. Biochem. Biotechnol. 2010, 160, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lan, C.Q. Biomass production and nitrogen and phosphorus removal by the green alga Neochloris oleoabundans in simulated wastewater and secondary municipal wastewater effluent. Bioresour. Technol. 2011, 102, 5639–5644. [Google Scholar] [CrossRef] [PubMed]
- Higgins, B.; Gennity, I.; Fitzgerald, P.; Ceballos, S.; Fiehn, O.; VanderGheynst, J. Algal–bacterial synergy in treatment of winery wastewater. NPJ Clean Water 2018, 1, 6. [Google Scholar] [CrossRef]
- Philipps, G.; Happe, T.; Hemschemeier, A. Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 2012, 235, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Pancha, I.; Chokshi, K.; George, B.; Ghosh, T.; Paliwal, C.; Maurya, R.; Mishra, S. Nitrogen stress triggered biochemical and morphological changes in the microalgae Scenedesmus sp. CCNM 1077. Bioresour. Technol. 2014, 156, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Araujo, G.S.; Silva, J.W.A.; Viana, C.A.S.; Fernandes, F.A.N. Effect of sodium nitrate concentration on biomass and oil production of four microalgae species. Int. J. Sustain. Energy 2019, 39, 41–50. [Google Scholar] [CrossRef]
- Gour, R.S.; Bairagi, M.; Garlapati, V.K.; Kant, A. Enhanced microalgal lipid production with media engineering of potassium nitrate as a nitrogen source. Bioengineered 2018, 9, 98–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiran, B.; Pathak, K.; Kumar, R.; Deshmukh, D.; Rani, N. Influence of varying nitrogen levels on lipid accumulation in Chlorella sp. Int. J. Environ. Sci. Technol. 2016, 13, 1823–1832. [Google Scholar] [CrossRef] [Green Version]
- Zarrinmehr, M.J.; Farhadian, O.; Heyrati, F.P.; Keramat, J.; Koutra, E.; Kornaros, M.; Daneshvar, E. Effect of nitrogen concentration on the growth rate and biochemical composition of the microalga, Isochrysis galbana. Egypt. J. Aquat. Res. 2020, 46, 153–158. [Google Scholar] [CrossRef]
- Sánchez-García, D.; Resendiz-Isidro, A.; Villegas-Garrido, T.L.; Flores-Ortiz, C.M.; Chávez-Gómez, B.; Cristiani-Urbina, E. Effect of nitrate on lipid production by T. suecica, M. contortum, and C. minutissima. Cent. Eur. J. Biol. 2013, 8, 578–590. [Google Scholar] [CrossRef]
- Cabello-Pasini, A.; Figueroa, F.L. Effect of nitrate concentration on the relationship between photosynthetic oxygen evolution and electron transport rate in Ulva rigida (Chlorophyta). J. Phycol. 2005, 41, 1169–1177. [Google Scholar] [CrossRef]
- Li, Y.; Horsman, M.; Wang, B.; Wu, N.; Lan, C.Q. Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl. Microbiol. Biotechnol. 2008, 81, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, J.; Parmar, V.; Vavilala, S.L. Nitrate stress-induced bioactive sulfated polysaccharides from Chlamydomonas reinhardtii. Biomed. Res. J. 2019, 6, 7–16. [Google Scholar] [CrossRef]
- Patel, V.K.; Sundaram, S.; Patel, A.K.; Kalra, A. Characterization of seven species of Cyanobacteria for high-quality biomass production. Arab. J. Sci. Eng. 2018, 43, 109–121. [Google Scholar] [CrossRef]
- Cataldo, D.A.; Haroon, M.; Schrader, L.E.; Youngs, V.L. Rapid colorimetric determination of nitrate in plant tissue by nitration in salicylic acid. Commun. Soil. Sci. Plant Anal. 1975, 6, 71–80. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Liu, S.; Liu, B.; Gao, Y.; Wu, Z. Generation of reactive oxygen species in cyanobacteria and green algae induced by allelochemicals of submerged macrophytes. Chemosphere 2011, 85, 977–982. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.L. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- OECD. Test No. 209: Activated Sludge, Respiration Inhibition Test (Carbon and Ammonium Oxidation), OECD Guidelines for the Testing of Chemicals, Section 2; OECD Publishing: Paris, France, 2010. [Google Scholar]
- Jeanfils, J.; Canisius, M.-F.; Burlion, N. Effect of high nitrate concentrations on growth and nitrate uptake by free-living and immobilized Chlorella vulgaris cells. J. Appl. Phycol. 1993, 5, 369–374. [Google Scholar] [CrossRef]
- Sayadi, M.H.; Ahmadpour, N.; Fallahi Capoorchali, M.; Rezaei, M.R. Removal of nitrate and phosphate from aqueous solutions by microalgae: An experimental study. Glob. J. Environ. Sci. Manag. 2016, 2, 357–364. [Google Scholar] [CrossRef]
- Fernandez, E.; Galvan, A. Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 2007, 58, 2279–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, E.; Galvan, A. Nitrate assimilation in Chlamydomonas. Eukaryotic Cell. 2008, 7, 555–559. [Google Scholar] [CrossRef] [Green Version]
- Bellido-Pedraza, C.M.; Calatrava, V.; Sanz-Luque, E.; Tejada-Jiménez, M.; Llamas, Á.; Plouviez, M.; Guieysse, B.; Fernández, E.; Galván, A. Chlamydomonas reinhardtii, an Algal Model in the Nitrogen Cycle. Plants 2020, 9, 903. [Google Scholar] [CrossRef] [PubMed]
- Galván, A.; Fernández, E. Eukaryotic nitrate and nitrite transporters. Cell. Mol. Life Sci. 2001, 58, 225–233. [Google Scholar] [CrossRef]
- Tischner, R.; Lorenzen, H. Nitrate uptake and nitrate reduction in synchronous Chlorella. Planta 1979, 146, 287–292. [Google Scholar] [CrossRef]
- Ugya, A.Y.; Imam, T.; Li, A.; Ma, J.; Hua, X. Antioxidant response mechanism of freshwater microalgae species to reactive oxygen species production: A mini review. Chem. Ecol. 2020, 36, 174–193. [Google Scholar] [CrossRef]
- Mallick, N.; Mohn, F.H. (2000). Reactive oxygen species: Response of algal cells. J. Plant Physiol. 2000, 157, 183–193. [Google Scholar] [CrossRef]
- Çelekli, A.; Balcı, M. The influence of different phosphate and nitrate concentrations on growth, protein and chlorophyll a content of Scenedesmus obliquus. Fresenius Environ. Bull. 2009, 18, 1363–1366. [Google Scholar]
- Xie, T.; Xia, Y.; Zeng, Y.; Li, X.; Zhang, Y. Nitrate concentration-shift cultivation to enhance protein content of heterotrophic microalga Chlorella vulgaris: Over- compensation strategy. Bioresour. Technol. 2017, 233, 247–255. [Google Scholar] [CrossRef]
- Rückert, G.V.; Giani, A. Effect of nitrate and ammonium on the growth and protein concentration of Microcystis viridis Lemmermann (Cyanobacteria). Rev. Bras. Bot. 2004, 27, 325–331. [Google Scholar] [CrossRef] [Green Version]
- Brányiková, I.; Maršálková, B.; Doucha, J.; Brányik, T.; Bišová, K.; Zachleder, V.; Vítová, M. Microalgae—novel highly efficient starch producers. Biotechnol. Bioeng. 2011, 108, 766–776. [Google Scholar] [CrossRef]
- Morsy, F.M. Acetate versus sulfur deprivation role in creating anaerobiosis in light for hydrogen production by Chlamydomonas reinhardtii and Spirulina platensis: Two different organisms and two different mechanisms. Photochem. Photobiol. 2011, 87, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Converti, A.; Casazza, A.A.; Ortiz, E.Y.; Perego, P.; Borghi, M.D. Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production, Chem. Eng. Process. 2009, 48, 1146–1151. [Google Scholar] [CrossRef]
- Rodolfi, L.; Chini Zittelli, G.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef]
- Juneja, A.; Ceballos, R.M.; Murthy, G.S. Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: A review. Energies 2013, 6, 4607–4638. [Google Scholar] [CrossRef] [Green Version]
- Sydney, E.B.; da Silva, T.E.; Tokarski, A.; Novak, A.C.; de Carvalho, J.C.; Woiciecohwski, A.L.; Larroche, C.; Soccol, C.R. Screening of microalgae with potential for biodiesel production and nutrient removal from treated domestic sewage. Appl. Energy 2011, 88, 3291–3294. [Google Scholar] [CrossRef]
- Kim, J.; Liu, Z.; Lee, J.-Y.; Lu, T. Removal of nitrogen and phosphorus from municipal wastewater effluent using Chlorella vulgaris and its growth kinetics. Desalin. Water Treat. 2013, 51, 7800–7806. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, Z.; Shu, Q.; Takala, J.; Hiltunen, E.; Feng, P.; Yuan, Z. Nutrient removal and biodiesel production by integration of freshwater algae cultivation with piggery wastewater treatment. Water Res. 2013, 47, 4294–4302. [Google Scholar] [CrossRef] [PubMed]
- McGaughy, K.; Hajer, A.A.; Drabold, E.; Bayless, D.; Reza, M.T. Algal remediation of wastewater produced from hydrothermally treated septage. Sustainability 2019, 11, 3454. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Srivastava, P.; Yadav, A.K. Simultaneous removal of organic matters and nutrients from high-strength wastewater in constructed wetlands followed by entrapped algal systems. Environ. Sci. Pollut. Res. 2020, 27, 1112–1117. [Google Scholar] [CrossRef] [PubMed]

| Medium/Sample | Chlamydomonas sp. MACC-216 | Chlorella sp. MACC-360 | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of Generations (n) | Mean Generation Time (g). | Specific Growth Rate Day−1 (µ) | Cell Size (µm) | Number of Generations (n) | Mean Generation Time (g) | Specific Growth Rate Day−1 (µ) | Cell Size (µm) | |
| TAP | 3.2 ± 0.0 | 0.6 ± 0.0 | 1.1 ± 0.0 | 24.2 ± 2.1 | 2.3 ± 0.1 | 0.9 ± 0.0 | 0.8 ± 0.0 | 12.9 ± 1.2 |
| TAP-M5 | 3.5 ± 0.01 | 0.6 ± 0.0 | 1.2 ± 0.0 | 25.5 ± 2.8 | 2.1 ± 0.5 | 1 ± 0.2 | 0.7 ± 0.2 | 13.9 ± 2 |
| TAP-M10 | 3.4 ± 0.5 | 0.6 ± 0.1 | 1.2 ± 0.1 | 25.1 ± 2.9 | 1.9 ± 0.0 | 1.1 ± 0.0 | 0.7 ± 0.0 | 15.2 ± 2.5 |
| TAP-M15 | 2.9 ± 0.3 | 0.7 ± 0.1 | 1 ± 0.1 | 25.3 ± 3.3 | 1.7 ± 0.1 | 1.2 ± 0.1 | 0.6 ± 0.1 | 14.5 ± 1.7 |
| Medium/Sample | Total Nitrate Removal | |||||
|---|---|---|---|---|---|---|
| Chlamydomonas sp. MACC-216 | Chlorella sp. MACC-360 | |||||
| mM | mg L−1 | % | mM | mg L−1 | % | |
| TAP-M5 | 5 ± 0.0 | 425 ± 0.0 | 100 ± 0.0 | 5 ± 0.0 | 425 ± 0.0 | 100 ± 0.0 |
| TAP-M10 | 8.4 ± 0.2 | 714 ± 18.2 | 84 ± 2.1 | 7.2 ± 0.3 | 613.8 ± 21.4 | 72.2 ± 2.5 |
| TAP-M15 | 8 ± 0.3 | 683 ± 23.5 | 53.6 ± 1.8 | 7.7 ± 0.6 | 652.8 ± 54.3 | 51.2 ± 4.3 |
| Medium/Sample | Removal Rate (nmol Cell−1 h−1) | |||||
|---|---|---|---|---|---|---|
| Chlamydomonas sp. MACC-216 | Chlorella sp. MACC-360 | |||||
| 3 h | 6 h | 9 h | 3 h | 6 h | 9 h | |
| TAP-M5 | 36.7 ± 2.8 | 61 ± 1.7 | 47.4 ± 1.3 | 6.6 ± 0.9 | 11.3 ± 0.0 | 10.7 ± 0.1 |
| TAP-M10 | 30.1 ± 13.4 | 55.9 ± 2.8 | 53.9 ± 1.1 | 3.8 ± 0.1 | 11.4 ± 0.4 | 12.3 ± 0.0 |
| TAP-M15 | 25.6 ± 5.2 | 55 ± 5.3 | 59 ± 3.6 | 10 ± 0.3 | 12.3 ± 0.01 | 13.3 ± 0.2 |
| Medium/Sample | Total Nitrate Removal | |||||
|---|---|---|---|---|---|---|
| Chlamydomonas sp. MACC-216 | Chlorella sp. MACC-360 | |||||
| mM | mg L−1 | % | mM | mg L−1 | % | |
| 5 mM | 1.8 ± 0.2 | 151.9 ± 20.2 | 35.8 ± 4.8 | 1.6 ± 0.1 | 134.2 ± 11.8 | 31.9 ± 2.8 |
| 10 mM | 2.8 ± 0.2 | 237.1 ± 14.5 | 27.9 ± 1.7 | 3.7 ± 0.3 | 315.1± 25.8 | 37.1 ± 3.1 |
| 25 mM | 9.5 ± 0.9 | 806.2 ± 75.4 | 38 ± 4 | 8.1 ± 0.8 | 692.3 ± 64.9 | 32.6 ± 3.1 |
| 50 mM | 17.3 ± 0.8 | 1469.6 ± 66.5 | 34.6 ± 1.6 | 13.8 ± 0.8 | 1171 ± 65.5 | 27.6 ± 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rani, V.; Maróti, G. Assessment of Nitrate Removal Capacity of Two Selected Eukaryotic Green Microalgae. Cells 2021, 10, 2490. https://doi.org/10.3390/cells10092490
Rani V, Maróti G. Assessment of Nitrate Removal Capacity of Two Selected Eukaryotic Green Microalgae. Cells. 2021; 10(9):2490. https://doi.org/10.3390/cells10092490
Chicago/Turabian StyleRani, Vaishali, and Gergely Maróti. 2021. "Assessment of Nitrate Removal Capacity of Two Selected Eukaryotic Green Microalgae" Cells 10, no. 9: 2490. https://doi.org/10.3390/cells10092490
APA StyleRani, V., & Maróti, G. (2021). Assessment of Nitrate Removal Capacity of Two Selected Eukaryotic Green Microalgae. Cells, 10(9), 2490. https://doi.org/10.3390/cells10092490