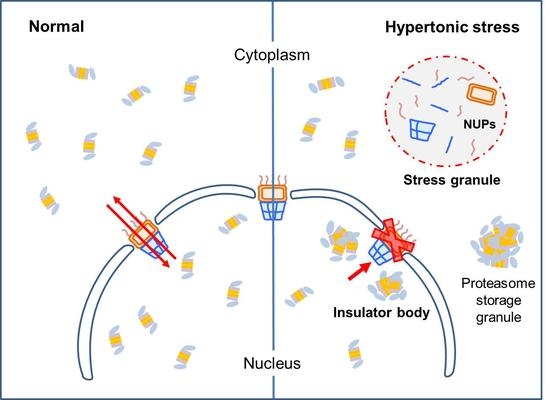
Formation of Non-Nucleoplasmic Proteasome Foci during the Late Stage of Hyperosmotic Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids, Antibodies and Reagents
2.2. Cell Culture and Osmotic Stress
2.3. Western Blot Analysis and Subcellular Fractionation
2.4. Nondenaturing Gel Electrophoresis
2.5. Immunofluorescence Microscopy and Fluorescence Recovery after Photobleaching (FRAP) Analysis
2.6. Live-Cell Imaging
2.7. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assay
2.8. Purification of 26S Human Proteasomes
2.9. Size-Exclusion Chromatography
3. Results
3.1. NaCl-Mediated Hyperosmotic Stress Induced the Formation of Nuclear Proteasome Foci
3.2. The Formation of the Hyperosmotic Stress-Induced Nuclear Proteasome Foci Was Not Mediated by General Osmotic Stress Responses but via Liquid-Liquid Phase Separation
3.3. Formation of Nuclear Proteasome Foci Was Affected by Proteasome Transport between the Nucleus and Cytosol
3.4. Stress Granules and Damaged Nuclear Pore Complexes Were Linked to Nuclear Proteasome Foci under Hyperosmotic Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, B.; Retzlaff, M.; Roos, T.; Frydman, J. Cellular Strategies of Protein Quality Control. Cold Spring Harb. Perspect. Biol. 2011, 3, a004374. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Park, S.; Lee, J.H.; Mun, J.Y.; Choi, W.H.; Yun, Y.; Lee, J.; Kim, J.H.; Kang, M.-J.; Lee, M.J. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Park, S.; Kim, E.; Lee, M.J. Negative-feedback coordination between proteasomal activity and autophagic flux. Autophagy 2019, 15, 726–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Leeuwen, W.; Rabouille, C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 2019, 20, 623–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.H.; Yun, Y.; Park, S.; Jeon, J.H.; Lee, J.; Lee, J.H.; Yang, S.-A.; Kim, N.-K.; Jung, C.H.; Kwon, Y.T.; et al. Aggresomal sequestration and STUB1-mediated ubiquitylation during mammalian proteaphagy of inhibited proteasomes. Proc. Natl. Acad. Sci. USA 2020, 117, 19190–19200. [Google Scholar] [CrossRef] [PubMed]
- Kroschwald, S.; Maharana, S.; Mateju, D.; Malinovska, L.; Nüske, E.; Poser, I.; Richter, D.; Alberti, S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 2015, 4, e06807. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Protter, D.S.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Park, Y.; Yoon, S.K.; Yoon, J.-B. Osmotic Stress Inhibits Proteasome by p38 MAPK-dependent Phosphorylation. J. Biol. Chem. 2010, 285, 41280–41289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, S.; Fukuda, Y.; Beck, F.; Aufderheide, A.; Förster, F.; Danev, R.; Baumeister, W. A molecular census of 26S proteasomes in intact neurons. Science 2015, 347, 439–442. [Google Scholar] [CrossRef]
- Pack, C.G.; Yukii, H.; Toh-e, A.; Kudo, T.; Tsuchiya, H.; Kaiho, A.; Sakata, E.; Murata, S.; Yokosawa, H.; Sako, Y.; et al. Quan-titative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 2014, 5, 3396. [Google Scholar] [CrossRef] [Green Version]
- Wendler, P.; Enenkel, C. Nuclear Transport of Yeast Proteasomes. Front. Mol. Biosci. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef]
- Schoborg, T.; Rickels, R.; Barrios, J.; Labrador, M. Chromatin insulator bodies are nuclear structures that form in response to osmotic stress and cell death. J. Cell Biol. 2013, 202, 261–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, Y.; Lee, S.-Y.; Choi, W.H.; Park, J.-C.; Lee, D.H.; Kim, Y.K.; Lee, J.H.; Lee, J.-Y.; Lee, M.J.; Kim, Y.H. Proteasome Activity in the Plasma as a Novel Biomarker in Mild Cognitive Impairment with Chronic Tinnitus. J. Alzheimer’s Dis. 2020, 78, 195–205. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, E.; Choi, W.H.; Lee, J.; Lee, J.H.; Lee, H.; Kim, D.-E.; Suh, Y.H.; Lee, M.J. Inhibitory RNA Aptamers of Tau Oligomerization and Their Neuroprotective Roles against Proteotoxic Stress. Mol. Pharm. 2016, 13, 2039–2048. [Google Scholar] [CrossRef]
- Lee, J.H.; Shin, S.K.; Jiang, Y.; Choi, W.H.; Hong, C.; Kim, D.-E.; Lee, M.J. Facilitated Tau Degradation by USP14 Aptamers via Enhanced Proteasome Activity. Sci. Rep. 2015, 5, 10757. [Google Scholar] [CrossRef]
- Shin, S.K.; Kim, J.H.; Lee, J.H.; Son, Y.H.; Lee, M.W.; Kim, H.J.; Noh, S.A.; Kim, K.P.; Kim, I.-G.; Lee, M.J. Docosahexaenoic acid-mediated protein aggregates may reduce proteasome activity and delay myotube degradation during muscle atrophy in vitro. Exp. Mol. Med. 2017, 49, e287. [Google Scholar] [CrossRef] [Green Version]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Naguro, I.; Ichijo, H.; Watanabe, K. Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta (BBA) Gen. Subj. 2016, 1860, 2037–2052. [Google Scholar] [CrossRef]
- Liu, Y.; Deisenroth, C.; Zhang, Y. RP-MDM2-p53 Pathway: Linking Ribosomal Biogenesis and Tumor Surveillance. Trends Cancer 2016, 2, 191–204. [Google Scholar] [CrossRef] [Green Version]
- Fu, A.; Cohen-Kaplan, V.; Avni, N.; Livneh, I.; Ciechanover, A. p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc. Natl. Acad. Sci. USA 2021, 118, e2107321118. [Google Scholar] [CrossRef] [PubMed]
- Soderholm, J.F.; Bird, S.L.; Kalab, P.; Sampathkumar, Y.; Hasegawa, K.; Uehara-Bingen, M.; Weis, K.; Heald, R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem. Biol. 2011, 6, 700–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.-L.; Tarn, W.-Y. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 2009, 37, 6600–6612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Daigle, J.G.; Cunningham, K.; Coyne, A.N.; Ruan, K.; Grima, J.C.; Bowen, K.; Wadhwa, H.; Yang, P.; Rigo, F.; et al. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 2018, 173, 958–971.e17. [Google Scholar] [CrossRef] [Green Version]
- Sampuda, K.M.; Riley, M.; Boyd, L. Stress induced nuclear granules form in response to accumulation of misfolded proteins in Caenorhabditis elegans. BMC Cell Biol. 2017, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic shuttling of TIA-1 ac-companies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Mazroui, R.; Di Marco, S.; Kaufman, R.J.; Gallouzi, I.-E. Inhibition of the Ubiquitin-Proteasome System Induces Stress Granule Formation. Mol. Biol. Cell 2007, 18, 2603–2618. [Google Scholar] [CrossRef] [Green Version]
- Stoecklin, G.; Gross, B.; Ming, X.-F.; Moroni, C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene 2003, 22, 3554–3561. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Le, L.T.H.L.; Kim, E.; Lee, M.J. Formation of Non-Nucleoplasmic Proteasome Foci during the Late Stage of Hyperosmotic Stress. Cells 2021, 10, 2493. https://doi.org/10.3390/cells10092493
Lee J, Le LTHL, Kim E, Lee MJ. Formation of Non-Nucleoplasmic Proteasome Foci during the Late Stage of Hyperosmotic Stress. Cells. 2021; 10(9):2493. https://doi.org/10.3390/cells10092493
Chicago/Turabian StyleLee, Jeeyoung, Ly Thi Huong Luu Le, Eunkyoung Kim, and Min Jae Lee. 2021. "Formation of Non-Nucleoplasmic Proteasome Foci during the Late Stage of Hyperosmotic Stress" Cells 10, no. 9: 2493. https://doi.org/10.3390/cells10092493
APA StyleLee, J., Le, L. T. H. L., Kim, E., & Lee, M. J. (2021). Formation of Non-Nucleoplasmic Proteasome Foci during the Late Stage of Hyperosmotic Stress. Cells, 10(9), 2493. https://doi.org/10.3390/cells10092493