In Vitro Anticancer Screening and Preliminary Mechanistic Study of A-Ring Substituted Anthraquinone Derivatives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Cell Lines
2.2. Cell Viability
2.3. Flow Cytometry Analysis
2.3.1. Cell Cycle Analysis
2.3.2. Apoptosis, Caspase Production and Autophagy Analysis
2.3.3. Cell Division Analysis
2.3.4. Investigation of ROS/RNS and NO Production
2.4. Western Blot Analysis
2.5. Autophagy Inhibitor Assay
2.6. Topoisomerase I Assay
3. Results
3.1. Impact of Anthraquinone Compounds on the Cell Viability
3.2. Compound 4 Induces Trappment of PC3 Cells in G2/M Phase as well as Activation of Caspase Dependent Apoptosis
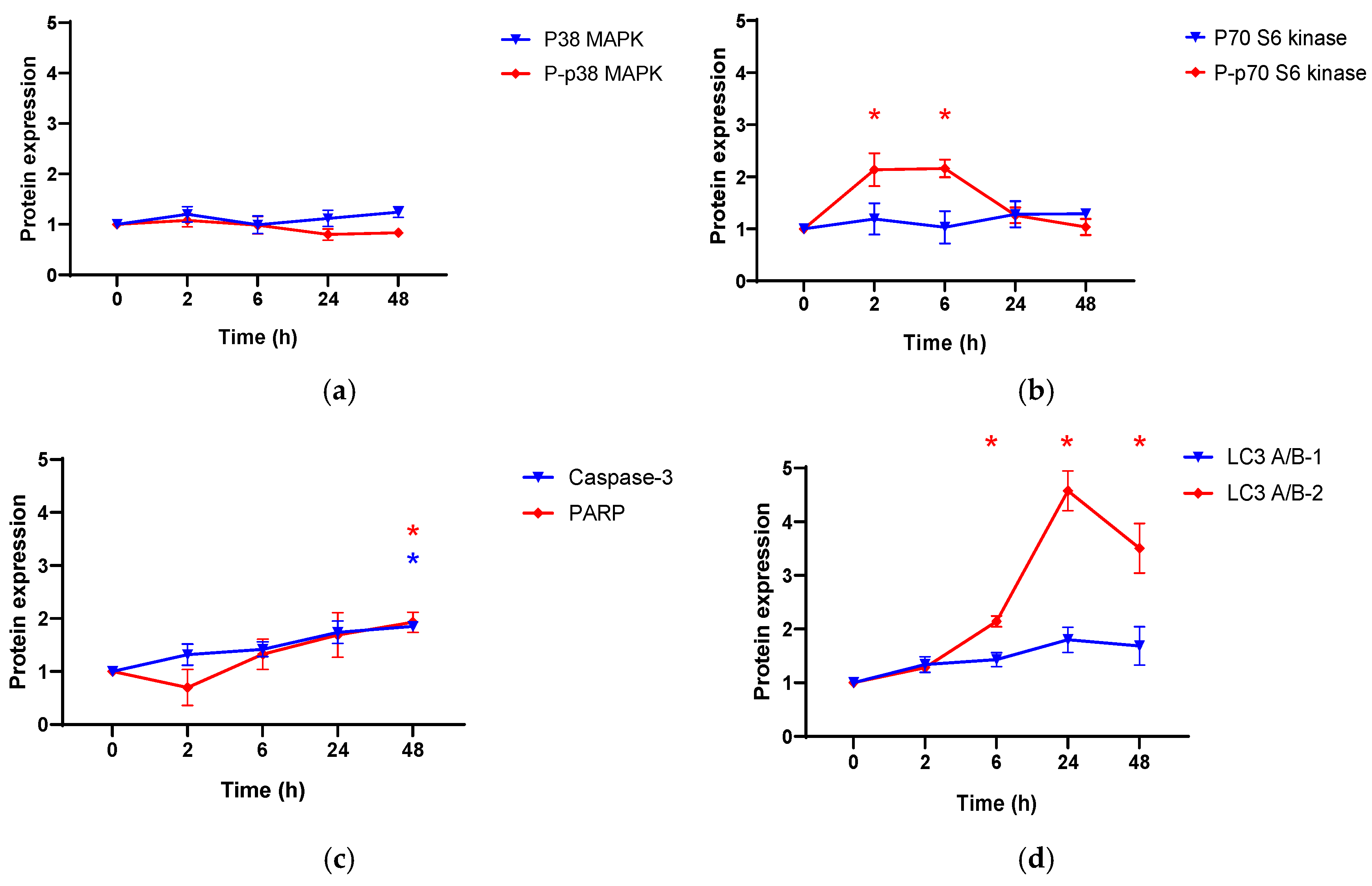
3.3. The Effect of Compound 4 on Apoptosis and Autophagy Related Proteins in PC3 Cell Line
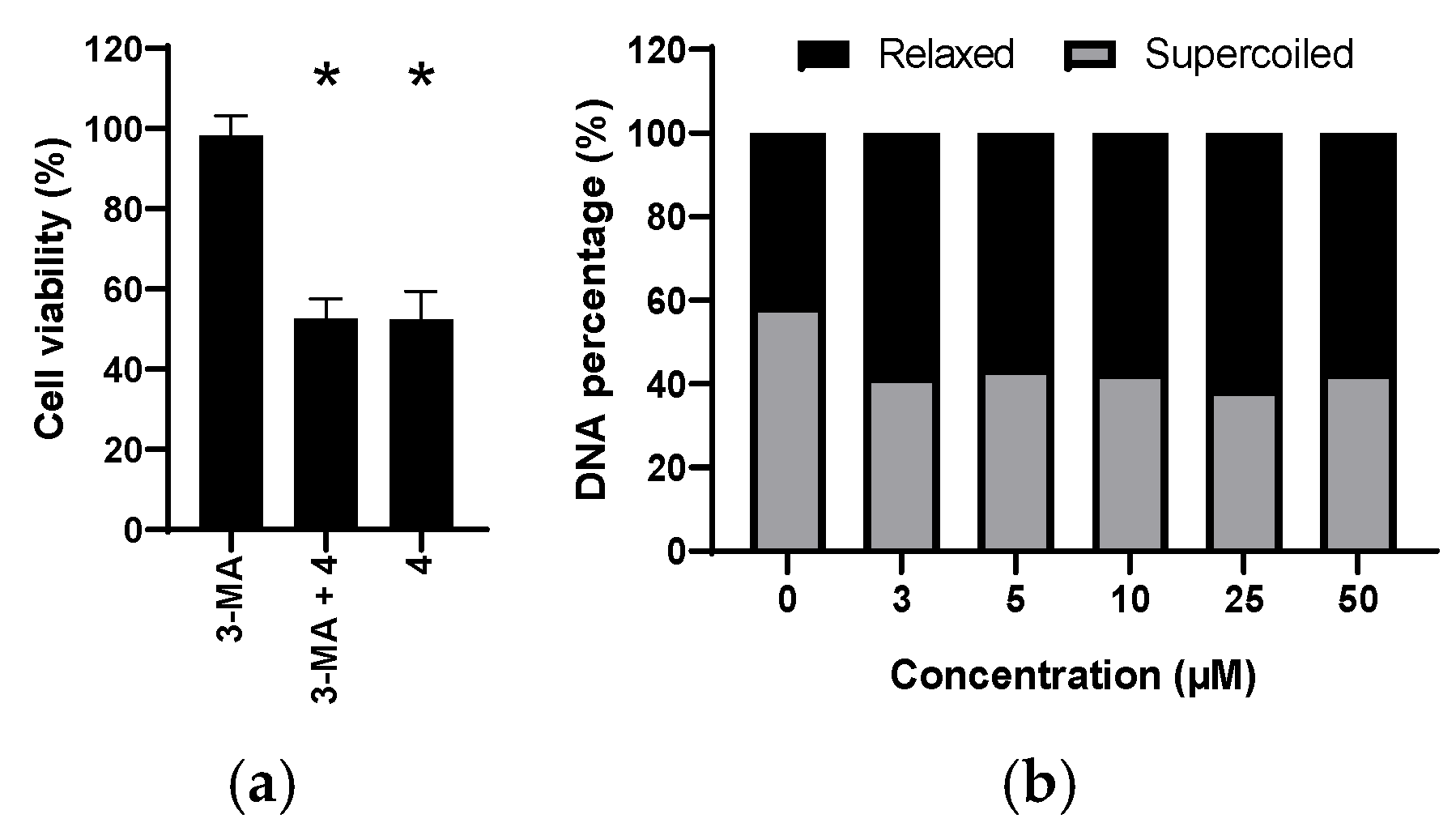
3.4. The Role of Autophagy
3.5. Influence of Compound 4 on the Topoisomerase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Causes of Death by Chapters of the ICD-10 and Gender. Available online: https://www.destatis.de/EN/Themes/Society-Environment/Health/Causes-Death/Tables/number-of-death.html (accessed on 9 April 2020).
- Miller, A.B.; Hoogstraten, B.; Staquet, M.; Winkler, A. Reporting Results of Cancer Treatment. Cancer 1981, 47, 207–214. [Google Scholar] [CrossRef]
- Texas Cancer Treatment Options|Prostate, Breast Cancer Doctors. Available online: https://choicecancercare.com/technology-treatments/treatments/ (accessed on 13 February 2018).
- Lind, M.J. Principles of Cytotoxic Chemotherapy. Medicine 2011, 39, 711–716. [Google Scholar] [CrossRef]
- Fulda, S. Autophagy in Cancer Therapy. Front. Oncol. 2017, 7, 128. [Google Scholar] [CrossRef]
- Badadani, M. Autophagy Mechanism, Regulation, Functions, and Disorders. ISRN Cell Biol. 2012, 2012, e927064. [Google Scholar] [CrossRef] [Green Version]
- Denton, D.; Kumar, S. Autophagy-Dependent Cell Death. Cell Death Differ. 2019, 26, 605–616. [Google Scholar] [CrossRef] [Green Version]
- Mathiassen, S.G.; De Zio, D.; Cecconi, F. Autophagy and the Cell Cycle: A Complex Landscape. Front. Oncol. 2017, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Gewirtz, D.A. Cytoprotective and Nonprotective Autophagy in Cancer Therapy. Autophagy 2013, 9, 1263–1265. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-T.; Tan, H.-L.; Shui, G.; Bauvy, C.; Huang, Q.; Wenk, M.R.; Ong, C.-N.; Codogno, P.; Shen, H.-M. Dual Role of 3-Methyladenine in Modulation of Autophagy via Different Temporal Patterns of Inhibition on Class I and III Phosphoinositide 3-Kinase. J. Biol. Chem. 2010, 285, 10850–10861. [Google Scholar] [CrossRef] [Green Version]
- Vogel, A. Anthraquinone. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2000; ISBN 978-3-527-30673-2. [Google Scholar]
- Goor, G.; Glenneberg, J.; Jacobi, S. Hydrogen Peroxide. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; ISBN 978-3-527-30673-2. [Google Scholar]
- Benamar, H.; Rarivoson, E.; Tomassini, L.; Frezza, C.; Marouf, A.; Bennaceur, M.; Nicoletti, M. Phytochemical Profiles, Antioxidant and Anti-Acetylcholinesterasic Activities of the Leaf Extracts of Rhamnus lycioides Subsp. Oleoides (L.) Jahand. & Maire in Different Solvents. Nat. Prod. Res. 2018, 33, 1456–1462. [Google Scholar] [CrossRef]
- Sakulpanich, A.; Gritsanapan, W. Laxative Anthraquinone Contents in Fresh and Cooked Senna Siamea Leaves. Southeast Asian J. Trop. Med. Public Health 2009, 40, 835–839. [Google Scholar]
- Rizzo, P.; Altschmied, L.; Stark, P.; Rutten, T.; Gündel, A.; Scharfenberg, S.; Franke, K.; Bäumlein, H.; Wessjohann, L.; Koch, M.; et al. Discovery of Key Regulators of Dark Gland Development and Hypericin Biosynthesis in St. John’s Wort (Hypericum perforatum). Plant Biotechnol. J. 2019, 17, 2299–2312. [Google Scholar] [CrossRef] [Green Version]
- Pankewitz, F.; Zöllmer, A.; Gräser, Y.; Hilker, M. Anthraquinones as Defensive Compounds in Eggs of Galerucini Leaf Beetles: Biosynthesis by the Beetles? Arch. Insect Biochem. Physiol. 2007, 66, 98–108. [Google Scholar] [CrossRef]
- Laub, A.; Sendatzki, A.-K.; Palfner, G.; Wessjohann, L.A.; Schmidt, J.; Arnold, N. HPTLC-DESI-HRMS-Based Profiling of Anthraquinones in Complex Mixtures—A Proof-of-Concept Study Using Crude Extracts of Chilean Mushrooms. Foods 2020, 9, 156. [Google Scholar] [CrossRef] [Green Version]
- Chakiri, A.B.; Hodge, P. Synthesis of Isopropyl-Substituted Anthraquinones via Friedel–Crafts Acylations: Migration of Isopropyl Groups. R. Soc. Open Sci. 2017, 4, 170451. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Yang, Z.-J.; He, J.-H.; Lu, A.-P.; Liu, S.; Hou, T.-J.; Cao, D.-S. Benchmarking the Mechanisms of Frequent Hitters: Limitation of PAINS Alerts. Drug Discov. Today 2021, 26, 1353–1358. [Google Scholar] [CrossRef]
- Michels, K.; Heinke, R.; Schöne, P.; Kuipers, O.P.; Arnold, N.; Wessjohann, L.A. A Fluorescence-Based Bioassay for Antibacterials and Its Application in Screening Natural Product Extracts. J. Antibiot. 2015, 68, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, S.K.; Singh, S.S.; Verma, S.; Kumar, S. Antifungal Activity of Anthraquinone Derivatives from Rheum Emodi. J. Ethnopharmacol. 2000, 72, 43–46. [Google Scholar] [CrossRef]
- Comini, L.R.; Núñez Montoya, S.C.; Páez, P.L.; Argüello, G.A.; Albesa, I.; Cabrera, J.L. Antibacterial Activity of Anthraquinone Derivatives from Heterophyllaea pustulata (Rubiaceae). J. Photochem. Photobiol. B Biol. 2011, 102, 108–114. [Google Scholar] [CrossRef]
- DeLiberto, S.T.; Werner, S.J. Review of Anthraquinone Applications for Pest Management and Agricultural Crop Protection. Pest Manag. Sci. 2016, 72, 1813–1825. [Google Scholar] [CrossRef] [Green Version]
- Zvarych, V.; Stasevych, M.; Novikov, V.; Rusanov, E.; Vovk, M.; Szweda, P.; Grecka, K.; Milewski, S. Anthra [1, 2-d][1, 2, 3] Triazine-4, 7, 12 (3H)-Triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation. Molecules 2019, 24, 4581. [Google Scholar] [CrossRef] [Green Version]
- Mohamadzadeh, M.; Zarei, M.; Vessal, M. Synthesis, In vitro Biological Evaluation and in Silico Molecular Docking Studies of Novel β-Lactam-Anthraquinone Hybrids. Bioorganic Chem. 2020, 95, 103515. [Google Scholar] [CrossRef]
- Sydiskis, R.J.; Owen, D.G.; Lohr, J.L.; Rosler, K.H.; Blomster, R.N. Inactivation of Enveloped Viruses by Anthraquinones Extracted from Plants. Antimicrob. Agents Chemother. 1991, 35, 2463–2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnard, D.L.; Huffman, J.H.; Morris, J.L.; Wood, S.G.; Hughes, B.G.; Sidwell, R.W. Evaluation of the Antiviral Activity of Anthraquinones, Anthrones and Anthraquinone Derivatives against Human Cytomegalovirus. Antivir. Res. 1992, 17, 63–77. [Google Scholar] [CrossRef]
- Stasevich, M.V.; Zvarich, V.I.; Novikov, V.P.; Zagorodnyaya, S.D.; Povnitsa, O.Y.; Chaika, M.A.; Nesterkina, M.V.; Kravchenko, I.A.; Druzhilovskii, D.S.; Poroikov, V.V. 9,10-Anthraquinone Dithiocarbamates as Potential Pharmaceutical Substances with Pleiotropic Actions: Computerized Prediction of Biological Activity and Experimental Validation. Pharm. Chem. J. 2020, 53, 905–913. [Google Scholar] [CrossRef]
- Malik, E.M.; Müller, C.E. Anthraquinones As Pharmacological Tools and Drugs. Med. Res. Rev. 2016, 36, 705–748. [Google Scholar] [CrossRef]
- Vilanova-Sanchez, A.; Gasior, A.C.; Toocheck, N.; Weaver, L.; Wood, R.J.; Reck, C.A.; Wagner, A.; Hoover, E.; Gagnon, R.; Jaggers, J.; et al. Are Senna Based Laxatives Safe When Used as Long Term Treatment for Constipation in Children? J. Pediatr. Surg. 2018, 53, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Diaby, V.; Alqhtani, H.; van Boemmel-Wegmann, S.; Wang, C.-Y.; Ali, A.A.; Balkrishnan, R.; Ko, Y.; Palacio, S.; de Lima Lopes, G. A Cost-Effectiveness Analysis of Trastuzumab-Containing Treatment Sequences for HER-2 Positive Metastatic Breast Cancer Patients in Taiwan. Breast 2019, 49, 141–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Cao, S.; Brodie, P.J.; Callmander, M.W.; Ratovoson, F.; Rakotobe, E.A.; Rasamison, V.E.; Ratsimbason, M.; Alumasa, J.N.; Roepe, P.D.; et al. Antiproliferative and Antimalarial Anthraquinones of Scutia Myrtina from the Madagascar Forest. Bioorg. Med. Chem. 2009, 17, 2871–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campora, M.; Francesconi, V.; Schenone, S.; Tasso, B.; Tonelli, M. Journey on Naphthoquinone and Anthraquinone Derivatives: New Insights in Alzheimer’s Disease. Pharmaceuticals 2021, 14, 33. [Google Scholar] [CrossRef]
- Siddamurthi, S.; Gutti, G.; Jana, S.; Kumar, A.; Singh, S.K. Anthraquinone: A Promising Scaffold for the Discovery and Development of Therapeutic Agents in Cancer Therapy. Future Med. Chem. 2020, 12, 1037–1069. [Google Scholar] [CrossRef]
- Adnan, M.; Rasul, A.; Hussain, G.; Shah, M.A.; Sarfraz, I.; Nageen, B.; Riaz, A.; Khalid, R.; Asrar, M.; Selamoglu, Z.; et al. Physcion and Physcion 8-O-β-D-Glucopyranoside: Natural Anthraquinones with Potential Anticancer Activities. Curr. Drug Targets 2021, 22, 488–504. [Google Scholar] [CrossRef]
- Parker, C.; Waters, R.; Leighton, C.; Hancock, J.; Sutton, R.; Moorman, A.V.; Ancliff, P.; Morgan, M.; Masurekar, A.; Goulden, N.; et al. Effect of Mitoxantrone on Outcome of Children with First Relapse of Acute Lymphoblastic Leukaemia (ALL R3): An Open-Label Randomised Trial. Lancet 2010, 376, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Borchmann, P.; Morschhauser, F.; Parry, A.; Schnell, R.; Harousseau, J.L.; Gisselbrecht, C.; Rudolph, C.; Wilhelm, M.; Günther, H.; Pfreundschuh, D.M.; et al. Phase-II Study of the New Aza-Anthracenedione, BBR 2778, in Patients with Relapsed Aggressive Non-Hodgkin’s Lymphomas. Haematologica 2003, 88, 888–894. [Google Scholar]
- Chatterjee, K.; Zhang, J.; Honbo, N.; Karliner, J.S. Doxorubicin Cardiomyopathy. Cardiology 2010, 115, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, J.; Hu, J.; Zheng, J.; Zheng, Z.; Liu, T.; Lin, Z.; Lin, M. Emodin Enhances ATRA-Induced Differentiation and Induces Apoptosis in Acute Myeloid Leukemia Cells. Int. J. Oncol. 2014, 45, 2076–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayasuriya, H.; Koonchanok, N.M.; Geahlen, R.L.; McLaughlin, J.L.; Chang, C.J. Emodin, a Protein Tyrosine Kinase Inhibitor from Polygonum Cuspidatum. J. Nat. Prod. 1992, 55, 696–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lau, Y.-K.; Xia, W.; Hortobagyi, G.N.; Hung, M.-C. Tyrosine Kinase Inhibitor Emodin Suppresses Growth of HER-2/Neu-Overexpressing Breast Cancer Cells in Athymic Mice and Sensitizes These Cells to the Inhibitory Effect of Paclitaxel. Clin. Cancer Res. 1999, 5, 343–353. [Google Scholar]
- Li, W. Emodin Potentiates the Antitumor Effects of Gemcitabine in PANC-1 Pancreatic Cancer Xenograft Model In Vivo via Inhibition of Inhibitors of Apoptosis. Int. J. Oncol. 2012, 40, 1849–1857. [Google Scholar] [CrossRef] [Green Version]
- Mijatovic, S.; Maksimovic-Ivanic, D.; Radovic, J.; Miljkovic, D.; Harhaji, L.; Vuckovic, O.; Stosic-Grujicic, S.; Mostarica Stojkovic, M.; Trajkovic, V. Anti-Glioma Action of Aloe Emodin: The Role of ERK Inhibition. Cell. Mol. Life Sci. CMLS 2005, 62, 589–598. [Google Scholar] [CrossRef]
- Krohn, K.; Radeloff, M. Stereoselektive Totalsynthese von Anthracyclinonen. Chem. Ber. 1978, 111, 3823–3837. [Google Scholar] [CrossRef]
- Krohn, K.; Behnke, B. Synthetische Anthracyclinone, XI. Synthese von 4-Desoxy-β1-rhodomycinon über eine intramolekulare Marschalk-Reaktion. Liebigs Ann. Der Chem. 1979, 1979, 2011–2017. [Google Scholar] [CrossRef]
- Krohn, K.; Hemme, C. Synthetische Anthracyclinone, VI Synthese strukturanaloger Anthracyclinone. Liebigs Ann. Der Chem. 1979, 1979, 35–42. [Google Scholar] [CrossRef]
- Krohn, K.; Müller, U.; Priyono, W.; Sarstedt, B.; Stoffregen, A. Intramolekulare Addition von Carbanionen an Anthrachinone. Liebigs Ann. Der Chem. 1984, 1984, 306–318. [Google Scholar] [CrossRef]
- Krohn, K.; Behnke, B. Synthetische Anthracyclinone, XV. Regio- und stereoselektive Synthese der α-, β- und γ-Rhodomycinone über intramolekulare Marschalk-Cyclisierung. Chem. Ber. 1980, 113, 2994–3009. [Google Scholar] [CrossRef]
- Cory, A.H.; Owen, T.C.; Barltrop, J.A.; Cory, J.G. Use of an Aqueous Soluble Tetrazolium/Formazan Assay for Cell Growth Assays in Culture. Cancer Commun. 1991, 3, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Sladowski, D.; Steer, S.J.; Clothier, R.H.; Balls, M. An Improved MTT Assay. J. Immunol. Methods 1993, 157, 203–207. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (3rd Edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herceg, Z.; Wang, Z.-Q. Functions of Poly (ADP-Ribose) Polymerase (PARP) in DNA Repair, Genomic Integrity and Cell Death. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2001, 477, 97–110. [Google Scholar] [CrossRef]
- Kaluđerović, G.N.; Krajnović, T.; Momcilovic, M.; Stosic-Grujicic, S.; Mijatović, S.; Maksimović-Ivanić, D.; Hey-Hawkins, E. Ruthenium(II) p-Cymene Complex Bearing 2,2′-Dipyridylamine Targets Caspase 3 Deficient MCF-7 Breast Cancer Cells without Disruption of Antitumor Immune Response. J. Inorg. Biochem. 2015, 153, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Loveless, J.; Shay, C.; Teng, Y. Targeting ROS-Mediated Crosstalk between Autophagy and Apoptosis in Cancer. Rev. New Drug Targets Age-Relat. Disord. 2020, 1260, 1–12. [Google Scholar] [CrossRef]
- Snyder, C.M.; Shroff, E.H.; Liu, J.; Chandel, N.S. Nitric Oxide Induces Cell Death by Regulating Anti-Apoptotic BCL-2 Family Members. PLoS ONE 2009, 4, e7059. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Korolchuk, V.I.; Renna, M.; Imarisio, S.; Fleming, A.; Williams, A.; Garcia-Arencibia, M.; Rose, C.; Luo, S.; Underwood, B.R.; et al. Complex Inhibitory Effects of Nitric Oxide on Autophagy. Mol. Cell 2011, 43, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Krajnović, T.; Kaluđerović, G.N.; Wessjohann, L.A.; Mijatović, S.; Maksimović-Ivanić, D. Versatile Antitumor Potential of Isoxanthohumol: Enhancement of Paclitaxel Activity In Vivo. Pharmacol. Res. 2016, 105, 62–73. [Google Scholar] [CrossRef]
- Maksimovic-Ivanic, D.; Mijatovic, S.; Harhaji, L.; Miljkovic, D.; Dabideen, D.; Cheng, K.F.; Mangano, K.; Malaponte, G.; Al-Abed, Y.; Libra, M.; et al. Anticancer Properties of the Novel Nitric Oxide-Donating Compound (S,R)-3-Phenyl-4,5-Dihydro-5-Isoxazole Acetic Acid-Nitric Oxide In vitro and In Vivo. Mol. Cancer Ther. 2008, 7, 510–520. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.H.; Cheng, S.; Zheng, Z.; Wang, L.; Shen, Y.; Shen, Z.X.; Chen, S.J.; Zhao, W.L. Histone Deacetylase Inhibitor Potentiated the Ability of MTOR Inhibitor to Induce Autophagic Cell Death in Burkitt Leukemia/Lymphoma. J. Hematol. Oncol. 2013, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen Peroxide—Production, Fate and Role in Redox Signaling of Tumor Cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Jeremić, S.; Amić, A.; Stanojević-Pirković, M.; Marković, Z. Selected Anthraquinones as Potential Free Radical Scavengers and P-Glycoprotein Inhibitors. Org. Biomol. Chem. 2018, 16, 1890–1902. [Google Scholar] [CrossRef]
- Sahin, F.; Kannangai, R.; Adegbola, O.; Wang, J.; Su, G.; Torbenson, M. MTOR and P70 S6 Kinase Expression in Primary Liver Neoplasms. Clin. Cancer Res. 2004, 10, 8421–8425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, H.; Andersen, J.S.; Mann, M.; Terada, N.; Korsmeyer, S.J. P70S6 Kinase Signals Cell Survival as Well as Growth, Inactivating the pro-Apoptotic Molecule BAD. Proc. Natl. Acad. Sci. USA 2001, 98, 9666–9670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and Functions of P38 MAPK Signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krehl, S.; Loewinger, M.; Florian, S.; Kipp, A.P.; Banning, A.; Wessjohann, L.A.; Brauer, M.N.; Iori, R.; Esworthy, R.S.; Chu, F.-F.; et al. Glutathione Peroxidase-2 and Selenium Decreased Inflammation and Tumors in a Mouse Model of Inflammation-Associated Carcinogenesis Whereas Sulforaphane Effects Differed with Selenium Supply. Carcinogenesis 2012, 33, 620–628. [Google Scholar] [CrossRef] [Green Version]
- Rahn, J.; Lennicke, C.; Kipp, A.P.; Müller, A.S.; Wessjohann, L.A.; Lichtenfels, R.; Seliger, B. Altered Protein Expression Pattern in Colon Tissue of Mice upon Supplementation with Distinct Selenium Compounds. Proteomics 2017, 17, 1600486. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan, I.; Wessjohann, L.A.; Kaluđerović, G.N. In Vitro Anticancer Screening and Preliminary Mechanistic Study of A-Ring Substituted Anthraquinone Derivatives. Cells 2022, 11, 168. https://doi.org/10.3390/cells11010168
Morgan I, Wessjohann LA, Kaluđerović GN. In Vitro Anticancer Screening and Preliminary Mechanistic Study of A-Ring Substituted Anthraquinone Derivatives. Cells. 2022; 11(1):168. https://doi.org/10.3390/cells11010168
Chicago/Turabian StyleMorgan, Ibrahim, Ludger A. Wessjohann, and Goran N. Kaluđerović. 2022. "In Vitro Anticancer Screening and Preliminary Mechanistic Study of A-Ring Substituted Anthraquinone Derivatives" Cells 11, no. 1: 168. https://doi.org/10.3390/cells11010168





