Nicotinamide Mononucleotide Administration Amends Protein Acetylome of Aged Mouse Liver
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. Fractionation of Nuclear and Cytosolic Protein
2.3. Western Blot
2.4. Acetylated Peptide Enrichment
2.5. Mass Spectrometry Analysis
2.6. Acetylated Peptides Searches
2.7. Quantitative Proteomic Analysis
2.8. Gene Ontology Analysis
2.9. Ingenuity Pathway Analysis
2.10. Statistical Analysis
3. Results
3.1. Protein Acetylation Was Globally Enhanced and NMN Decreased This Acetylation in Aged Livers
3.2. Enrichment and Identification of the Liver Acetylome
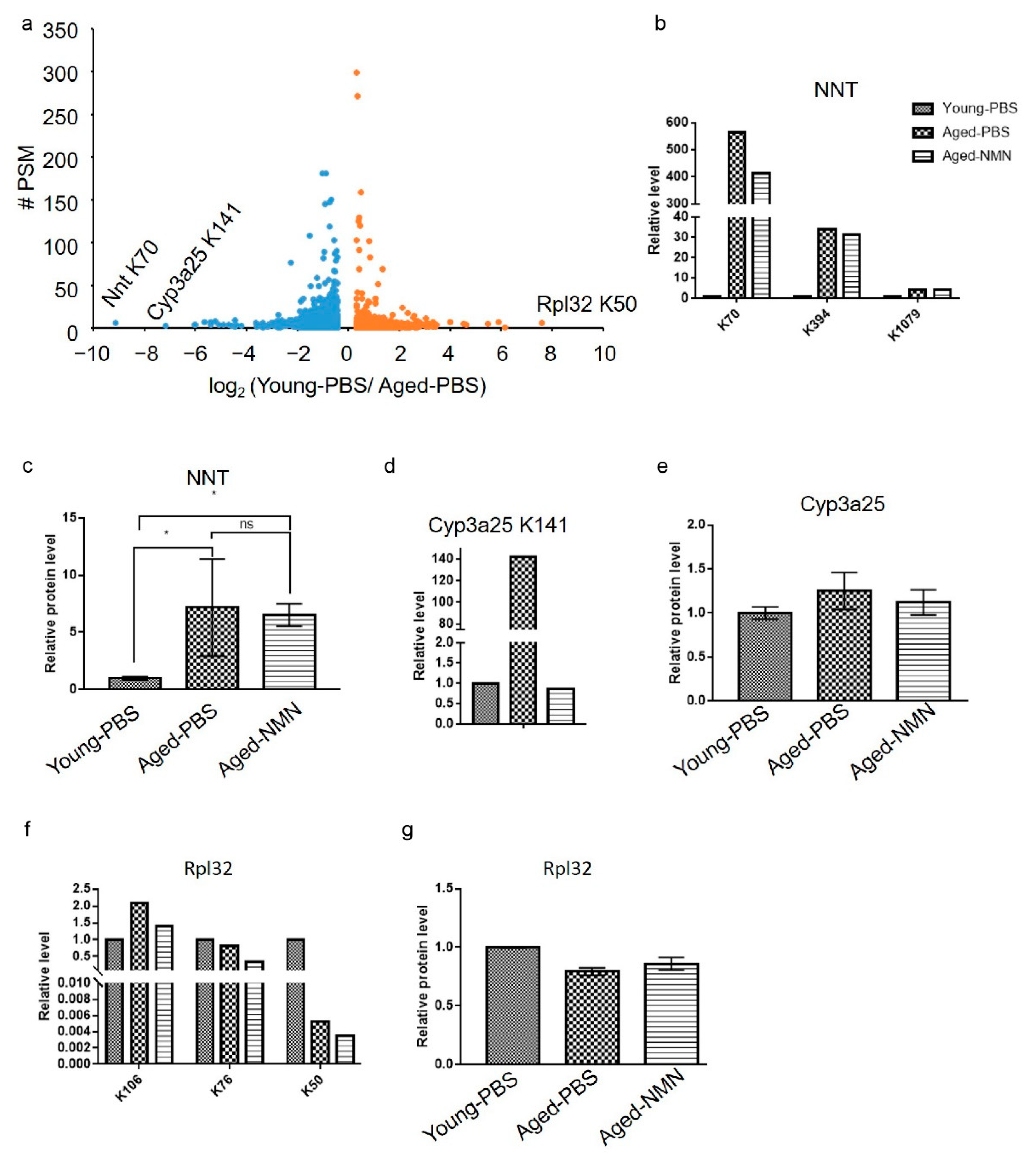
3.3. Aging-Induced Changes in the Acetylomic Landscape
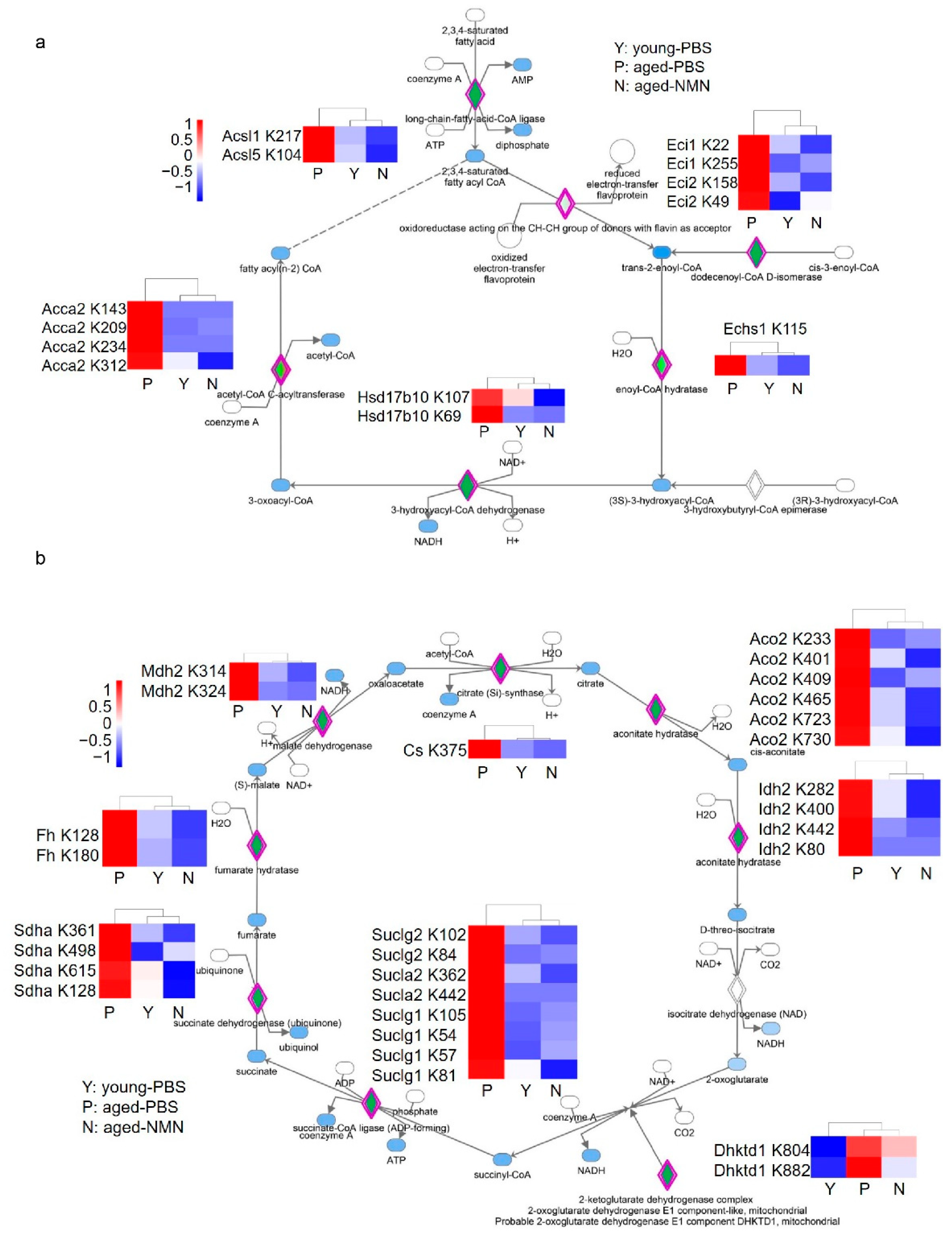
3.4. NMN Changed Aging-Related Acetylome Involved in Key Metabolic Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, D. The presence of acetyl groups in histones. Biochem. J. 1963, 87, 258–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggs, M.G.; Whittaker, R.G.; Neumann, J.R.; Ingram, V.M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 1977, 268, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Waddell, J.; Banerjee, A.; Kristian, T. Acetylation in Mitochondria Dynamics and Neurodegeneration. Cells 2021, 10, 3031. [Google Scholar] [CrossRef]
- Ghosh, A.K. Acetyltransferase p300 is a Putative Epidrug Target for Amelioration of Cellular Aging-Related Cardiovascular Disease. Cells 2021, 10, 2839. [Google Scholar] [CrossRef]
- McCay, C.M.; Maynard, L.A.; Sperling, G.; Barnes, L.L. Retarded growth, life span, ultimate body size and age changes in the albino rat after feeding diets restricted in calories. Nutr. Rev. 1975, 33, 241–243. [Google Scholar] [CrossRef]
- Imai, S.-i.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef]
- Rine, J.; Herskowitz, A.I. Four Genes Responsible for a Position Effect on Expression from HML and HMR in Saccharomyces cerevisiae. Genetics 1987, 116, 9–22. [Google Scholar] [CrossRef]
- Hershberger, K.A.; Martin, A.S.; Hirschey, M.D. Role of NAD+ and mitochondrial sirtuins in cardiac and renal diseases. Nat. Rev. Nephrol. 2017, 13, 213. [Google Scholar] [CrossRef] [Green Version]
- Gerhart-Hines, Z.; Dominy, J.E., Jr.; Blattler, S.M.; Jedrychowski, M.P.; Banks, A.S.; Lim, J.H.; Chim, H.; Gygi, S.P.; Puigserver, P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol. Cell 2011, 44, 851–863. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.; Seok, S.; Yau, P.; Li, X.; Kemper, B.; Kemper, J.K. Obesity and aging diminish sirtuin 1 (SIRT1)-mediated deacetylation of SIRT3, leading to hyperacetylation and decreased activity and stability of SIRT3. J. Biol. Chem. 2017, 292, 17312–17323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bong-Hyun, A.; Kim, H.-S.; Song, S.; Lee, I.H.; Liu, J.; Athanassios Vassilopoulos; Deng, C.-X.; Finkel, T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. PNAS 2008, 105, 6. [Google Scholar]
- Benigni, A.; Cassis, P.; Conti, S.; Perico, L.; Corna, D.; Cerullo, D.; Zentilin, L.; Zoja, C.; Perna, A.; Lionetti, V.; et al. Sirt3 Deficiency Shortens Life Span and Impairs Cardiac Mitochondrial Function Rescued by Opa1 Gene Transfer. Antioxid. Redox Signal. 2019, 31, 1255–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caton, P.W.; Kieswich, J.; Yaqoob, M.M.; Holness, M.J.; Sugden, M.C. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia 2011, 54, 3083–3092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wan, T.; Ye, M.; Qiu, Y.; Pei, L.; Jiang, R.; Pang, N.; Huang, Y.; Liang, B.; Ling, W.; et al. Nicotinamide riboside attenuates alcohol induced liver injuries via activation of SirT1/PGC-1alpha/mitochondrial biosynthesis pathway. Redox Biol. 2018, 17, 89–98. [Google Scholar] [CrossRef]
- Cantó, C.; Houtkooper, R.H.; Pirinen, E.; Youn, D.Y.; Oosterveer, M.H.; Cen, Y.; Fernandez-Marcos, P.J.; Yamamoto, H.; Andreux, P.A.; Cettour-Rose, P.; et al. The NAD+ Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metab. 2012, 15, 838–847. [Google Scholar] [CrossRef] [Green Version]
- Hong, G.; Zheng, D.; Zhang, L.; Ni, R.; Wang, G.; Fan, G.-C.; Lu, Z.; Peng, T. Administration of nicotinamide riboside prevents oxidative stress and organ injury in sepsis. Free. Radic. Biol. Med. 2018, 123, 125–137. [Google Scholar] [CrossRef]
- Forero-Baena, N.; Sánchez-Lancheros, D.; Buitrago, J.C.; Bustos, V.; Ramírez-Hernández, M.H. Identification of a nicotinamide/nicotinate mononucleotide adenylyltransferase in Giardia lamblia (GlNMNAT). Biochim. Open 2015, 1, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Magni, G.; Amici, A.; Emanuelli, M.; Raffaelli, N.; Ruggieri, S. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1999, 73, 135–182. [Google Scholar] [PubMed]
- Camacho-Pereira, J.; Tarrago, M.G.; Chini, C.C.S.; Nin, V.; Escande, C.; Warner, G.M.; Puranik, A.S.; Schoon, R.A.; Reid, J.M.; Galina, A.; et al. CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metab. 2016, 23, 1127–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frederick, D.W.; Loro, E.; Liu, L.; Davila, A.; Chellappa, K.; Silverman, I.M.; Quinn, W.J.; Gosai, S.J.; Tichy, E.D.; Davis, J.G.; et al. Loss of NAD Homeostasis Leads to Progressive and Reversible Degeneration of Skeletal Muscle. Cell Metab. 2016, 24, 269–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, B.; Pan, Y.; Vempati, P.; Zhao, W.; Knable, L.; Ho, L.; Wang, J.; Sastre, M.; Ono, K.; Sauve, A.A.; et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-γ coactivator 1α regulated β-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 2013, 34, 1581–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhassan, Y.S.; Kluckova, K.; Fletcher, R.S.; Schmidt, M.S.; Garten, A.; Doig, C.L.; Cartwright, D.M.; Oakey, L.; Burley, C.V.; Jenkinson, N.; et al. Nicotinamide Riboside Augments the Aged Human Skeletal Muscle NAD+ Metabolome and Induces Transcriptomic and Anti-inflammatory Signatures. Cell Rep. 2019, 28, 1717–1728.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S.-I. Nicotinamide Mononucleotide, a Key NAD+ Intermediate, Treats the Pathophysiology of Diet- and Age-Induced Diabetes in Mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Zong, Z.; Liu, J.; Wang, N.; Yang, C.; Wang, Q.; Zhang, W.; Chen, Y.; Liu, X.; Deng, H. Nicotinamide mononucleotide inhibits hepatic stellate cell activation to prevent liver fibrosis via promoting PGE2 degradation. Free Radic. Biol. Med. 2021, 162, 571–581. [Google Scholar] [CrossRef]
- Bertoldo, M.J.; Listijono, D.R.; Ho, W.-H.J.; Riepsamen, A.H.; Goss, D.M.; Richani, D.; Jin, X.L.; Mahbub, S.; Campbell, J.M.; Habibalahi, A.; et al. NAD+ Repletion Rescues Female Fertility during Reproductive Aging. Cell Rep. 2020, 30, 1670–1681.e7. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.S.; Abraham, D.M.; Hershberger, K.A.; Bhatt, D.P.; Mao, L.; Cui, H.; Liu, J.; Liu, X.; Muehlbauer, M.J.; Grimsrud, P.A.; et al. Nicotinamide mononucleotide requires SIRT3 to improve cardiac function and bioenergetics in a Friedreich’s ataxia cardiomyopathy model. JCI Insight 2017, 2, e93885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimova, N.; Long, A.; Kristian, T. Nicotinamide mononucleotide alters mitochondrial dynamics by SIRT3-dependent mechanism in male mice. J. Neurosci. Res. 2019, 97, 975–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschey, M.D.; Shimazu, T.; Goetzman, E.; Jing, E.; Schwer, B.; Lombard, D.B.; Grueter, C.A.; Harris, C.; Biddinger, S.; Ilkayeva, O.R.; et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010, 464, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Yu, W.; Dittenhafer-Reed, K.E.; Denu, J.M. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J. Biol. Chem. 2012, 287, 14078–14086. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Coleman, M.C.; Pennington, J.D.; Ozden, O.; Park, S.H.; Jiang, H.; Kim, H.S.; Flynn, C.R.; Hill, S.; Hayes McDonald, W.; et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 2010, 40, 893–904. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, L.; Li, C.Y.; Pei, Z.; Zhou, M.; Li, N. SIRT3 protects cells from hypoxia via PGC-1alpha- and MnSOD-dependent pathways. Neuroscience 2015, 286, 109–121. [Google Scholar] [CrossRef]
- Hallows, W.C.; Lee, S.; Denu, J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 2006, 103, 10230–10235. [Google Scholar] [CrossRef] [Green Version]
- Schmucker, D.L. Age-related changes in liver structure and function: Implications for disease? Exp. Gerontol. 2005, 40, 650–659. [Google Scholar] [CrossRef]
- Huang, P.; Zhou, Y.; Tang, W.; Ren, C.; Jiang, A.; Wang, X.; Qian, X.; Zhou, Z.; Gong, A. Long-term treatment of Nicotinamide mononucleotide improved age-related diminished ovary reserve through enhancing the mitophagy level of granulosa cells in mice. J. Nutr. Biochem. 2022, 101, 108911. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronchi, J.A.; Figueira, T.R.; Ravagnani, F.G.; Oliveira, H.C.; Vercesi, A.E.; Castilho, R.F. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic. Biol. Med. 2013, 63, 446–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, K.N.S.; Shen, X.; Pardue, S.; Krzywanski, D.M. Nicotinamide nucleotide transhydrogenase (NNT) regulates mitochondrial ROS and endothelial dysfunction in response to angiotensin II. Redox Biol. 2020, 36, 101650. [Google Scholar] [CrossRef] [PubMed]
- Oldford, C.; Kuksal, N.; Gill, R.; Young, A.; Mailloux, R.J. Estimation of the hydrogen peroxide producing capacities of liver and cardiac mitochondria isolated from C57BL/6N and C57BL/6J mice. Free Radic. Biol. Med. 2019, 135, 15–27. [Google Scholar] [CrossRef]
- Smith, C.D.; Schmidt, C.A.; Lin, C.T.; Fisher-Wellman, K.H.; Neufer, P.D. Flux through mitochondrial redox circuits linked to nicotinamide nucleotide transhydrogenase generates counterbalance changes in energy expenditure. J. Biol. Chem. 2020, 295, 16207–16216. [Google Scholar] [CrossRef]
- Kaludercic, N.; Di Lisa, F. The energetic cost of NNT-dependent ROS removal. J. Biol. Chem. 2020, 295, 16217–16218. [Google Scholar] [CrossRef]
- Ho, H.Y.; Lin, Y.T.; Lin, G.; Wu, P.R.; Cheng, M.L. Nicotinamide nucleotide transhydrogenase (NNT) deficiency dysregulates mitochondrial retrograde signaling and impedes proliferation. Redox Biol. 2017, 12, 916–928. [Google Scholar] [CrossRef]
- Gameiro, P.A.; Laviolette, L.A.; Kelleher, J.K.; Iliopoulos, O.; Stephanopoulos, G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2013, 288, 12967–12977. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Xia, S.; Zhang, R.; Li, Y.; Famulare, C.A.; Fan, H.; Wu, R.; Wang, M.; Zhu, A.C.; Elf, S.E.; et al. Lysine acetylation restricts mutant IDH2 activity to optimize transformation in AML cells. Mol. Cell 2021, 81, 3833–3847.e11. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, Y.; Park, S.H.; Liu, G.; O’Brien, J.; Jiang, H.; Gius, D. SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth. Cancer Res. 2017, 77, 3990–3999. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, C.; Ding, W.; Zhu, S.; Chen, Y.; Liu, X.; Deng, H. Nicotinamide Mononucleotide Administration Amends Protein Acetylome of Aged Mouse Liver. Cells 2022, 11, 1654. https://doi.org/10.3390/cells11101654
Luo C, Ding W, Zhu S, Chen Y, Liu X, Deng H. Nicotinamide Mononucleotide Administration Amends Protein Acetylome of Aged Mouse Liver. Cells. 2022; 11(10):1654. https://doi.org/10.3390/cells11101654
Chicago/Turabian StyleLuo, Chengting, Wenxi Ding, Songbiao Zhu, Yuling Chen, Xiaohui Liu, and Haiteng Deng. 2022. "Nicotinamide Mononucleotide Administration Amends Protein Acetylome of Aged Mouse Liver" Cells 11, no. 10: 1654. https://doi.org/10.3390/cells11101654
APA StyleLuo, C., Ding, W., Zhu, S., Chen, Y., Liu, X., & Deng, H. (2022). Nicotinamide Mononucleotide Administration Amends Protein Acetylome of Aged Mouse Liver. Cells, 11(10), 1654. https://doi.org/10.3390/cells11101654







